Abstract
Human T-cell leukemia virus type 1 (HTLV-1), the cause of adult T-cell leukemia, stimulates the growth of infected T cells in cultures and in nonleukemic patients. In the latter, HTLV-1 is found in long-term persisting T-cell clones. The persistence of normal T cells is controlled by the growth-stimulating and antiapoptotic functions of costimulatory receptors, while the growth-stimulating HTLV-1 functions are mediated by the viral oncoprotein Tax. Here we analyzed the impact of Tax on costimulatory receptors in T cells with repressible Tax and found that among these receptors 4-1BB (TNFRSF9/CD137/ILA) was induced most strongly. Up-regulated 4-1BB expression was a consistent feature of all HTLV-1–infected cell lines, whether patient-derived or in vitro transformed. Tax was sufficient to induce the expression of the endogenous 4-1BB gene in uninfected T cells, and it strongly activated (45-fold) the 4-1BB promoter via a single NF-κB site. The ligand of 4-1BB was also found on transformed T-cell lines, opening up the possibility of autostimulation. Moreover, 4-1BB expression in patients' lymphocytes ex vivo correlated with Tax expression, strongly suggesting Tax-mediated 4-1BB activation in vivo. Thus, 4-1BB up-regulation by Tax could contribute to growth, survival, and clonal expansion of the infected cells during persistence and disease.
Introduction
The human T-cell leukemia virus type 1 (HTLV-1), a delta-retrovirus, is the etiologic agent of a severe and fatal lymphoproliferative disorder of CD4+ T cells, the adult T-cell leukemia (ATL), and the neurodegenerative, inflammatory disease HTLV-1–associated myelopathy/tropical spastic paraparesis (HAM/TSP).1-4 These diseases develop as a consequence of prolonged viral persistence in T cells.
As emerged from many observations, HTLV-1 has developed a unique strategy for lifelong persistence in the presence of an active immune system. It is characterized by (a) replication of the virus mainly in its provirus form, (b) stimulation of cell division by the virus, and, as a consequence, (c) clonal amplification of infected cells. Evidence that HTLV-1 rarely replicates via reverse transcription but mostly by division of infected cells using cellular DNA polymerase is provided by the high HTLV-1 reverse transcriptase error rate, which is comparable to other retroviruses,5 and the low mutation rate of the HTLV-1 genome.6 The stimulation of T-cell proliferation in patients by viral gene expression was substantiated recently by cell dynamic studies.7 They revealed the correlation of the in vivo proliferation rate of CD4+CD45RO+ cells with viral expression (ex vivo). Another indication that HTLV-1 stimulates growth and survival of T lymphocytes is provided by their expansion to detectable clones, which can persist over many years even in nonleukemic individuals.8,9 Finally, the virus' ability to stimulate permanent T-lymphocyte growth in vitro, resulting in T-cell immortalization,10 completes the arguments in favor of viral gene functions as cause of host cell proliferation and clonal expansion of patients' lymphocytes.
Besides structural proteins, HTLV-1 encodes the regulatory proteins Tax and Rex, which are essential for viral replication,11 and the accessory proteins p12, p30, p13,12,13 and HBZ.14 While Tax strongly enhances viral mRNA synthesis by transactivating the HTLV-1 long terminal repeat promoter, Rex controls the synthesis of the structural proteins on a posttranscriptional level.15,16 Although the accessory proteins are important for viral infectivity and replication,12,17 p12, p13, p30, and HBZ are not required for lymphocyte immortalization.18-20
Tax is able to stimulate transcription by interacting with various signaling pathways. It activates nuclear factor kappa B (NF-κB) by 2 pathways. While the canonical pathway is induced by binding and stimulating IKKγ, a component of the inhibitor of kappa B kinase,10,17 the activation of the noncanonical pathway is less well understood. Transactivation of various cellular promoters is mediated by Tax via direct contact with transcriptional activators CREB and SRF and with the coactivators p300/CBP.11,21
Tax confers the transforming properties on HTLV-1.10 The protein can immortalize T lymphocytes22,23 and induce leukemia in transgenic mice.24 Biochemically, several Tax functions may contribute to its transforming capacity. At several checkpoints, Tax interferes with normal cell-cycle control.10,25 In particular, it is capable of stimulating the G1 to S phase transition through activation of cyclin-dependent kinase holoenzymes,26,27 and by disturbing cellular DNA repair and chromosomal segregation control pathways it can induce chromosomal damage.25 Importantly, Tax interferes with tumor suppressor functions, for instance, it inactivates p5328 and PDZ proteins.29,30 As a further means to promote cellular proliferation, Tax can stimulate the expression of cellular proteins controlling growth and survival.31 In particular, Tax-mediated modulation of cellular gene expression may explain the resistance of HTLV-1–positive cells to various proapoptotic stimuli.32-36 Moreover, the presence of Tax-stimulated antiapoptotic proteins can be essential for cellular survival. For example, RNAi-mediated knockdown of expression of the antiapoptotic protein HIAP1/CIAP2 induces apoptosis in HTLV-1–transformed T-cell lines.35
The persistence of HTLV-1 in T-cell clones detectable over many years suggested that proteins mediating survival and growth of long-lived T cells could be crucial for HTLV-1 persistence and, thus, be potential targets of its oncoprotein Tax. Such properties are characteristic of the costimulatory receptor subgroup of the tumor necrosis factor receptor (TNFR) superfamily. While being mostly absent from naive T cells, several TNFR superfamily members are present on long-lived T-lymphocyte populations like memory and regulatory T cells, where they can augment proliferation and survival.37
To identify Tax-dependent costimulatory receptors, we screened mRNA from T cells with repressible Tax expression and found that, among all costimulatory receptors' mRNAs, 4-1BB mRNA was most strongly increased. Up-regulation of 4-1BB was a consistent feature of HTLV-1–transformed cell lines and was caused by efficient transactivation of the 4-1BB promoter by Tax via the NF-κB pathway. In the presence of Tax, 4-1BB expression was strongly stimulated on the surface of CD4+ T cells isolated from patients. Thus, the costimulatory receptor 4-1BB is a target of Tax stimulation in cultured cells and in patients and is likely to support the survival of HTLV-1–infected T-cell clones.
Methods
Peripheral blood mononuclear cells (PBMCs) were isolated from venipuncture samples taken from HTLV-1–infected patients attending the National Center for Human Retrovirology, London, United Kingdom, with fully informed written consent. This study was approved by St Mary's National Health Service Trust Local Research Ethics committee. Informed consent was obtained in accordance with the Declaration of Helsinki. Microarray data are available at Gene Expression Omnibus under accession #GSE10508.
Cell culture
HTLV-1–negative acute lymphoblastic leukemia (ALL) T-cell lines Jurkat, HuT-78, and CCRF-CEM, the ATL patient-derived HTLV-1–positive (ATL) T-cell lines HuT-102, StEd, ATL-3, PaBe, JuanaW, Champ, and the HTLV-1 in vitro–immortalized, interleukin-2 (IL-2)–independent T-cell lines C91-PL and MT-2 were kept as described.35,38 The HAM/TSP patient-derived HTLV-1–positive T-cell lines Abgho, Nilu (both 40 U/mL IL-2), Eva and Xpos (both 20 U/mL IL-2) were cultured in RPMI 1640 containing 50% Panserin (PAN-Biotech, Aidenbach, Germany), 20% fetal calf serum (FCS), glutamine (0.35g/L,), streptomycin, and IL-2 (Roche Diagnostics, Mannheim, Germany) as indicated. The cell line Tesi23 was cultured as described.23,35 Tesi is a Tax in vitro–immortalized T-cell line featuring tetracycline-repressible Tax expression; for complete Tax repression, cells were grown in medium containing 1 μg/mL tetracycline for 10 days. JPX-9 and JPX-9M39 cells, which contain the Tax open reading frame under control of a heavy metal ion-inducible promoter, were grown like CCRF-CEM. For induction of Tax expression, 20 μM CdCl2 was added to the medium.
