Abstract
Chronic lymphocytic leukemia (CLL) has a variable clinical course. Presence of specific genomic aberrations has been shown to impact survival outcomes and can help categorize CLL into clinically distinct subtypes. We studied 178 CLL patients enrolled in a prospective study at the University of Michigan, of whom 139 and 39 were previously untreated and previously treated, respectively. We obtained unbiased, high-density, genome-wide measurements of subchromosomal copy number changes in highly purified DNA from sorted CD19+ cells and buccal cells using the Affymetrix 50kXbaI SNP array platform (Santa Clara, CA). Genomic complexity scores were derived and correlated with the surrogate clinical end points time to first therapy (TTFT) and time to subsequent therapy (TTST): measures of disease aggressiveness and/or therapy efficaciousness. In univariate analysis, progressively increasing complexity scores in previously untreated CLL patients identified patients with short TTFT at high significance levels. Similarly, TTST was significantly shorter in pretreated patients with high as opposed to low genomic complexity. In multivariate analysis, genomic complexity emerged as an independent risk factor for short TTFT and TTST. Finally, algorithmic subchromosomal complexity determination was developed, facilitating automation and future routine clinical application of CLL whole-genome analysis.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common form of adult leukemia in the Western world, and is characterized by a highly variable clinical course.1 The Rai and Binet staging systems were the first attempts at risk stratification in CLL.2,3 Although these systems have helped to identify patients with high-risk disease, they are less applicable in stratifying low- and intermediate-risk cohorts, which account for the vast majority of patients at diagnosis.
Recently, several disease-related factors have been identified that further classify CLL into biologically and clinically distinct subtypes. Lack of mutation in the immunoglobulin heavy chain variable region (IgVH) genes, increased expression of zeta-associated protein of 70 kDa (ZAP-70), increased expression of CD38, and mutations involving p53 and/or ataxia telangiectasia mutated (ATM) genes are some of the disease parameters that have been variously associated with an aggressive disease course, shortened disease-free intervals, and compromised survival.4-23
The description of chromosomal abnormalities has served an important role in understanding biologic properties and clinical behavior of various hematologic malignancies.24 In CLL, conventional cytogenetic analyses have revealed chromosomal aberrations in 40% to 50% of patients, but detection of abnormalities is limited by the low mitotic activity of CLL cells in vitro.25 Interphase fluorescent in situ hybridization (FISH) has identified chromosomal changes in approximately 80% of patients with CLL, and presence of specific chromosomal abnormalities has proven to be a prognostic indicator for disease progression and survival.26 However, FISH is limited to detection of chromosomal changes specific to the probes used and, thus, underestimates the extent of chromosomal aberrations present in CLL.
Although recent advances using in vitro immunostimulatory methods have substantially improved analysis via conventional metaphase cytogenetics,27-29 submicroscopic deletions of genetic material can escape detection, and thus a more quantitative, unbiased, sensitive, specific, and readily available method is ultimately desired.
High-density, single nucleotide polymorphism (SNP) arrays are commercially available technologies that allow genome-wide exploration for allelic copy number gains and losses30-33 that are commonly found in cancer genomes. Herein, we report the results of a prospective study using high-density 50k Affymetrix SNP microarrays (Santa Clara, CA) to quantify genome-wide allelic copy number losses, gains, and regions of loss-of-heterozygosity (LOH), which we incorporate into a genomic complexity score. The clinical relevance of this score was tested in a cohort of previously untreated and pretreated CLL patients, with TTFT and TTST as the primary end points, respectively.
From these data, we can firmly conclude that high genomic complexity is an independent risk factor for disease progression and treatment failure in CLL, warranting future investigations into the usefulness of this marker in clinical practice.
Methods
Patients
Between January 2005 and September 2007, 186 CLL patients evaluated at the University of Michigan Comprehensive Cancer Center were enrolled onto this study. The trial was approved by the University of Michigan Institutional Review Board (IRBMED no. 2004-0962) and written informed consent was obtained from all patients in accordance with the Declaration of Helsinki before enrollment. Regardless of whether the subjects were originally diagnosed at our institution or another, we used the same CLL diagnostic criteria, based on the National Cancer Institute-Working Group Guidelines for CLL.34 We validated that the diagnostic criteria were met for all subjects through review of both laboratory (complete blood count with differential) and pathology (flow cytometry, bone marrow biopsy/aspirate, lymph node biopsy) reports dated at the time of diagnosis. Eligible patients needed to have more than 5 × 109/L (5000/μL) mature lymphocytes, and lymphocytes needed to express CD19, CD23, sIg (weak), and CD5 in the absence of other pan-T-cell markers. Five patients enrolled on the study were excluded from analysis (diagnoses: large cell lymphoma, marginal zone lymphoma, small lymphocytic lymphoma [SLL], Crohn disease, and CLL with concurrent acute myeloid leukemia [AML]). Three posttreatment patient samples gave insufficient DNA for analysis and thus were excluded from analysis. This study is therefore based on 178 patients (139 previously untreated and 39 previously treated).
