Abstract
Recently, we identified a population of very small embryonic like (VSEL) SCs in adult bone marrow (BM) (
very small in size (~3.6 um);
Oct-4+CXCR4+SSEA-1+Sca-1+CD45−lin−;
possessing large nuclei containing unorganized chromatin (euchromatin); and
we learned that in co-cultures with C2C12 cells, VSELs form embryoid body-like spheres (VSEL-DSs) that contain primitive SCs capable to differentiate into all three germ layers (e.g., myocardium, neural tissue, and pancreas).
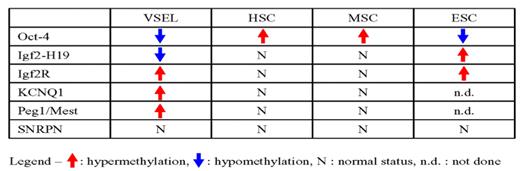
To better characterize this intriguing population of SCs, we employed bisulfite sequencing, combined bisulfite restriction analysis (COBRA), and methylation specific PCR (MSP) to analyze the methylation status of i) Oct-4 promoter and ii) differentially methylated regions (DMRs) of imprinted genes that are known to be crucial for maintaining the pluripotency of embryonic (E)SCs [e.g., insulin-like growth factor 2 (Igf2)-H19 locus, Igf2 receptor (Igf2R), paternally expressed gene 1(Peg1/Mest), small nuclear ribonucleoprotein polypeptide N (SNRPN), and KCNQ1 locus that includes cyclin-dependent kinase inhibitor 1c (CDKN1C/p57Kip2)]. The methylation status of these genes was evaluated in adult bone marrow (BM)-derived Sca+lin−CD45− VSELs, Sca+lin− CD45+ hematopoietic (H)SCs, mesenchymal stem cells (MSCs), and the murine ESC line ESC-D3 (Table 1). We noticed that Oct-4 promoter is hypomethylated in VSELs similarly to ESCs. In contrast, Oct-4 promoter was hypermethylated in HSCs and MSCs. This observation combined with our mRNA and protein expression data provides evidence that Oct-4 gene is transcribed in VSELs residing in adult tissues. We also found that the DMR of the Igf2-H19 locus, which is crucial for controlling SC pluripotency, was significantly hypomethylated in VSELs. Of note methylation of this locus is known to be erased in migrating primordial germ cells (PGC), and is envisioned as a crucial mechanism that controls “unleashed proliferation” of PGC and prevents them from forming teratomas. On other hand, Igf2-H19 locus was hypermethylated in ESCs and the proper somatic pattern of methylation (~50%) was observed in HSCs and MSCs (Table 1). Furthermore, VSEL similarly to ESCs showed hypermethylation of DMRs of Igf2R, KCNQ1, and Peg1/Mest – loci, which show proper somatic imprint (~50%) in HSCs and MSCs. Of note, SNRPN DMR methylation was normally maintained in all cells tested. Our methylation data were subsequently confirmed by mRNA expression studies. Accordingly, as predicted VSELs showed increased expression levels of mRNA for H19, Igf2R, and CDKN1C/p57Kip2, but reduced expression level of Igf2 as compared to HSCs. Finally, when VSELs were cultured in our “expansion” model over a C2C12 feeder layer, the methylation pattern of VSELs (hypomethylation of Igf2-H19, hypermethylation of Igf2R and Peg1/Mest) had been properly restored in cells inside VSEL-DSs to levels observed in normal somatic cells. At the same time, however, the Oct-4 promoter became hypermethylated and Oct-4 mRNA was downregulated. In conclusion, our methylation studies at Oct-4 promoter and Igf2-H19 locus provide additional evidence that VSELs show a similar methylation pattern to PGC, what supports their developmental origin directly form epiblast/germ line. We believe that they are deposited during embryogenesis in the adult tissues as a backup for tissue committed SCs and that their proliferative potential is tightly regulated/controlled by the status of Oct-4 promoter and Igf2-H19 DMR locus methylation. Erasure of methylation at Igf2-H19 locus on one hand prevents them from “unleashed proliferation” and formation of teratomas. Thus, identification of mechanisms that control and modify genomic imprinting in VSELs will be crucial for developing more powerful strategies to “unleash a power” of these cells and employ them in regenerative medicine.
Table 1. Methylation status of Oct-4 promoter and crucial somatic imprinted genes.
Disclosures: No relevant conflicts of interest to declare.
Author notes
Corresponding author