PBMC isolation
PBMCs from buffy coats of healthy donors (Institute for Transfusion Medicine, Suhl, Germany) were isolated by Ficoll Hypaque gradient centrifugation (Biocoll, Biochrom, Berlin, Germany). For analysis of 4-1BB expression, PBMCs were stimulated with 10 ng/mL phorbol myristate acetate (PMA), (Sigma, Hamburg, Germany) and 500 ng/mL ionomycin (Calbiochem, San Diego, CA) for 16 hours in RPMI 1640M, containing 10% FCS, glutamine, streptomycin, and penicillin.
Transcriptome analysis of Tesi cells
RNA from Tesi cells with and without Tax (Tesi + tet) was extracted (RNeasy, Qiagen, Hilden, Germany). Tax repression after tetracycline treatment was checked in real-time reverse transcriptase–polymerase chain reaction (RT-PCR). Transcriptome analysis was carried out on the Affymetrix HGU133plus 2.0 platform (Affymetrix, Santa Clara, CA). For each chip a biological replicate was performed. Differential expression analysis using ArrayAssist software (Stratagene, La Jolla, CA) compared the mean log-values of the replicated chips.
4-1BB and 4-1BBL mRNA detection
Total cellular RNA from cell lines and PBMCs was isolated (Trizol, Invitrogen, Karlsruhe, Germany) and reversely transcribed (Superscript II, Invitrogen) using random hexamer primers (Invitrogen). Primers and probes are listed in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) or have been published.35,40 Real-time RT-PCR was performed on an ABI Prism 7700 Sequence Analyzer (Applied Biosystems, Foster City, CA) from 200 ng cDNA. In RT-PCR, 500 ng cDNA were used as template. Expression levels were computed by interpolation from standard curves generated from plasmids and calculating the mean of triplicate samples. Expression in each cell line was measured in at least 3 biological replicates. β-Actin (ACTB) was used for normalization.
Analysis of 4-1BB and 4-1BBL surface expression
Expression of 4-1BB and 4-1BBL was analyzed by flow cytometry with monoclonal anti–human CD137-FITC (clone 4B4-1; AbD Serotec, Düsseldorf, Germany) and anti–human CDw137 Ligand-R-PE (clone C65-485; Becton Dickinson [BD], Heidelberg, Germany) antibodies. All analyses were controlled with isotype-matched control antibodies (BD). Cells were washed in phosphate-buffered saline (PBS) containing 5% FCS and 0.01% sodium azide (fluorescence-activated cell sorting [FACS] buffer), fixed with 2% paraformaldehyde in PBS, and stained on ice for 45 minutes. After 2 wash steps (FACS buffer), fluorescence was measured on a flow cytometer (FACSCalibur, BD). When analyzing PBMCs, cells were gated on peripheral blood lymphocytes (PBLs). Data were evaluated with FCS Express version 3 (De Novo Software, Los Angeles, CA). All analyses were performed at least in triplicate and reflect life cell gates.
Simultaneous analysis of 4-1BB surface expression and cell- cycle state
For simultaneous analysis of 4-1BB surface expression and DNA-content, 2*106 cells were stained with anti–human CD137-FITC, washed twice with PBS, permeabilized and propidium iodide (PI) stained for 10 minutes at 4°C with a buffer containing 0.3% saponin (Sigma), 10 μg/mL PI (Sigma), and 10 μg/mL RNaseA (Invitrogen). After fixation in 2% paraformaldehyde, cells were measured on a FACSCalibur. Absence of cell doublets was confirmed by FL-2A and FL-2W channels. To adjust quadrants, single stains with anti–human CD137-FITC, the respective isotype, and PI were performed. In double stains, gates were set for G1/G0, S, and G2/M phases. The proportion of 4-1BB high cells (and 4-1BB low cells, respectively) in S and G2/M phase was calculated as follows: [(S-phase + G2/M-phase)4-1BBhigh]/[(G1/G0-phase + S-phase + G2/M-phase)4-1BBhigh]. Each experiment was performed in triplicate.
4-1BB promoter analysis
A 1.5-kb fragment of genomic DNA (NC_000001.9 GI:89161185) upstream of the 4-1BB open reading frame (−1440 to +58 with the transcriptional start site, as given in NM_001561, referred to as +1) was cloned into pGL3basic (Promega, Mannheim, Germany) using Pwo polymerase (Roche Diagnostics) and primers containing NheI and HindIII restriction sites (5-TATTGTGCTAGCTTCATCTACAGCCTGGAG-3 [fwd], 5-TATTGTAAGCTTCCAGATGGACAGACCTTG-3 [rev]) producing pGL3basic:4-1BBprom. The NF-κB deletion mutant pGL3basic:4-1BBdelNF-κB was generated by overlap-extension PCR (same outer primers; inner primers: 5-CTGTCACAGAGCTGTGTGAGACCCCGCCCCTG-3 [fwd], 5-CAGGGGCGGGGTCTCACACAGCTCTGTGACAG -3 [rev]). All plasmids were sequenced. Tax impact on the 4-1BB promoter was tested in reporter gene assays in transfected Jurkat T cells as described.35,38 Briefly, Jurkat T cells were cotransfected by electroporation (EasyJect plus, Equibio, Ashford, United Kingdom, at 290V and 1500 μF) with 20 μg of the reporter construct and either 20 μg pcDNA3, pcTax,41 pM7, pM22, pM47,42 pLMP-1, or 2 μg pIκBDN43 . Luciferase activity was measured after 48 hours (Orion Microplate Luminometer; Berthold, Pforzheim, Germany) and normalized to protein concentration. Each plasmid combination was tested at least 3 times. Values are given as multiple of the control (pGL3basic:4-1BBprom plus pcDNA3).
Ex vivo analysis of Tax effects on 4-1BB expression in patients' T cells
PBMCs were isolated from venipuncture samples taken from HTLV-1–infected patients attending the National Center for Human Retrovirology, London, United Kingdom, with fully informed written consent, and cryopreserved until use. PBMCs were thawed and cultivated for 18 hours at 37°C, 5% CO2 in RPMI-1640 medium (Sigma) supplemented with FCS (Sigma), l-glutamine (Gibco, Gaithersburg, MD), penicillin (Gibco), and streptomycin (Gibco) in presence of concanamycin A (CMA; Sigma) at a final concentration of 20 nM. Then the cells were stained with monoclonal antibodies to CD4, CD8 (Beckman Coulter, Fullerton, CA), and the monoclonal antibody to 4-1BB (BD). Intracellular staining of Tax protein was done as previously described44 using anti-Tax antibody Lt-4. Flow cytometry was done on a Coulter Epics XL flow cytometer using Coulter Expo32 software (Beckman Coulter).
Statistical analysis
For evaluation of differences in expression levels of HTLV-1–positive and –negative samples, the Mann-Whitney test was applied using SPSS version 12.0.2 (SPSS, Chicago, IL).