Time to first therapy (TTFT) was defined from date (month/year) of diagnosis of CLL to date of first therapy (month/year). Time to subsequent therapy (TTST) was defined from date (month/year) of completion of the chemotherapy immediately before enrollment to the start date of next therapy (month/year) and was calculated for the previously treated patients. Nine patients who were untreated at enrollment and proceeded to receive therapy required subsequent therapy during the follow-up of this trial. TTST for these 9 patients is defined as the date (month/year) of completion of the first chemotherapy to the start date of second therapy (month/year), and data were combined with the TTST data for the previously treated group of 39 patients. In total, TTST data are therefore based on 48 patients.
CLL treatment was defined as cytotoxic chemotherapy and/or monoclonal antibody therapy for CLL and/or associated autoimmune cytopenias. For the majority of patients, this included purine analog–based therapy combined with rituximab. Clinical information including Rai stage was collected on all patients.2
Cell isolation
Cell purification.
Peripheral blood mononuclear cells from CLL patients were isolated by Ficoll-paque gradient centrifugation (GE Healthcare, Little Chalfont, United Kingdom), aliquoted into fetal calf serum with 10% dimethylsulfoxide, and cryopreserved in liquid nitrogen.
Flow cytometry.
Cryopreserved CLL cells were washed and stained with phycoerythrin (PE)–conjugated anti-CD19 and fluorescein isothiocyanate (FITC)–conjugated anti-CD3 antibodies (eBioscience, San Diego, CA). Propidium iodide was added to a concentration of 1 μg/mL to discriminate dead cells, and viable CD19+ single-positive cells were sorted on a high-speed fluorescence-activated cell sorter, FACSAria (Becton Dickinson, Mountain View, CA).
Array data analysis
Affymetrix data files were generated from SNP chips after hybridization via scanners at the University of Michigan Microarray Core facility and imported into the Affymetrix GCOS and GDAS software suites. The freely available software dChip used native Affymetrix .CEL files and text formatted .CHP files from GDAS to generate copy number heatmap displays.35 Copy number data for all SNPs, for all patients, were exported from dChip and used to generate the algorithmic complexity score.
Determination of immunoglobulin heavy chain variable gene (IgVH) mutation status
Determination of IgVH mutation status was done as described.36
Determination of percentage of ZAP-70–positive cells using multiparameter FACS analysis
Determination of percentage of ZAP-70–positive cells using multiparameter FACS analysis was done as described.36
p53 genomic DNA sequencing
p53 genomic exon DNA resequencing was done as described.36
CD38 expression in CLL cells by flow cytometry
Cryopreserved aliquots of CLL cells were washed, and between 105 and 106 cells per aliquot were first blocked for 10 minutes with 200 μg/mL mouse IgG, then stained in duplicate with anti-CD38 allophycocyanin (APC), anti-CD19 PE, and anti-CD5 FITC (all from eBioscience). A third aliquot of cells was stained with isotype-matched controls. Jurkat cells were used as a positive control for CD38 and CD5 staining; normal human peripheral blood mononuclear cells were also used with each run to verify reproducibility. Cells were washed after antibody staining, fixed in 1% paraformaldehyde, and analyzed on a Becton Dickinson FACSCalibur flow cytometer. Isotype controls were used to set positive-negative threshold markers for each sample, and then the percentage of CD19+/CD5+ lymphocytes also positive for CD38 was determined.37
Fluorescence in situ hybridization
FISH was performed for all patient samples at the Mayo Clinic (Rochester, MN) as a routine clinical test, with the following published chromosomal target regions: 6cen (D6Z1), 6q23.3 (c-myb locus), 11 cen (D11Z1), 11q13 (CCND1-XT), 11q22.3 (ATM), 12 cen (D12Z3), 12q15 (MDM2), 13q14 (D13S319), 13q34 (LAMP1), 14q32 (IGH-XT), 17 cen (D17Z1), and 17p13.1 (p53). Two hundred cells were scored for these probes per patient.
Preparation of sample DNA for hybridization to Affymetrix 50kXbaI mapping arrays and assay characteristics
50k SNP chip–based analysis using DNA purified from FACS-sorted CD19+ cells or buccal swabs were done as described.36
Derivation of a genomic complexity score
Visual method.
Two of the authors (P.O. and S.N.M.) independently reviewed dChipSNP-based copy number displays for all patients and all chromosomes, and visually scored lesions that were (1) at least 8 consecutive SNP positions in length and (2) either all blue (indicating less than 2N copy estimates = losses) or all red (indicating greater than 2N copy estimates = gains). A length of 8 consecutive SNPs was chosen as it allowed clear distinction from background. Losses or gains of entire chromosomes were excluded from analysis as they are not the products of double-strand DNA breaks. Polymorphic copy number variations were detected through review of data derived from buccal DNA, available for 100% of the patients. Subchromosomal uniparental disomy (UPD) was detected by comparing LOH data analyzed in the LOH tool with dChipSNP-based copy number displays.38 A total of 11 instances of subchromosomal UPD were detected in 10 CLL cases of 178 CLL genomes analyzed.
