In this issue of Blood, Oerlemans and colleagues present a fascinating report, detailing a mechanism by which cells acquire resistance to therapy with the proteasome inhibitor, bortezomib.
Proteasome inhibition represents one of the most successful anticancer strategies of this decade, improving the outcomes of many patients. The ubiquitin proteasome pathway is critical to normal cellular functioning and is involved in signal transduction, transcriptional regulation, and response to stress, among other pathways. The 26S proteasome consists of a core 20S catalytic complex and a 19S regulatory complex, forming 2 outer and 2 inner rings that are stacked to form a cylindrical structure.1 The 19S complex is responsible for selecting the ubiquitinated proteins for catalytic degradation by the 20S complex, which possesses chymotryptic, tryptic, and peptidylglutamyl-like activities. This critical cellular function has been successfully targeted for cancer therapy, as highlighted by the efficacy of proteasome inhibitor bortezomib in a wide spectrum of hematological and solid tumors.2 In fact, the introduction of bortezomib resulted in a paradigm shift in the treatment of multiple myeloma, and has undoubtedly contributed to the improved survival seen among patients with this incurable malignancy.3 The mechanism of antimyeloma activity of bortezomib is the subject of intense study. Bortezomib is currently believed to exert its effects through multiple pathways that target both the tumor cell and its microenvironment. For example, inhibition of the NFkB pathway leading to decreased cell proliferation and induction of apoptosis is one of the major effects of bortezomib therapy. Treatment with bor-tezomib prolongs survival in relapsed myeloma as well as newly diagnosed disease, leading to its regulatory approval for clinical use in both situations.4 However, resistance to therapy develops inevitably. Furthermore, nearly a third of the patients with multiple myeloma never respond to treatment with bortezomib, depending on the clinical situation. While some resistance mechanisms may be reversible in a small proportion of patients following withdrawal of the drug, as demonstrated by the efficacy of retreatment, the majority need to switch therapy. Understanding the mechanisms of resistance to proteasome inhibition will not only allow better use of proteasome inhibitors such as bortezomib, but should also allow the rational design of synergistic drug combinations.
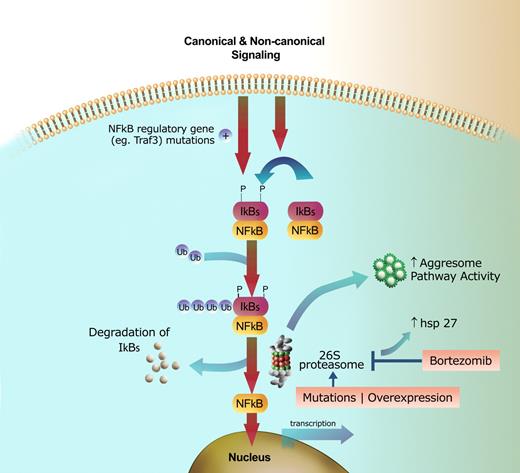
Mechanisms of resistance and susceptibility to proteasome inhibition. Signaling through canonical or noncanonical pathways leads to phosphorylation, ubiquitination, and subsequent degradation of the IkB kinases through the proteasome pathway, resulting in NFkB translocation to the nucleus and transcription of target genes. Constitutive NFkB signaling can result from mutations in regulatory genes, such as Traf3, leading to increased sensitivity of the cell to proteasome inhibition. Mechanisms of resistance include: (1) the 26S proteasome acquiring resistance due to mutations or overexpression of the PSMB5 subunit; (2) proteasome inhibitors being antagonized by upregulation of heat shock proteins, such as Hsp27; and (3) increased activity of the aggresome pathway. These resistance mechanisms can be targeted to increase the efficacy of proteasome inhibition.
Mechanisms of resistance and susceptibility to proteasome inhibition. Signaling through canonical or noncanonical pathways leads to phosphorylation, ubiquitination, and subsequent degradation of the IkB kinases through the proteasome pathway, resulting in NFkB translocation to the nucleus and transcription of target genes. Constitutive NFkB signaling can result from mutations in regulatory genes, such as Traf3, leading to increased sensitivity of the cell to proteasome inhibition. Mechanisms of resistance include: (1) the 26S proteasome acquiring resistance due to mutations or overexpression of the PSMB5 subunit; (2) proteasome inhibitors being antagonized by upregulation of heat shock proteins, such as Hsp27; and (3) increased activity of the aggresome pathway. These resistance mechanisms can be targeted to increase the efficacy of proteasome inhibition.
Malignant cells may develop several mechanisms to escape the effects of proteasome inhibition, including alterations in the proteasome complex itself leading to decreased function, increasing the efficiency of alternate mechanisms of protein degradation (the aggresome pathway), or modulation of cell signaling pathways that are affected by proteasome inhibition. Oerlemans et al, in this issue of Blood, report on a mutation involving the β5 unit of the proteasome catalytic unit (PSMB5) that leads to impaired binding of bortezomib and thus decreased proteosome inhibition. These investigators also noted a significant upregulation of the PSMB5 subunit following exposure to bortezomib and other proteasome inhibitors, an effect that wanes with time off-therapy, but reappears rapidly after re-exposure to the inhibitors. These studies were carried out using human monocytic/macrophage THP1 cells, and whether these findings are applicable to malignant cells in diseases like myeloma is unclear. However, these findings do highlight the susceptibility of proteasome units to genetic modifications under constant selection pressure, as can occur with continued treatment in patients. Mutation and overexpression of PSMB5 can lead to bortezomib resistance in lymphoma cells lines. While mutations such as this may explain development of resistance, baseline differences in susceptibility may be due to polymorphisms involving the PSMB5 locus.5
An alternate mechanism used by the cell for ubiquitinated protein degradation and disposal is the aggresome pathway, which can potentially compensate for proteasome pathway inhibition and contribute to drug resistance. This physiological compensatory mechanism has been targeted for enhancing the efficacy of proteasome inhibitors. Use of HDAC6 specific inhibitors, such as tubacin, can shut down the aggresome pathway and can synergize with and enhance the effect of proteasome inhibition on the tumor cell.6 Upregulation of the heat shock protein Hsp27 is yet another mechanism of proteasome inhibitor resistance and has been targeted as an avenue for enhancing proteasome inhibition as well as reversing resistance to this class of drugs.7
Finally, identification of mechanisms that confer sensitivity to proteasome inhibition is as important as understanding mechanisms of resistance. Recent studies have identified mutations involving genes associated with regulation of NFkB pathways that result in constitutive activation of the NFkB pathway.8 Cells that carry these mutations appear to be particularly sensitive to the effects of proteasome inhibition, a finding that could allow us to tailor the use of this class of drugs in the future.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■