Results
Expression profile of costimulatory receptors in Tax-transformed human T lymphocytes
To analyze the expression pattern of the TNFR superfamily genes in transformed T cells and its dependence on Tax, microarray analysis was conducted. For this end, RNA was prepared from Tesi cells under conditions of Tax expression and Tax repression (Tesi + Tet). The Tesi cell line was established by transforming human cord blood lymphocytes (CBLs) with a tetracycline-repressible HTLV-1 Tax gene.23 This system was chosen since it allows analysis of Tax's impact on the transcriptome of a human CD4+ T cell that is not derived from leukemia but directly from normal human lymphocytes. Table 1 shows the comparison of the expression pattern in the presence and absence of Tax. In Tesi cells, mRNAs of several costimulatory receptors (OX40, CD40, CD30, 4-1BB, HVEM, and GITR) and ligands (OX40L, CD27L, and 4-1BBL) could be detected. The other costimulatory receptors from the TNFR superfamily as well as the 2 costimulatory receptors from another protein family (CD28 and ICOS) were not expressed. While 4-1BB, 4-1BBL, and HVEM expression has not been previously observed in HTLV-1–transformed or Tax-expressing cells, the detection of the others was expected since they were already described to be expressed in HTLV-1–infected cell lines.45-47 The expression of most of these genes (CD27L, OX40, GITR) was verified by FACS/PCR analysis in HTLV-1–infected cells (data not shown). Tax had no or little impact on most of these genes, with 2 exceptions: Among the ligands, the Tesi system reproduced Tax induction of OX40L48 and, remarkably, as the only costimulatory receptor, 4-1BB was highly induced by Tax (11-fold). This induction was verified by quantitative RT-PCR (Figure 1A,B).
Expression of costimulatory TNF(R) superfamily genes in the presence and absence of Tax in a transformed T-cell line (Tesi)
| HGNC nomenclature . | Gene alias . | Average signals Tesi* . | Average signals Tesi without Tax* . | Fold change Tesi vs Tesi without Tax† . | Regulation Tesi vs Tesi without Tax . | P for fold change . | Affymetrix probe set . |
|---|---|---|---|---|---|---|---|
| TNFRSF4 | OX40 | 416 | 208 | 2 | Up | .06605 | 214228_x_at |
| TNFSF4 | OX40L | 512 | 24 | 22 | Up | .00492 | 207426_s_at |
| TNFRSF5 | CD40 | 169 | 97 | 2 | Up | .04191 | 35150_at |
| TNFSF5 | CD40L | (2) | (6) | NA | NA | NA | 207892_at |
| TNFRSF7 | CD27 | (20) | (28) | NA | NA | NA | 206150_at |
| TNFSF7 | CD27L | 2896 | 3566 | 1 | NA | .21953 | 206508_at |
| TNFRSF8 | CD30 | 549 | 256 | 2 | Up | .06542 | 206729_at |
| TNFSF8 | CD30L | (4) | (6) | NA | NA | NA | 235735_at |
| TNFRSF9 | 4–1BB | 362 | 34 | 11 | Up | .0022 | 207536_s_at |
| TNFSF9 | 4–1BBL | 256 | 362 | 1 | NA | .02614 | 206907_at |
| TNFRSF14 | HVEM | 119 | 111 | 1 | NA | .607 | 209354_at |
| TNFSF14 | LIGHT | (2) | (3) | NA | NA | NA | 207907_at |
| TNFRSF18 | GITR | 588 | 478 | 1 | NA | .31609 | 224553_s_at |
| TNFSF18 | GITRL | (4) | (6) | NA | NA | NA | 221371_at |
| HGNC nomenclature . | Gene alias . | Average signals Tesi* . | Average signals Tesi without Tax* . | Fold change Tesi vs Tesi without Tax† . | Regulation Tesi vs Tesi without Tax . | P for fold change . | Affymetrix probe set . |
|---|---|---|---|---|---|---|---|
| TNFRSF4 | OX40 | 416 | 208 | 2 | Up | .06605 | 214228_x_at |
| TNFSF4 | OX40L | 512 | 24 | 22 | Up | .00492 | 207426_s_at |
| TNFRSF5 | CD40 | 169 | 97 | 2 | Up | .04191 | 35150_at |
| TNFSF5 | CD40L | (2) | (6) | NA | NA | NA | 207892_at |
| TNFRSF7 | CD27 | (20) | (28) | NA | NA | NA | 206150_at |
| TNFSF7 | CD27L | 2896 | 3566 | 1 | NA | .21953 | 206508_at |
| TNFRSF8 | CD30 | 549 | 256 | 2 | Up | .06542 | 206729_at |
| TNFSF8 | CD30L | (4) | (6) | NA | NA | NA | 235735_at |
| TNFRSF9 | 4–1BB | 362 | 34 | 11 | Up | .0022 | 207536_s_at |
| TNFSF9 | 4–1BBL | 256 | 362 | 1 | NA | .02614 | 206907_at |
| TNFRSF14 | HVEM | 119 | 111 | 1 | NA | .607 | 209354_at |
| TNFSF14 | LIGHT | (2) | (3) | NA | NA | NA | 207907_at |
| TNFRSF18 | GITR | 588 | 478 | 1 | NA | .31609 | 224553_s_at |
| TNFSF18 | GITRL | (4) | (6) | NA | NA | NA | 221371_at |
HGNC indicates Human Genome Organization Gene Nomenclature Committee; vs, versus; and NA, not applicable.
Average fluorescence intensities were rounded to integers; parenthesized signal values were deemed absent calls.
Values were rounded to integers.
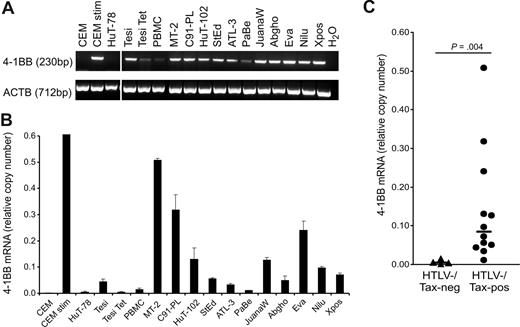
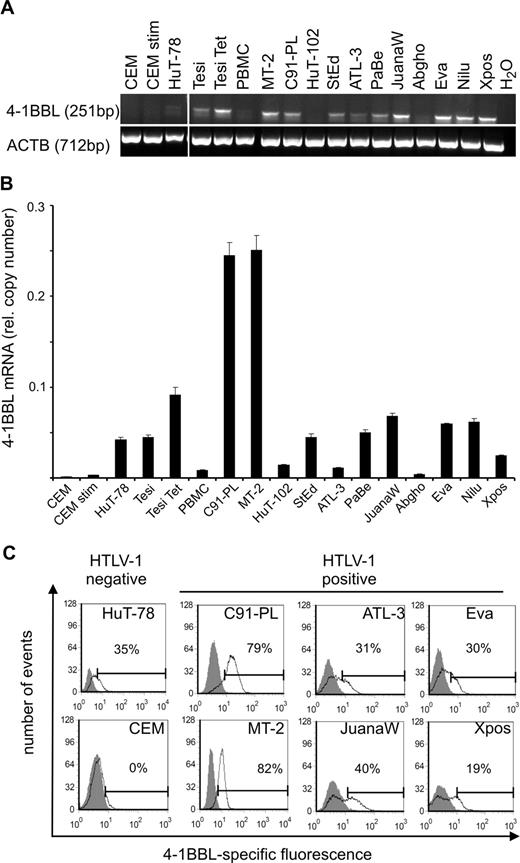
Up-regulation of 4-1BB mRNA in HTLV-1/Tax–positive cells. (A) Transcripts of 4-1BB and β-actin (ACTB) were detected by RT-PCR. Vertical lines have been inserted to indicate a repositioned gel lane. (B) Transcripts of 4-1BB were quantified by real-time RT-PCR. Relative copy number was determined by normalizing the 4-1BB transcripts to those of β-actin. The means of 3 independent experiments plus or minus standard error are shown. (C) Relative copy numbers of 4-1BB mRNA in HTLV-1/Tax–positive cell lines (Tesi, MT-2, C91-PL, HuT-102, StEd, ATL-3, PaBe, JuanaW, Abgho, Eva, Nilu, Xpos) were compared with those of HTLV-1/Tax–negative controls (CCRF-CEM, HuT-78, Tesi + tet, PBMCs) using a 2-tailed Mann-Whitney test. Horizontal bars indicate the median; •, HTLV-1/Tax–positive cell lines; and ▴, the HTLV-1/Tax–negative controls.