The lesion calls were then used to produce a total genomic complexity score by adding the number of losses, gains, and UPD. We also considered the score resulting from counting only the losses. Pearson correlations between the complexity scores from P.O. and S.N.M. were calculated and demonstrated an overall correlation of greater than 0.95.
Sensitivities and specificities for the visual lesion calling method were calculated against the clinical FISH data (Mayo Clinic) for del17p, del13q14, del11q, and trisomy 12 as the gold standard. Percentage of positive FISH results for the lesions listed in the first paragraph of this subsection in our CLL cohort ranged from 4% to 96% in individual cases, reflective of the known fact that these lesions in CLL occur in subclones of various sizes and proportions. We therefore selected 2 alternative cutoffs for FISH positivity for comparative calculations: (1) all positive FISH findings as defined by the Mayo clinic laboratory (all FISH) and (2) FISH categories using a cutoff of 25% FISH positive cells (FISH25; FISH results found in > 25% of cells were grouped as positive and < 25% of cells as negative), as we wished to test the performance of the visual lesion calling method under routine conditions and under conditions where a sizable portion of the respective CLL clones harbored a lesion. Sensitivities/specificities for SNP array–based compared with all FISH-positive lesions (S.N.M./P.O.), respectively, were (85%/88%) and (99%/96%) for del13q14, (87%/90%) and (100%/100%) for trisomy 12, (93%/100%) and (99%/99%) for del17p, and (61%/61%) and (99%/99%) for del11q.
Sensitivities/specificities for SNP array–based compared with greater than 25% FISH-positive lesions (S.N.M./P.O.), respectively, were (94%/95%) and (95%/94%) for del13q14, (91%/97%) and (97%/98%) for trisomy 12, (100%/100%) and (99%/99%) for del17p, and (85%/85%) and (99%/99%) for del11q.
Algorithmic method.
We optimized sliding window algorithms that score a lesion as “copy loss” when a fixed SNP window of 8 consecutive SNPs had at least N SNPs with a copy number estimate of X or less (the 8/N/X rule). Optimizations were based on calculations of sensitivities and specificities for detection of del13q14, del17p, and del11q for multiple rules in parallel using the Mayo Clinic FISH results (all FISH) as the gold standard. Sensitivities and specificities were also calculated for FISH categories using a cutoff of 25% FISH-positive cells. To minimize false-positive lesion calling, only rules with specificities of 95% or more measured against all FISH were considered further. The rules were trained and validated based only on the array data, without reference to any clinical data. One rule (8/7/1.45) achieved a sensitivity/specificity for del17p/del13q/del11q detection of (93%/98%)/(82%/95%)/67%/96%), respectively (for FISH25 these results were [93%/98%]/[92%/94%]/[86%/96%], respectively), and was subsequently applied to dChipSNP-based copy number estimates for all CLL cases and all SNP positions.
To avoid calling the same genetic lesion multiple times (as most lesions are significantly longer than 8 consecutive SNPs), and in an effort to fully automate algorithmic lesion calling, thus obviating visual editing, we combined all calls that were less than 3 Mb apart into one lesion. The Pearson correlation coefficient between the resulting algorithmic genomic loss calls and the visual loss calls was 0.87 (range: 0.82-0.91).
Assessment of association between genomic complexity and clinical end point
TTFT and TTST were the clinical end points used. For univariate analysis, we considered each prognostic factor (SNP array–derived complexity, IgVH, p53, ZAP-70, individual FISH categories, CD38) separately. Visual and algorithmic genomic complexity scores were divided into bins (eg, < 3 lesions vs ≥ 3 lesions). The Kaplan-Meier method was used to estimate the survivor function to first treatment for each subgroup, and the log-rank test was used to calculate 2-sided P values testing significant differences in the survivor function between subgroups.
For bivariate analysis, we considered individual risk factors together with the dichotomized visual complexity score of 3 or more or fewer than 3 lesions. Based on these 2 dichotomous factors, the patient cohort was divided into 4 subgroups.
For multivariate analysis, we used Cox proportional hazards models. Modeling was performed separately using the 139 first treatment events (TTFT), or using the 48 subsequent treatment events (TTST). For TTFT analysis, we included as binary covariates ZAP-70, VH status, and Rai stage (coded as stage 0 vs stage 1-2). These variables were included in all models for TTFT because they were all highly significant in univariate analysis for TTFT. We also always included a factor coding for the presence of either del11q or del17p, which was borderline significant in univariate analysis for TTFT (and significant for TTST). Except for a baseline model, one of the genomic complexity scores was always included. Further, we considered models with and without p53 mutations, and with and without CD38 expression.
These same factors were considered in the multivariate analysis for TTST, with the exception that Rai stage was omitted. Models were interpreted based on the hazard ratio estimates for factors of interest and their P values. A threshold of .05 was used to define statistical significance, and no adjustments for multiple comparisons were performed. All statistical calculations were performed using R version 2.5.1.