Up-regulation of 4-1BB mRNA in HTLV-1/Tax–positive cells. (A) Transcripts of 4-1BB and β-actin (ACTB) were detected by RT-PCR. Vertical lines have been inserted to indicate a repositioned gel lane. (B) Transcripts of 4-1BB were quantified by real-time RT-PCR. Relative copy number was determined by normalizing the 4-1BB transcripts to those of β-actin. The means of 3 independent experiments plus or minus standard error are shown. (C) Relative copy numbers of 4-1BB mRNA in HTLV-1/Tax–positive cell lines (Tesi, MT-2, C91-PL, HuT-102, StEd, ATL-3, PaBe, JuanaW, Abgho, Eva, Nilu, Xpos) were compared with those of HTLV-1/Tax–negative controls (CCRF-CEM, HuT-78, Tesi + tet, PBMCs) using a 2-tailed Mann-Whitney test. Horizontal bars indicate the median; •, HTLV-1/Tax–positive cell lines; and ▴, the HTLV-1/Tax–negative controls.
Consistent expression of 4-1BB in HTLV-1–infected cell lines
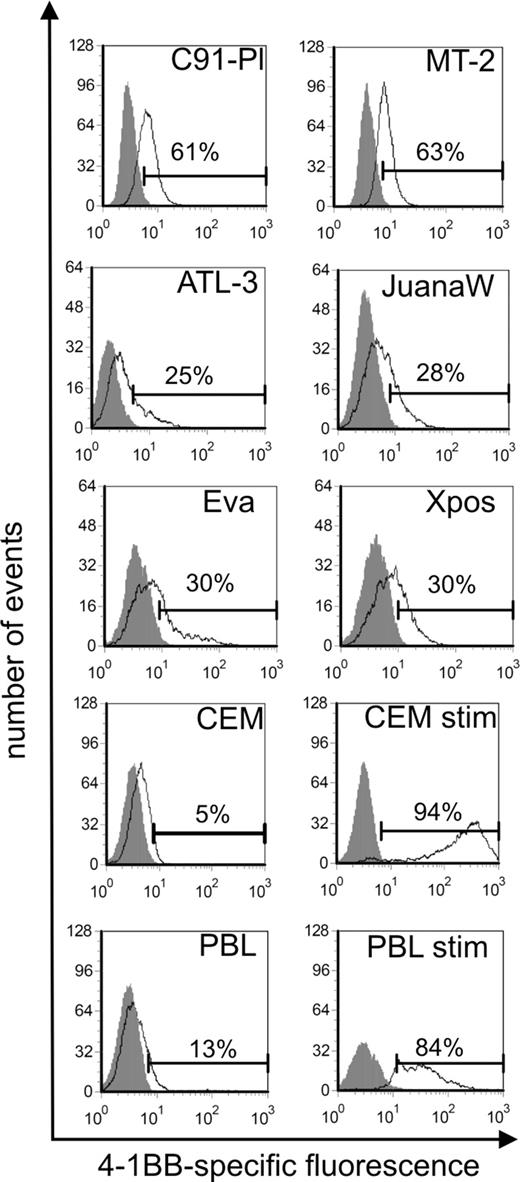
To verify the up-regulation of 4-1BB gene in HTLV-1–infected cell cultures, RT and real-time RT-PCR were performed. RNA from a spectrum of HTLV-1–transformed cultures and controls was analyzed. HTLV-1–positive cell cultures were derived from in vitro transformation, ATL, and HAM/TSP patients. HTLV-1–negative controls included the CD4+ T-cell ALL lines CCRF-CEM and HuT-78, and fresh PBMCs. As a positive control, CEM cells were stimulated with PMA/ionomycin, which is known to induce high amounts of 4-1BB expression.49 Specific transcripts were detected by RT-PCR and quantified by real-time PCR (Figure 1A,B). As a result, all of the HTLV-1–infected cells expressed 4-1BB mRNA, mostly at high levels. In all HTLV-1–positive cells the content of specific RNA was higher than in the CD4+ leukemic T-cell lines and, with one exception, exceeded even the 4-1BB RNA content of PBMCs. The increased expression was depending neither on the origin of the cell (in vitro transformed, HAM/TSP derived, ATL derived) nor on the cultures' requirement for exogenous IL-2. Statistical analysis using the Mann-Whitney 2-tailed test revealed that the 4-1BB mRNA content in HTLV-1–positive cells was significantly increased compared with HTLV-1/Tax-negative cells (Figure 1C). Cytometric staining with a specific antibody and FACS analysis (Figure 2) demonstrated the receptor on the cell surface of all types of HTLV-1–infected cultured T cells with amounts exceeding the negative controls and unstimulated PBMCs, which should be at least partially positive. As an additional positive control, PMA/ionomycin-stimulated PBMCs gated on lymphocytes were included, the expression of which is much higher than in long-lived T cells. The protein expression of 4-1BB on HTLV-1–positive cells clearly exceeds the expression in noninfected control T cells. The surface expression is intermediate and does not reach the levels of freshly stimulated PBMCs. In this respect, HTLV-1–infected cells resemble memory and regulatory T cells, which also express moderate levels of 4-1BB. In summary, the up-regulation of the costimulatory 4-1BB receptor on RNA and protein level is a consistent feature of HTLV-1–infected cells in culture.
Surface expression of 4-1BB protein on HTLV-1–transformed human T cells. Expression of 4-1BB was determined in ATL-derived (ATL-3, JuanaW), HAM/TSP-derived (Eva, Xpos), and HTLV-1 in vitro–transformed cell lines (MT-2, C91-PL). Uninfected CCRF-CEM and PBL cells from a healthy donor served as negative controls. For positive control, CCRF-CEM and PBLs were treated with PMA/ionomycin (stim). Extracellular 4-1BB was stained with specific antibodies (black curves) and analyzed by flow cytometry; gray curves represent an unspecific isotype-matched control antibody. One representative experiment of at least 3 is shown.
Surface expression of 4-1BB protein on HTLV-1–transformed human T cells. Expression of 4-1BB was determined in ATL-derived (ATL-3, JuanaW), HAM/TSP-derived (Eva, Xpos), and HTLV-1 in vitro–transformed cell lines (MT-2, C91-PL). Uninfected CCRF-CEM and PBL cells from a healthy donor served as negative controls. For positive control, CCRF-CEM and PBLs were treated with PMA/ionomycin (stim). Extracellular 4-1BB was stained with specific antibodies (black curves) and analyzed by flow cytometry; gray curves represent an unspecific isotype-matched control antibody. One representative experiment of at least 3 is shown.