Results
Patient characteristics
Included in this analysis are 139 patients with previously untreated and 39 patients with previously treated CLL. Their clinical characteristics are shown in Table 1. Median patient age at enrollment was 63 years for previously untreated patients. Forty-one percent of patients had low-risk (Rai stage 0) and 55% intermediate-risk (Rai stage I-II) disease at enrollment. The median time from diagnosis to enrollment was 12 months, and the median follow-up time from enrollment to clinical outcome analysis was 18 months. Treatment status was verified for all patients. Of the 139 previously untreated patients, 38 patients had received therapy for their CLL and 9 of these moved on to receive subsequent therapy at the time of this analysis. Of the 39 previously treated patients, 21 had received subsequent therapy. The median number of prior therapies received in the previously treated group of patients was 1 (range: 1-7). All analyses described in the next subsection are based solely on biologic specimens collected at the time of enrollment.
Internal validation of the study cohort using established biomarkers
At enrollment, IgVH mutational status was successfully determined for 98% of patients. Interphase FISH was performed at the Mayo Clinic and categorized using the Dohner hierarchic model.26 ZAP-70 expression, CD38 expression, and p53 mutational status were determined in our laboratory for 100% of patients, and results are summarized in Table 1.
Figure S1A-I (available on the Blood website; see the Supplemental Materials link at the top of the online article) depicts Kaplan-Meier curves for previously untreated patients for univariate analyses of presence/absence of each of these prognostic markers in relationship to TTFT.4-7,10-12 Table S1 contains estimates of median TTFT, univariate hazard ratios, and significance levels. Presence of established high-risk prognostic features, including higher Rai stage, unmutated IgVH (using either a 97% or 98% homology to germ line as the cutoff), ZAP-70 positivity, CD38 positivity (using either 7% or 30% positive cells as the cutoff17 ), and mutated p53 resulted in significantly shorter TTFT in comparison with lower-risk counterparts. Individual hierarchic FISH categories did not reach statistical significance, although a trend for shorter TTFT for a combined group of patients with del17p/del11q (P = .06) was evident.
Given that FISH-defined chromosomal aberrations occur in CLL subclones of various proportions (range: 4%-96% in this study), we also tested FISH categories defined by a cutoff of 25% (FISH25) positivity. Under these conditions, del11q/del17p (FISH25) was a significant risk factor (P = .01) for short TTFT (Figure S1H).
Subsequently, we performed a multivariate analysis of TTFT using Cox proportional hazard modeling with ZAP-70, IgVH, CD38, p53 mutations, del11q/del17p, and Rai stage included in the model. ZAP-70 status, IgVH mutations (unmutated), and higher Rai stage emerged as independent predictors for shortened TTFT.
Analysis of previously treated patients using univariate analyses for presence/absence of each of the prognostic markers in relationship to time to subsequent therapy (TTST) is depicted in Kaplan-Meier plots in Figure S2A-H, with median TTST, univariate hazard ratios, and significance levels shown in Table S1.
Presence of higher Rai stage, unmutated IgVH, ZAP-70 positivity, CD38 positivity, and mutated p53 resulted in significantly shorter TTST in comparison with lower-risk counterparts. Further, del17p alone was significantly associated with a very short TTST (P = .008) as was the combined variable del17p/del11q (P = .02).
SNP arrays can detect single copy differences in the CLL genome
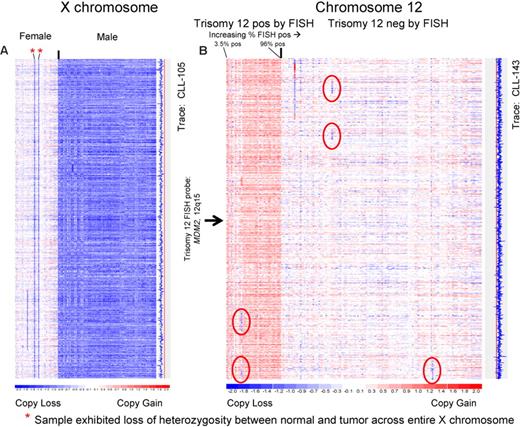
To obtain experimental verification that the SNP chip platform would reliably detect single copy number changes, we initially analyzed the X-chromosome across all patients using dChipSNP. In Figure 1A, we have vertically displayed the color-coded copy number estimates for the X-chromosome at all SNP positions for all patients who were analyzed, grouped by sex. All but 2 female patients were identified as 2N and all males were recognized as 1N. The 2 female patients with 1N estimates demonstrated LOH across the entire X-chromosome. Therefore, we achieved 100% sensitivity and 100% specificity for detecting 1 versus 2 X-chromosomes in our patient cohort.
The 50k SNP array platform can detect single copy chromosomal differences in the CLL genome with high sensitivity and specificity. DNA from FACS-sorted CD19+ cells and buccal cells was extracted and prepared for hybridization to the 50kXbaI SNP chips as per manufacturer's recommendation. Copy number estimates for each SNP position were calculated using dChipSNP and displayed for all SNP positions, for all patients, for the indicated chromosomes. Copy losses are displayed with blue colors; copy gains, with red colors. The physical position of SNPs is not linear along the displayed portions of the chromosome. (A) Chromosome X. Color-coded copy number estimates are grouped by the known sex of the patients. Red asterisks indicate females with LOH at chromosome X indicating loss. The estimated copy numbers for all SNP positions for CLL no. 105 are displayed along the entire chromosome to the right of the X chromosome display, with the red line indicating the 2N state. (B) Chromosome 12. All patients with FISH results positive for trisomy 12 are grouped on the left of the image. The estimated copy numbers for all SNP positions for CLL no. 143 is shown along the entire chromosome to the right of the chromosome 12 display, with the red line indicating the 2N state. Sporadic subchromosomal copy losses are encircled with red ovals. The approximate position of the clinically used trisomy 12 FISH probe is indicated.