Stimulation of 4-1BB gene expression by the viral transactivator Tax
HTLV-transformed cells display several features of activated T cells, including nuclear NFAT transcription factor.50 To identify a potential role of NFAT in the 4-1BB activation, cyclosporin A, an inhibitor of the cyclophilin/calcineurin pathway of NFAT activation, was added to Eva, JuanaW, and StEd cells and RNA was harvested after 48 hours. Real-time RT-PCR revealed no significant change of the cellular content of 4-1BB transcripts in any of these cells (Figure S1A). Additionally, a kinetic analysis of the CsA activity on 4-1BB mRNA levels was conducted in JuanaW cells, yielding the same result (Figure S1B), thus excluding NFAT activity as a mechanism for 4-1BB up-regulation. These experiments confirm earlier findings that show that an NFAT element is not present in the promoter region mediating induction in response to T-cell activation.49
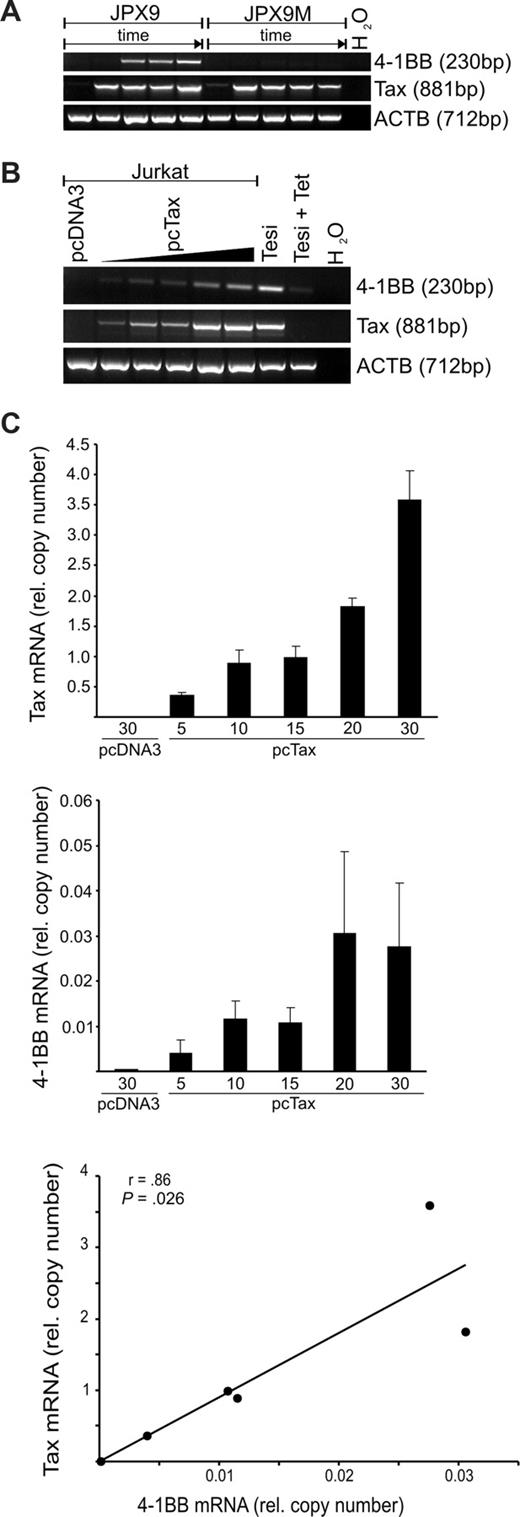
To determine whether the viral transactivator Tax is responsible for 4-1BB up-regulation in the HTLV-1–positive cultures, its impact on endogenous 4-1BB in T cells was tested. To this end, the Tax-inducible T-cell line JPX9, which expresses wild-type (wt) Tax after addition of heavy metal ions, and a variant, JPX9-M, with a nonfunctional mutated Tax allele, were treated with Cd2+ ions. After increasing time intervals, RNA was extracted and tested for the presence of 4-1BB and Tax transcripts (Figure 3A). RT-PCR verified the induction of Tax mRNA expression in both cell lines. About 6 to 12 hours after the appearance of the wt Tax transcripts, the transcription of the JPX-9 endogenous 4-1BB gene could be demonstrated, whereas no 4-1BB transcription could be evoked in the control culture containing the nonfunctional Tax. To determine the influence of various Tax protein concentrations on 4-1BB expression, Jurkat T cells were transfected with increasing amounts of a Tax expression plasmid. RT-PCR revealed that Tax expression resulted in the synthesis of significant amounts of endogenous 4-1BB transcripts, whereas in empty vector–transfected Jurkat cells 4-1BB mRNA was undetectable (Figure 3B). Moreover, quantification of Tax and 4-1BB transcripts by real-time RT-PCR (Figure 3C) indicated a direct correlation between Tax mRNA and endogenous 4-1BB mRNA synthesis. According to Pearson analysis (2-tailed), this correlation was significant (P = .026). Thus, as could be shown in 3 different T-cell systems (Tesi, JPX-9, transfected Jurkat), the viral transactivator Tax is sufficient to induce the expression of the costimulatory receptor 4-1BB in CD4+ T cells. These results suggest that Tax mediates the increased expression of the 4-1BB gene.
Induction of 4-1BB in T cells by HTLV-1 Tax. (A) Tax expression in JPX-9 (wt Tax) and JPX-9M (inactive Tax mutant) cells was induced by 20 μM CdCl2. RNA was harvested after 0, 6, 12, 24, and 48 hours, and transcripts of Tax, 4-1BB, and ACTB were detected by RT-PCR. (B) Jurkat cells were transfected with increasing amounts (5, 10, 15, 20, or 30 μg) of Tax expression plasmid or 30 μg of pcDNA3 as control. After 48 hours, Tax, 4-1BB and ACTB transcripts were detected by RT-PCR. (C) The Tax, 4-1BB, and ACTB mRNAs in the samples from panel B were quantified by real-time RT-PCR. Correlation of Tax and 4-1BB mRNA expression was tested using Pearson test.
Induction of 4-1BB in T cells by HTLV-1 Tax. (A) Tax expression in JPX-9 (wt Tax) and JPX-9M (inactive Tax mutant) cells was induced by 20 μM CdCl2. RNA was harvested after 0, 6, 12, 24, and 48 hours, and transcripts of Tax, 4-1BB, and ACTB were detected by RT-PCR. (B) Jurkat cells were transfected with increasing amounts (5, 10, 15, 20, or 30 μg) of Tax expression plasmid or 30 μg of pcDNA3 as control. After 48 hours, Tax, 4-1BB and ACTB transcripts were detected by RT-PCR. (C) The Tax, 4-1BB, and ACTB mRNAs in the samples from panel B were quantified by real-time RT-PCR. Correlation of Tax and 4-1BB mRNA expression was tested using Pearson test.
Stimulation of the 4-1BB promoter via NF-κB by the viral transactivator Tax
To test whether promoter transactivation by Tax causes 4-1BB overexpression, corresponding sequences of the 4-1BB promoter (nucleotides −1440 to +58) were PCR amplified from genomic DNA and cloned into a luciferase reporter plasmid (Figure 4A). Luciferase activity determined from transfected Jurkat T lymphocytes revealed a strong stimulation (45-fold) of the 4-1BB promoter in the presence of Tax (Figure 4B).
Transactivation of the 4-1BB promoter by HTLV-1 Tax via a single NF-κB site. (A) Schematic representation of the 4-1BB promoter sequence cloned into pGL3basic. Luc represents the open reading frame for the firely luciferase gene, +1 indicates the transcriptional start site. (B) To determine promoter activity, Jurkat T cells were cotransfected with pGL3basic:4-1BBprom (wt) and plasmids as indicated. Luciferase assays were performed 48 hours after transfection. At least 3 independent experiments were performed. Values were normalized to protein content and are given as mean plus or minus standard error. M7, M22, M47: Tax mutants; IκBDN: dominant active inhibitor of NF-κB (IκB); LMP-1: latent membrane protein of the Epstein-Barr virus (NF-κB activator). (C) The responsiveness of the 4-1BB NF-κB–deleted promoter to Tax was analyzed as described in panel B and compared with the 4-1BB wt promoter.