The 50k SNP array platform can detect single copy chromosomal differences in the CLL genome with high sensitivity and specificity. DNA from FACS-sorted CD19+ cells and buccal cells was extracted and prepared for hybridization to the 50kXbaI SNP chips as per manufacturer's recommendation. Copy number estimates for each SNP position were calculated using dChipSNP and displayed for all SNP positions, for all patients, for the indicated chromosomes. Copy losses are displayed with blue colors; copy gains, with red colors. The physical position of SNPs is not linear along the displayed portions of the chromosome. (A) Chromosome X. Color-coded copy number estimates are grouped by the known sex of the patients. Red asterisks indicate females with LOH at chromosome X indicating loss. The estimated copy numbers for all SNP positions for CLL no. 105 are displayed along the entire chromosome to the right of the X chromosome display, with the red line indicating the 2N state. (B) Chromosome 12. All patients with FISH results positive for trisomy 12 are grouped on the left of the image. The estimated copy numbers for all SNP positions for CLL no. 143 is shown along the entire chromosome to the right of the chromosome 12 display, with the red line indicating the 2N state. Sporadic subchromosomal copy losses are encircled with red ovals. The approximate position of the clinically used trisomy 12 FISH probe is indicated.
The assay's ability to detect a 3N state (complete or partial trisomies) was then tested using data from the analysis of chromosome 12. In Figure 1B, we have displayed vertically the color-coded copy number estimates for chromosome 12 at all SNP positions, grouped by FISH-based knowledge of trisomy 12. We achieved a sensitivity of 100% and a specificity of 88% for detecting trisomy 12.
Visual complexity score
Two visual complexity scores (described in “Derivation of a genomic complexity score, Visual method”) were calculated for each patient: (1) total subchromosomal copy number changes, and (2) losses alone. Overall, we detected 281 losses, 41 gains, and 11 instances of UPD (proportionally representing 85%, 12%, and 3% of total aberrations, respectively) in 178 CLL genomes. The mean number of lesions per patient was 1.7 and 2.4 for previously untreated and treated patients, respectively. Within the group of 139 untreated patients, 49 (35%), 27 (19%), 21 (15%), 15 (11%), and 11(8%) had total visual complexity scores of equal to or greater than 2, 2.5, 3, 3.5, or 4, respectively. The relationship of genomic complexity of 2.5 or more and other prognostic variables for previously untreated CLL patients is summarized in Table S2.
Univariate analysis of the effect of genomic complexity on TTFT or TTST estimates
Previously untreated patients.
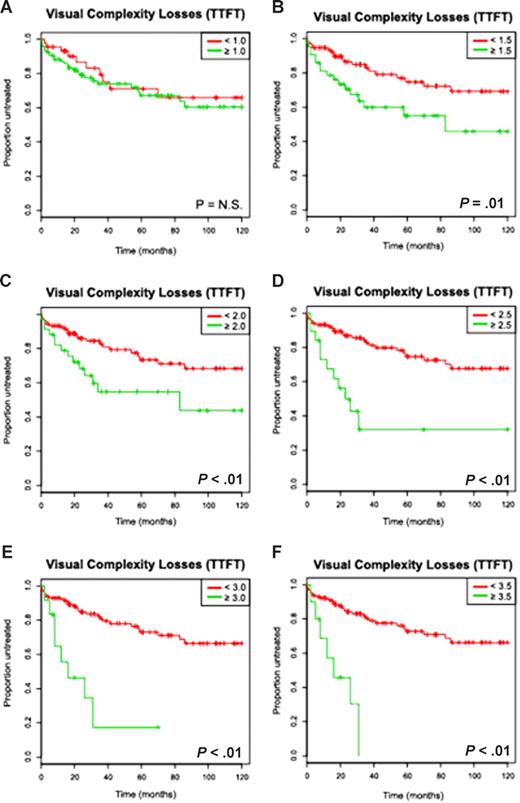
In univariate analysis, increasing binary cutoffs for a high versus low complexity score (analyzed separately for losses only and the total complexity score) were compared with TTFT in previously untreated patients and are depicted in Figure 2 and Figure S3. Genomic complexity based on losses attained significance as a prognostic marker at 1.5 or more lesions per genome with a sharp fall in TTFT estimates once 2.5 or more lesions were considered (score ≥ 2, TTFT 79 months; score ≥ 2.5, TTFT 23 months) and with progressively shorter TTFT estimates as higher cutoffs were analyzed (Table 2). Total complexity attained prognostic significance at 2.5 or more lesions per genome.