Transactivation of the 4-1BB promoter by HTLV-1 Tax via a single NF-κB site. (A) Schematic representation of the 4-1BB promoter sequence cloned into pGL3basic. Luc represents the open reading frame for the firely luciferase gene, +1 indicates the transcriptional start site. (B) To determine promoter activity, Jurkat T cells were cotransfected with pGL3basic:4-1BBprom (wt) and plasmids as indicated. Luciferase assays were performed 48 hours after transfection. At least 3 independent experiments were performed. Values were normalized to protein content and are given as mean plus or minus standard error. M7, M22, M47: Tax mutants; IκBDN: dominant active inhibitor of NF-κB (IκB); LMP-1: latent membrane protein of the Epstein-Barr virus (NF-κB activator). (C) The responsiveness of the 4-1BB NF-κB–deleted promoter to Tax was analyzed as described in panel B and compared with the 4-1BB wt promoter.
To narrow down transactivation-relevant signaling pathways, the impact of Tax mutants M7, M22, and M4742 on the 4-1BB promoter construct was analyzed. M22, which activates the CREB transcriptional pathway,42 but is incapable of stimulating NF-κB transcription factors, is greatly impaired in 4-1BB promoter transactivation. By contrast, Tax mutant M47 could transactivate the 4-1BB promoter like TaxWT. This mutant can activate NF-κB but is impaired in stimulating the CREB and SRF pathways. M7, which is inactive in transactivation, had no effect on the 4-1BB promoter. These results point to a Tax transactivation of the 4-1BB promoter via NF-κB. Similar to Tax, another viral oncogene (LMP-1), which is known to activate NF- κB, was able to stimulate the promoter. Moreover, the addition of a dominant active inhibitor of NF-κB signaling, IκBDN, completely abrogated the Tax effect on the 4-1BB promoter, confirming NF-κB–mediated Tax transactivation.
The promoter contains a single NF-κB binding site (Figure 4A) located at position −87 to −74 (−87GTGGGAATTTCCC-74) relative to the transcriptional start site (+1).49 To verify the role of NF-κB in transcriptional transactivation, a promoter mutant was constructed, in which the site is deleted (Figure 4A). This mutant lost its responsiveness to Tax stimulation (Figure 4C), indicating that this single sequence mediates Tax-transactivation. In summary, the viral transactivator Tax efficiently stimulated the 4-1BB promoter through NF-κB activation and, thus, could cause the observed 4-1BB overexpression.
Increased frequency of a premitotic state among HTLV-transformed cells with up-regulated 4-1BB expression
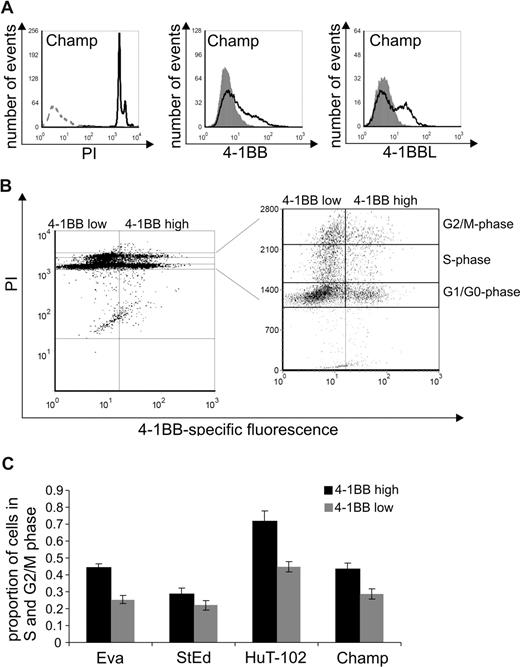
To determine whether 4-1BB expression in HTLV-infected cells is relevant to cell-cycle progression, surface expression levels of 4-1BB and cell-cycle phases were analyzed on individual cells by FACS. Cells of the cell lines Eva, StEd, HuT-102, and Champ were first stained using 4-1BB–specific antibodies, permeabilized, and subsequently treated with PI to stain the DNA content. As expected, all cultures contained cells in the G1/G0, S phase, and G2/M phase (Figure 5A). The cells were gated according to the 4-1BB expression level (4-1BB high/low) and cell-cycle phases G1/G0, S, G2/M (Figure 5B). Cells that have passed the restriction point within the G1-phase (ie, S, G2/M), and thus are committed to cell division, were taken as a measure of actively replicating cells (premitotic). The proportion of actively replicating cells was determined for both 4-1BB low and 4-1BB high cells. These analyses revealed a higher content of actively replicating cells within the 4-1BB high population (Figure 5C). This observation suggests that up-regulated 4-1BB expression increases the probability of a HTLV-infected cell to undergo cell division.
Increased frequency of a premitotic state among HTLV-transformed cells with high 4-1BB expression. (A) Expression of 4-1BB and ligand was determined in the ATL-derived cell line Champ. Extracellular 4-1BB and 4-BBL were stained with specific antibodies (black curves) and analyzed by flow cytometry; gray curves represent an unspecific isotype-matched control antibody. Intracellular PI staining is shown (black curves) in comparison to an unstained control (gray, dashed). One representative experiment of at least 3 is shown. (B) Simultaneous analysis of 4-1BB expression on the cell surface and staining of DNA-content by PI is shown in a representative dot plot of the ATL cell line Champ. Gates were set for cells in G1/G0, S, and G2/M phases. (C) Proportion of cells in the S and G2/M phase (premitotic cells) were calculated for 4-1BBhigh and 4-1BBlow cells of cell lines Eva, StEd, HuT-102, and Champ. The means of 3 independent experiments plus or minus standard error are shown.
Increased frequency of a premitotic state among HTLV-transformed cells with high 4-1BB expression. (A) Expression of 4-1BB and ligand was determined in the ATL-derived cell line Champ. Extracellular 4-1BB and 4-BBL were stained with specific antibodies (black curves) and analyzed by flow cytometry; gray curves represent an unspecific isotype-matched control antibody. Intracellular PI staining is shown (black curves) in comparison to an unstained control (gray, dashed). One representative experiment of at least 3 is shown. (B) Simultaneous analysis of 4-1BB expression on the cell surface and staining of DNA-content by PI is shown in a representative dot plot of the ATL cell line Champ. Gates were set for cells in G1/G0, S, and G2/M phases. (C) Proportion of cells in the S and G2/M phase (premitotic cells) were calculated for 4-1BBhigh and 4-1BBlow cells of cell lines Eva, StEd, HuT-102, and Champ. The means of 3 independent experiments plus or minus standard error are shown.
Expression of the ligand of 4-1BB (4-1BBL) on HTLV-1–transformed and ATL-derived cells
The ligand of 4-1BB (4-1BBL) is a transmembrane protein with a TNF homology domain in the carboxyl terminus that participates in binding to the respective receptor.51 4-1BBL protein expression has been described on stimulated B cells, macrophages, and dendritic cells and, at low levels, in T-cell lines,52 but not on resting or activated peripheral T cells. To determine whether the ligand is present and in principle can stimulate the 4-1BB receptor in HTLV-1–infected cultures, its expression was investigated in a series of HTLV-1–positive cell cultures by RT-PCR and quantitated by real-time RT-PCR (Figure 6A,B). Most of the HTLV-positive cells expressed moderate to high levels of 4-1BBL mRNA with copy numbers in the same order of magnitude as those of the receptor (Figure 6B). As the comparison of RNA contents of Tesi and Tesi + Tet indicates, Tax did not stimulate the expression of the ligand. Subsequent FACS analysis revealed that the RNA is translated and results in surface expression of the ligand (Figures 5A,6C). Double staining of both 4-1BB and its ligand showed the presence of double-positive cells, suggesting the possibility of autocrine receptor-ligand interactions. Interestingly, high expression of the ligand coincided with high expression of the receptor (data not shown), suggesting autostimulation at least in some of the HTLV-1–infected/–transformed cell lines.