The visual complexity score based on losses predicts for a short time to first therapy in univariate analysis in a cohort of previously untreated CLL patients (Kaplan-Meier plots). The numbers of subchromosomal genomic losses for each patient were determined using 50kXbaI SNP array technology as described in “Derivation of a genomic complexity score, Visual methods” and the mean number of losses as determined by 2 independent observers correlated against the clinical end point time to first therapy (TTFT). Depicted are Kaplan-Meier estimates (A-F) for increasing complexity score cutoffs (eg, < N vs ≥ N lesions per genome) and TTFT estimates (months).
The visual complexity score based on losses predicts for a short time to first therapy in univariate analysis in a cohort of previously untreated CLL patients (Kaplan-Meier plots). The numbers of subchromosomal genomic losses for each patient were determined using 50kXbaI SNP array technology as described in “Derivation of a genomic complexity score, Visual methods” and the mean number of losses as determined by 2 independent observers correlated against the clinical end point time to first therapy (TTFT). Depicted are Kaplan-Meier estimates (A-F) for increasing complexity score cutoffs (eg, < N vs ≥ N lesions per genome) and TTFT estimates (months).
For example, median TTFT was 14.5 months versus not reached at 10 years for patients with 3 or more as opposed to fewer than 3 losses (P < .001, hazard ratio: 5.4) and 25 months versus not reached at 10 years for patients with 3 or more as opposed to fewer than 3 total lesions (losses + gains + UPD, P < .001, hazard ratio: 4.3). Importantly, genomic complexity scores of 2.5, 3, 3.5, and 4 predicted for a high fraction of previously untreated patients in need of therapy (52%, 57%, 66%, and 73%, respectively) even after the relatively short follow up of this study.
Previously treated patients.
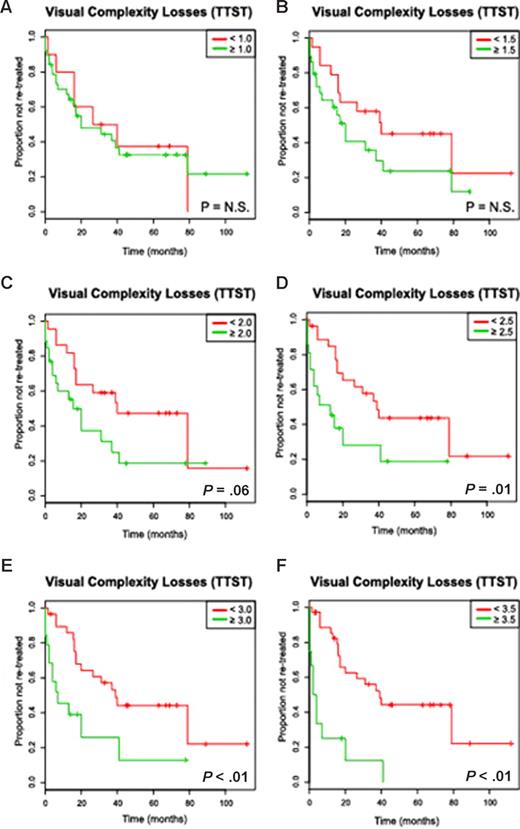
In univariate analysis, increasing binary cutoffs for a high versus low complexity score (losses only or total score) were compared with TTST in previously treated patients and are depicted in Figure 3 and Figure S4. Genomic complexity based on losses attained significance as a prognostic marker at 2.5 or more lesions per genome (Table 2) and with progressively shorter TTST estimates as higher cutoffs were analyzed. Total complexity attained significance at 3 or more lesions per genome. For example, median TTST was 6 months versus 38 months for patients with 3 or more as opposed to fewer than 3 losses (P < .001, hazard ratio: 3.1), and 7 months versus 39 months for patients with 3 or more as opposed to fewer than 3 total lesions (losses + gains + UPD, P < .001, hazard ratio: 3.4).
The visual complexity score based on losses predicts for a short time to subsequent therapy in univariate analysis in a cohort of previously treated CLL patients (Kaplan-Meier plots). The numbers of subchromosomal losses were determined using 50kXbaI SNP array technology for each patient as described in “Derivation of a genomic complexity score, Visual method,” and the mean number of losses as determined by 2 independent observers correlated against the clinical end point time to subsequent therapy (TTST). Depicted are Kaplan-Meier estimates (A-F) for increasing complexity score cutoffs (eg, < N vs ≥ N lesions per genome) and TTST estimates (months).
The visual complexity score based on losses predicts for a short time to subsequent therapy in univariate analysis in a cohort of previously treated CLL patients (Kaplan-Meier plots). The numbers of subchromosomal losses were determined using 50kXbaI SNP array technology for each patient as described in “Derivation of a genomic complexity score, Visual method,” and the mean number of losses as determined by 2 independent observers correlated against the clinical end point time to subsequent therapy (TTST). Depicted are Kaplan-Meier estimates (A-F) for increasing complexity score cutoffs (eg, < N vs ≥ N lesions per genome) and TTST estimates (months).
The occurrence of genomic complexity of 2.5 or more and other important prognostic variables for previously treated CLL patients is summarized in Table S3.
Multivariate analysis of the effect of genomic complexity on TTFT or TTST estimates
Previously untreated patients.
Multivariate analysis using Cox proportional hazard modeling was performed including Rai stage, ZAP-70, IgVH mutations status, del17p/del11q, and genomic complexity at a cutoff of 3 or more and incorporating either total complexity or losses only into the model. Parallel modeling was performed by adding the p53 mutation status, the CD38 status, or both to the models as outlined in “Assessment of a association between genomic complexity and clinical end point.” Data are summarized in Table 3.
In the model including all variables and genomic complexity based on losses only, ZAP-70, IgVH, Rai stage, and genomic complexity (hazard ratio of 10 at P = .01) emerged as independent risk factors for short TTFT. In the model including all variables and total complexity, variables with significant effects on TTFT were ZAP-70, IgVH, Rai stage, and total complexity (hazard ratio of 3.3 at P = .04). Both total complexity and the losses-only complexity measure remained significant regardless of whether p53 and/or CD38 were included in the model (all P values < .05 for the losses-only score and the total complexity score).
Incorporating del11q/del17p (FISH25) into the models instead of all FISH findings did not meaningfully change the fitted models.
Previously treated patients.
Multivariate analysis using Cox proportional hazard modeling was performed including ZAP-70, IgVH mutations status, del17p/del11q, and genomic complexity at a cutoff of 3 or more and incorporating either total complexity or losses only into the model. Parallel modeling was performed by adding the p53 mutation status, the CD38 status, or both to the models as outlined. Data are summarized in Table 3.
Importantly, in the model including all variables and complexity based on losses only, genomic complexity emerged as an independent risk factor for short TTST (hazard ratio of 2.8 at P = .02). Similar results were obtained when total complexity was analyzed. Both genomic complexity scores were significantly associated with TTST regardless of whether p53 and/or CD38 were included in the model (all P values < .02).
Selected bivariate analysis
Based on results using univariate and multivariate analysis, we selected a total complexity score at a cutoff of 3 or more for the following bivariate analysis. Bivariate displays were selected to display situations that one may encounter in clinical practice in the future, where one or more standard risk factors (Rai stage, ZAP-70, or IgVH) were available in addition to SNP array–based genomic profiles.
IgVH mutational status.
Patients with IgVH unmutated/high genomic complexity (n = 16) had a shorter median TTFT of 23 months versus 70 months in IgVH unmutated/low complexity patients (n = 42); this is compared with the median TTFT of 39 months noted in the IgVH unmutated group as a whole.
ZAP-70 expression.
Within the ZAP-70–positive group, the high complexity median TTFT was 26 months (n = 15) versus 58 months for low complexity (n = 44); this compared with the median TTFT of 35 months noted in the ZAP-70–positive group as a whole.
Rai staging system.
The TTFT was 25 months (n = 8) for intermediate Rai stage patients with high genomic complexity versus not reached at 10 years for low complexity patients.
Algorithmic complexity score
In an effort to automate detection of patients with high genomic complexity, we devised algorithmic lesion scoring for losses (described in “Derivation of a genomic complexity score, Algorithmic method”). The procedure was calibrated to 3 common CLL-associated lesions, without reference to the outcome data.
Univariate analysis
Previously untreated or pretreated patients.
As was demonstrated in univariate analysis using the visual complexity score, as the algorithmic genomic complexity score threshold defining the “high complexity” group was increased, greater statistical separation in TTFT and TTST between high and low complexity patients resulted (Figure S5A-H).
Multivariate analysis of the effect of algorithmic genomic complexity on TTFT or TTST estimates
Previously untreated patients.
Multivariate analysis using Cox proportional hazard modeling was performed including Rai stage, ZAP-70, IgVH mutations status, del17p/del11q, and algorithmic genomic complexity at a cutoff of 3 or more. Parallel modeling was performed by adding the p53 mutation status or the CD38 status or both to the models as outlined. Data are summarized in Table 4.
In the model including all variables and algorithmic genomic complexity, only ZAP-70, IgVH, Rai stage, and genomic complexity (hazard ratio of 4 at a P = .02) emerged as independent risk factors for short TTFT.
Previously treated patients.
Multivariate analysis using Cox proportional hazard modeling was performed including ZAP-70, IgVH mutations status, del17p/del11q, and algorithmic genomic complexity at a cutoff of 3 or more. Parallel modeling was performed by adding the p53 mutation status or the CD38 status or both to the models as outlined above. Data are summarized in Table 4.
Similar to the analysis using visual complexity estimates, in the model including all variables, algorithmic genomic complexity emerged as an independent risk factor for short TTST (hazard ratio of 3 at P = .01).
Discussion
High-density SNP arrays allow for an essentially unbiased evaluation of cancer genomes through the assessment of allelic losses/gains and LOH. In our study population of 178 patients with CLL, genome-wide SNP array–based copy number analysis was used to risk-stratify patients into low- and high-risk cohorts based on their degree of genomic complexity.