Presence of the 4-1BB ligand (4-1BBL) in HTLV-1/Tax-positive cells. (A) Transcripts of 4-1BBL and β-actin (ACTB) were detected by RT-PCR. Vertical lines have been inserted to indicate a repositioned gel lane. (B) Transcripts of 4-1BBL were quantified by real-time RT-PCR. Relative copy number was determined by normalizing the 4-1BBL transcripts to those of β-actin. The means of 3 independent experiments plus or minus standard error are shown. (C) Surface expression of 4-1BBL was analyzed in HTLV-1–positive cell lines and in uninfected controls (HuT-78 and CEM). Extracellular 4-1BBL was stained with specific antibodies (black curves) and analyzed by flow cytometry; gray curves represent an unspecific isotype-matched control antibody. One representative experiment of at least 3 is shown.
Presence of the 4-1BB ligand (4-1BBL) in HTLV-1/Tax-positive cells. (A) Transcripts of 4-1BBL and β-actin (ACTB) were detected by RT-PCR. Vertical lines have been inserted to indicate a repositioned gel lane. (B) Transcripts of 4-1BBL were quantified by real-time RT-PCR. Relative copy number was determined by normalizing the 4-1BBL transcripts to those of β-actin. The means of 3 independent experiments plus or minus standard error are shown. (C) Surface expression of 4-1BBL was analyzed in HTLV-1–positive cell lines and in uninfected controls (HuT-78 and CEM). Extracellular 4-1BBL was stained with specific antibodies (black curves) and analyzed by flow cytometry; gray curves represent an unspecific isotype-matched control antibody. One representative experiment of at least 3 is shown.
Expression of 4-1BB in CD4+ and CD8+ T cells from HTLV-1–infected individuals
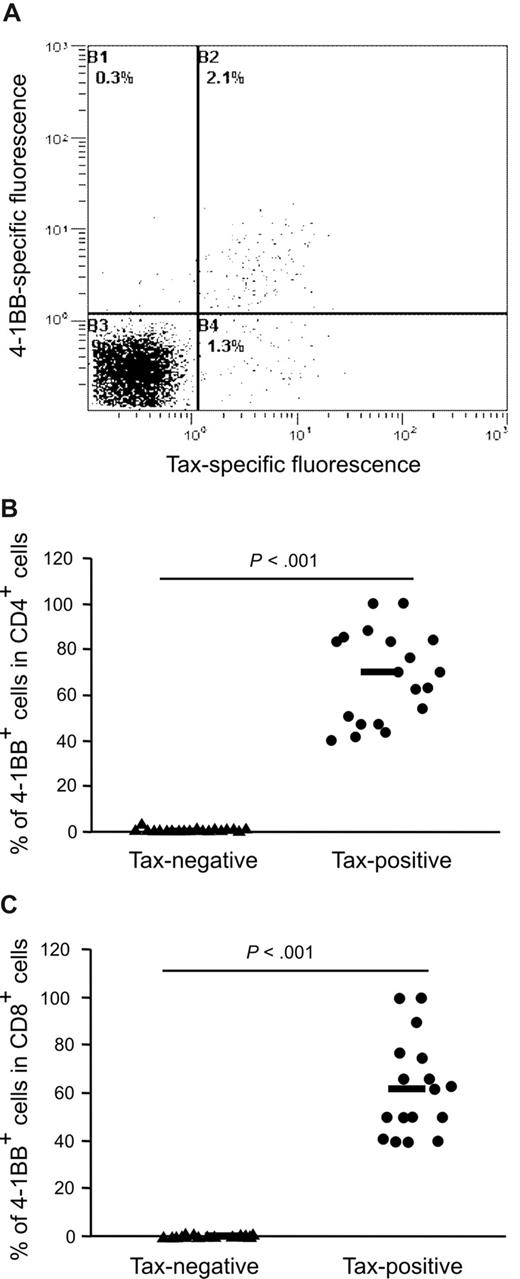
To analyze whether 4-1BB expression is activated in T lymphocytes from HTLV-1–infected patients, fresh PBMCs from an HTLV-1–seropositive subject were collected and cultivated for 18 hours in the presence of concanamycin A (to inhibit cytotoxic T-lymphocyte [CTL] activity). Subsequently, expression of 4-1BB and intracellular Tax within the CD4+ T-lymphocyte population was measured by flow cytometry. Figure 7A shows a representative example of a dot plot. The expression of Tax was strongly associated with the expression of the costimulatory molecule 4-1BB. To determine whether this is a general feature of HTLV-1–infected lymphocytes, we analyzed the PBMCs of 19 infected individuals: 9 asymptomatic HTLV-1 carriers, 8 patients with HAM/TSP, and 2 with other HTLV-1–associated inflammatory diseases. The percentage of 4-1BB expression was determined in the Tax-positive fraction and in the Tax-negative fraction of CD4+ T cells for each sample. Depending on the individual, 40% to 100% of the Tax-positive cells expressed surface 4-1BB, with no systematic difference between HAM patients and asymptomatic carriers at a given level of Tax expression. By contrast, the percentage of 4-1BB positivity in the Tax-negative cells was much lower (less than 3%). According to statistical analysis (2-tailed Mann-Whitney test) the percentage of expression of 4-1BB on the Tax expressing CD4+ cells was significantly higher (P = 1.3 × 10−7; n = 19) than in the Tax-negative cells (Figure 7B). Although HTLV-1 infects predominantly CD4+ T cells in vivo, CD8+ T cells can also be infected by HTLV-1. Flow cytometric analysis showed that Tax expression was accompanied by highly significant up-regulation of 4-1BB expression in CD8+ T cells incubated ex vivo for 18 hours (Figure 7C; P = 2.9 × 10−4; n = 17), as in the CD4+ T cells (Figure 7B). This result is a clear indication that the 4-1BB up-regulation by Tax is a feature of T lymphocytes in HTLV-1–infected individuals.
Association of Tax and 4-1BB expression on CD4+ and CD8+ lymphocytes from HTLV-1–infected patients. (A) PBMCs were cultivated in the presence of concanamycin A for 18 hours. Subsequently, they were stained for CD4, Tax, and 4-1BB and analyzed by flow cytometry. Cells were gated on the CD4+ T-cell population. One representative dot plot is shown. (B) Expression of 4-1BB in the Tax-positive (•) and Tax-negative (▴) CD4+ T cells was compared using a 2-tailed Mann-Whitney test. PBMCs from 19 subjects were analyzed by flow cytometry as exemplified in panel A. Horizontal bars indicate the median. (C) Expression of 4-1BB in the Tax-positive (•) and Tax-negative (▴) CD8+ T cells from 17 subjects was compared in analogy to panel B. Horizontal bars indicate the median.
Association of Tax and 4-1BB expression on CD4+ and CD8+ lymphocytes from HTLV-1–infected patients. (A) PBMCs were cultivated in the presence of concanamycin A for 18 hours. Subsequently, they were stained for CD4, Tax, and 4-1BB and analyzed by flow cytometry. Cells were gated on the CD4+ T-cell population. One representative dot plot is shown. (B) Expression of 4-1BB in the Tax-positive (•) and Tax-negative (▴) CD4+ T cells was compared using a 2-tailed Mann-Whitney test. PBMCs from 19 subjects were analyzed by flow cytometry as exemplified in panel A. Horizontal bars indicate the median. (C) Expression of 4-1BB in the Tax-positive (•) and Tax-negative (▴) CD8+ T cells from 17 subjects was compared in analogy to panel B. Horizontal bars indicate the median.