SNP array analysis proved to be a sensitive and highly specific method for determining subchromosomal copy number losses and gains in CLL, detecting genomic lesions that were found by FISH in 25% or more of cells with approximately 90% sensitivity and approximately 95% specificity. Approximately 35%, 19%, and 15% of patients in this study had 2 or more, 2.5 or more, or 3 or more genomic lesions (range, 0-14), and a large proportion of genomic lesions apparently arose spontaneously and were not induced by chemotherapeutics.
Pioneering research in colorectal carcinomas provided some of the earliest insights into the prognostic significance of fractional allelic losses as a marker for genomic complexity in cancer.39 Similar results have been reported in acute myeloid leukemia, where the presence of a complex karyotype (≥ 3 chromosomal abnormalities) has been associated with poor-prognosis disease,40,41 as well as in CLL, where a negative prognostic impact of mostly unbalanced chromosomal translocations has been reported.27,28,42 Finally, innovative analysis of multiple gene expression data sets, indirectly measuring allelic gains and losses, demonstrated that chromosomal complexity as a consequence of genomic instability negatively impacts on prognosis in many common human cancers.43
The overriding observation from our results is that genomic complexity emerged as an independent risk factor for disease progression before (TTFT analysis) and after (TTST analysis) therapy.
Recent data published in the setting of fludarabine or fludarabine/cyclophosphamide-based therapies uncovered increased risk for relapse in CLL patients with del17p or del11q.19 Using a multivariate Cox model applied to our dataset, which incorporated del17p, del11q, and other risk factors (ZAP-70, IgVH mutation status, CD38, and p53 mutations), as well as genomic complexity, we found that genomic complexity reached significance as an independent risk factor. These results imply that the relapse risk imparted on individuals through del17p or del11q is measurable through genomic complexity assessments. Clinically, SNP array–based analysis of genomic complexity could therefore help identify poor-risk cohorts of patients who might benefit from risk-adapted therapies44-46 ; such applications may necessitate enrichment of CD19+ cells from blood or marrow specimens through automated devices (magnetic beads/column devices or tabletop FACS machines).
It appears rational to assume that there are underlying defects in DNA double-strand break repair systems present in the CLL group with high genomic complexity,8,13,47 which could explain the relative therapeutic ineffectiveness (short TTST) of purine analogs in these patients. Fludarabine has been demonstrated to induce a p53-dependent set of genes in CLL, and ATM mutations have been associated with shorter overall survival and relative resistance to fludarabine in vitro.13,48 Work is in progress to define the contributions of individual components of the DNA ds-break response to genomic complexity in CLL.
With regard to the observed shortened TTFT estimates in patients with high genomic complexity, it remains mechanistically unclear why CLL progresses more rapidly in these patients than in genomically stable patients.47,49 Defects in various cell cycle checkpoints required to recognize and repair DNA damage or promote apoptosis, resulting in untimely cell-cycle progression, may be implicated.50
One limitation of our study derives from the often long natural history of CLL and the resulting need to assay most patients after the initial diagnosis date. This caveat is mitigated by the fact that 50% of all previously untreated patients in this study were enrolled and analyzed within 1 year of diagnosis. Few if any quantitative measurements of CLL genome stability over time are available to measure the possible bias introduced in analyzing patients for genomic complexity after the initial diagnosis date.20 Longitudinal measurements of acquired genomic instability are therefore a research goal for future investigations using the SNP array platform.
Both the visual and algorithmic methods of determining the genomic complexity score are feasible methods for detection of subchromosomal losses, and a complexity score based on losses alone performed better than a score incorporating losses, gains, and UPD. The algorithmic method for determination of the genomic complexity score is automated and rapid, making it a practical option for implementation in the clinical setting.
In conclusion, our data suggest that a high degree of genomic complexity can serve as a strong negative prognostic marker in CLL by identifying patients with aggressive disease. Larger prospective clinical trials with longer follow-up periods, determination of treatment response outcomes based on degree of complexity, and changes in individual complexity score values over time while on observation versus therapy20 will help to determine the definitive clinical significance of genomic complexity measurements and how best to use this information to guide treatment of CLL patients.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to the staff of the Cell Kinetics Laboratory (Dr Katzmann) at the Mayo Clinic for assistance in establishing the ZAP-70 assays in our laboratory.
This work was supported by a Leukemia & Lymphoma Society of America Special Fellow Award (S.N.M.), a Leukemia Research Foundation New Investigator Award (S.N.M.), the National Institutes of Health through 1 R21 CA124420-01A1 (S.N.M.), and (in part) by the National Institutes of Health through the University of Michigan's Cancer Center Support Grant (5 P30 CA46592).
National Institutes of Health
Authorship
Contribution: P.O., C.S., and S.N.M. performed the laboratory research; K.S. assisted with statistical methods; L.K., H.E., A.J., M.K., and S.N.M. enrolled patients and contributed and analyzed the clinical data; S.N.M. conceived the study; and L.K., P.O., K.S., and S.N.M. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sami N. Malek, Department of Internal Medicine, Division of Hematology and Oncology, University of Michigan, 1500 E Medical Center Drive, Ann Arbor, MI 48109-0936; e-mail: smalek@med.umich.edu.
References
Author notes
L.K. and P.O. contributed equally to this work.