Discussion
Here we show that the antiapoptotic and growth-stimulating costimulatory receptor 4-1BB is up-regulated in HTLV-1–infected cell lines and ex vivo and is strongly stimulated by the HTLV-1 oncoprotein Tax. The coexpression of its ligand in HTLV-1–transformed cells suggests an autostimulation of transformed cells by 4-1BB, which may be important for growth and survival.
Costimulatory receptors of the TNFR superfamily are crucial mediators of proliferation and survival signals in T-cell activation and the maintenance of long-lived T-cell subtypes. Therefore, they provide potential targets for viruses relying on long-term persistence of infected T-cell clones in their replication, like HTLV-1 does. Our systematic analyses of Tax's impact on the T-cell transcriptome revealed 4-1BB as the costimulatory receptor whose expression was most strongly stimulated in the presence of Tax in the Tesi system. Moreover, 4-1BB mRNA was among the transcripts most strongly stimulated in the presence of Tax in the whole microarray. These observations are in accordance with the conclusion that 4-1BB is a major Tax target with relevance to proviral replication in amplifying T-cell clones.
4-1BB was found to be consistently up-regulated in a broad array of HTLV-1/Tax -transformed T-cell cultures, whether the cells were derived from in vitro immortalization or from HAM/TSP or ATL patients. Also, the requirement of exogenous interleukin-2 as growth factor was irrelevant to the presence of 4-1BB. This uniform phenotype suggested that the virus is directly or indirectly causing the phenotype.
Transfection of Tax expression plasmids into Jurkat T cells, which did not express 4-1BB, revealed that the signaling generated by Tax is sufficient to induce the expression of the gene. This indicates that Tax replaces all necessary cellular signals for its induction. Promoter analyses indicated that the gene is strongly stimulated by Tax (45-fold). The strong impact on the promoter provides an explanation for the strong Tax stimulation of the 4-1BB expression observed in the microarray and the enhanced 4-1BB expression in HTLV-1–transformed cells. Several different experimental approaches, including the use of Tax-mutants and inhibitors, each revealed that the promoter is transactivated via the NF-κB pathway. A single NF-κB site could be identified as mediating the Tax effect. This site has been previously shown to bind NF-κB factors p65 and p50 in gel-shift experiments and is required for the stimulation of the 4-1BB gene during T-cell activation by T-cell receptor (TCR) (anti-CD3 treatment) or PMA/ionomycin treatment. Two AP-1 sites, which are also important for the promoter stimulation by these signals, were not required for the Tax transactivation. Tax also induces activated protein-1 (AP-1), a transcription factor complex composed of members of the Fos/Jun family.53 It is still unclear why the 4-1BB promoter is much more highly induced by Tax than other promoters of antiapoptotic genes that have been described to be activated by Tax via the NF-κB pathway, including Bcl-XL and OX40, although their promoters contain 2 and more NF-κB sites.49 In summary, these studies indicate that the enhanced 4-1BB expression in HTLV-1–transformed cells is mediated by Tax transactivation of the 4-1BB promoter through NF-κB.
In contrast to normal uninfected T cells, the HTLV-1–infected cell lines expressed moderate to high levels of the 4-1BB ligand, with a tendency to higher expression in faster growing cells. This unusual up-regulation could not be directly linked to Tax; it may be due to other viral effector proteins, which have been shown to affect cellular transcription. An alternative explanation for the coincidence of increased 4-1BBL expression and Tax-mediated 4-1BB up-regulation is that cellular transcription factors that stimulate the HTLV promoter in HTLV-transformed cell lines may also be responsible for the 4-1BBL up-regulation. cAMP-stimulated factors, which are able to stimulate 4-1BBL in transformed cells, may provide an example.54 The presence of both receptor and ligand in the same culture and on the same cell (data not shown) is a strong indication of autostimulation of the cultured HTLV-1–infected cells by 4-1BB. This notion is corroborated by results obtained from analysis of 4-1BB levels in premitotic cells. These suggest that up-regulated 4-1BB increases the probability of an HTLV-1–infected cell to undergo cell division. Autostimulation of the receptor via TRAF-2 may result in canonical and noncanonical NF-κB activation and JNK and p38 MAPK signaling.37 As an additional argument for this interaction, activation of these pathways has been described in HTLV-1–transformed cells.55,56
The 4-1BB receptor is present on long-lived T-cell populations, like memory57 and regulatory T cells. Thus, stimulation of 4-1BB by HTLV-1 Tax provides a causal linkage between Tax function and differentiation of infected lymphocytes into long-lived T-cell populations. In these cell types 4-1BB is important for the maintenance of specific clones. Like in some types of uninfected cells, 4-1BB may act as a potent factor controlling the persistence and clonal expansion of virally infected cells. Two mechanisms of clonal expansion have been proposed for HTLV-1–infected CD4+ and CD8+ subpopulation, resistance to apoptosis (CD8+ cells), and increased cell cycling (CD4+ cells).58 Both of these can be supported by 4-1BB signaling, which is antiapoptotic and growth stimulating.37,59 For instance, it is important for activation of proliferation in CD8+ and CD4+ memory T cells and induces the proliferation of the CD4+CD25+ regulatory T cells.60 Antiapoptotic signaling by 4-1BB may not only support clonal amplification but may also contribute to malignant transformation of CD4+ cells, which are more likely to accumulate chromosomal/nuclear damage.58 In these cells, 4-1BB could prevent damage-induced apoptotic pathways and hence conserve cells containing growth-stimulating mutations.
Since other viral oncoproteins of lymphotropic tumor viruses also activate cellular gene expression via NF-κB, it is possible that 4-1BB is a general target of lymphotropic tumor viruses and is up-regulated in transformed lymphocytes. As an example of this assumption, our results demonstrate that the Epstein-Barr virus latent membrane protein (LMP-1) and the herpesvirus ateles Tio protein (data not shown) are capable of activating the 4-1BB promoter.
4-1BB was strongly stimulated together with the expression of Tax in CD4+ T cells isolated from asymptomatic carriers and HAM/TSP patients. This is strong evidence that 4-1BB expression is also activated by Tax in patients. Although expression of the ligand is low or absent on HTLV-1–infected lymphocytes ex vivo (data not shown), 4-1BB signals could be generated by interaction with antigen presenting cells like macrophages, B cells, dendritic, or endothelial cells, which are all expressing the ligand.57 Moreover, the soluble form of the ligand is able to induce signaling and growth in T cells. Soluble 4-1BB ligand is released constitutively at low levels from leukocytes and at higher levels following cellular activation, resulting in variable serum levels (ranging from high in patients with hematologic malignancies to low in healthy donors).61 Thus, 4-1BB signals could be generated in Tax-expressing cells in patients and contribute to the amplification of the infected cells, including the chromosomally inserted provirus. Consequently, 4-1BB and the availability of the ligand may be determinants of the proviral load during viral persistence and (HAM/TSP) pathogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the staff and patients of the HTLV-1 clinic at the National Center for Human Retrovirology (NCHR), St Mary's NHS Trust, London. The NCHR is supported by a grant from the Department of Health, United Kingdom. The technical assistance of Grit Schneider is greatly appreciated.
This work was supported by the European Union (INCA, LSHC-CT-2005-018704) and by Deutsche Forschungsgemeinschaft (DFG-GRK1071, GR 1224/3-1).
Authorship
Contribution: K.P., T.K., J.G., and A.K.K. performed experiments, analyzed the results, and made the figures. C.B., G.P.T., and R.G. designed the research, analyzed the results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ralph Grassmann, Institute of Clinical and Molecular Virology, Friedrich-Alexander-Universität Erlangen-Nürnberg, Schlossgarten 4, 91054 Erlangen, Germany; e-mail: grassmann@viro.med.uni-erlangen.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal