Abstract
Variegated expression of 6 inhibitory HLA class I–specific receptors on primary NK cells was studied using high-dimension flow cytometry in 58 humans to understand the structure and function of NK-cell repertoires. Sixty-four subsets expressing all possible receptor com-binations were present in each repertoire, and the frequency of receptor-null cells varied among the donors. Enhancement in missing-self response between NK subsets varied substantially where subset responses were defined by donor KIR/HLA allotypes, reflecting the differences in interaction between inhibitory receptors and their ligands. This contrasted to the enhancement conferred by NKG2A, which was constant and of intermediate strength. We infer a mechanism that modulates frequencies of the NK subsets displaying diverse levels of missing-self response, a system that reduces the presence of KIR-expressing subsets that display either too strong or too weak a response and effectively replaces them with NKG2A-expressing cells in the repertoire. Through this high-resolution analysis of inhibitory receptor expression, 5 types of NK-cell repertoire were defined by their content of NKG2A+/NKG2A− cells, frequency of receptor-null cells, and degree of KIR receptor coexpression. The analyses provide new perspective on how personalized human NK-cell repertoires are structured.
Introduction
Natural killer (NK) cells are lymphocytes of innate immunity that secrete cytokines and kill cells in response to infection, cancer, or allogeneic tissue.1-3 A unique feature of NK-cell recognition is detection of the loss of self-major histocompatibility complex (MHC) class I expression.4-6 This “missing-self response” is determined by inhibitory MHC class I receptors, which in human comprise the conserved NKG2A receptor and the variable killer-cell immunoglobulin-like receptors (KIR).1,7-10 In mouse, the structurally divergent Ly49 receptors have functions analogous to human KIR.11-13 Variability in KIR gene content and allelic polymorphism are 2 factors that diversify human NK-cell repertoires.14-16 Further crucial factors are the HLA-A, -B, and -C polymorphisms, which are genetically unlinked to KIR and determine which inhibitory KIR will be engaged by self-MHC class I and contribute to the missing-self response.1,16
During development, NK cells first express NKG2A and then KIR, in a variegated manner, which creates an NK-cell repertoire in which cellular subsets express different combinations of receptors that are stable over time.17-21 However, few reports have comprehensively characterized the distribution of MHC class I–specific inhibitory receptors, particularly in freshly isolated NK-cell populations in either human or mouse.21,22
The “product rule” proposes that the frequency of an NK subset coexpressing multiple inhibitory receptors follows the probability of each receptor.21-24 Previous studies have suggested that coexpression between NKG2A and KIR/Ly49 is reduced in both mouse and human and that receptor expression tends to deviate from the product rule.21,25 Studies in mouse have proposed that presence of the cognate ligand reduces frequency of the corresponding Ly49 inhibitory receptor in NK cells mainly because of reduction of receptor coexpression.23,26 A more complex picture is emerging from analyses on human NK cells, where key questions remain as to why it is that presence of the cognate ligand increases KIR expression frequency but reduces their frequency when additional ligands are present.27
For a decade, the inhibitory NK-cell receptors for MHC class I were considered mainly as a mechanism for self-tolerance; thus, previous models of MHC-specific inhibitory receptor acquisition on NK cells have built on the concept that individual NK cells require an adequate number of inhibitory receptors to maintain self-tolerance, the “at-least-one” theory.12,13,21 Recent research has established that NK cells lacking MHC-specific inhibitory receptors “receptor-null” cells are both tolerant of autologous cells and hyporesponsive to missing-self. It is now seen that, to be strongly responsive to missing-self, an NK cell requires an inhibitory receptor that can interact with a self-MHC class I ligand. This phenomenon has been described by several groups of investigators and termed “education,” “arming,” or “licensing.”6,28-31 Such enhancement of the missing-self response calls for a fresh perspective to explain NK-cell repertoire formation.
The effects of KIR and HLA on NK-cell repertoire formation are complex and have eluded clarification. We sought to understand the mechanisms underlying human NK-cell repertoire formation through characterization and comparison of NK-cell repertoires by visualizing KIR, NKG2A, and LILRB1 expression in a panel of humans using 11-parameter flow cytometry on primary NK-cell populations. Assessment of individual NK subsets enabled us to evaluate the effects of KIR and HLA polymorphisms on the NK-cell response and how levels of missing-self response contribute to expression and coexpression of KIR and NKG2A and to the overall structure of human NK-cell repertoires.
Methods
Human subjects and blood samples
Blood samples were from 58 healthy persons (35 male, 23 female; age, 19-72 years; mean, 29.9 ± 10 years [SD]); written consent was obtained from all donors, and procedures were performed as approved by the Stanford University Institutional Review Board on Human Subjects. Peripheral blood mononuclear cells (PBMCs) were prepared with Ficoll gradient separation. This study was performed in accordance with the Declaration of Helsinki.
KIR and HLA class I characterization
KIR gene content typing was performed with sequence-specific polymorphism-polymerase chain reaction (SSP-PCR) based typing as reported previously, and allele typing for KIR2DL1/2DL3/3DL1 was performed by sequencing KIR transcripts of cDNA and using the SSP-PCR method as described.27,32 HLA-A, -B, and -C allele assignment was performed as described.27
High-dimension flow cytometry
The following monoclonal antibodies (mAbs) were used for flow cytometric analyses. Anti-2DL1/2DS1 (EB6) and anti-3DL1/3DS1 (Z27) from Beckman Coulter (Fullerton, CA); anti-2DL2/2DL3/2DS2 (CHL), anti-3DL1 (DX9), anti-NKG2A (Z199), and anti-LILRB1 (GHI/75) from BD Biosciences (San Jose, CA); and anti-3DL2 (DX31) was a gift of J. Phillips and L. Lanier (Schering Plough, Biopharma, Palo Alto, CA; and Department of Microbiology & Immunology, University of California–San Francisco). These mAbs were labeled with fluorescein isothiocyanate (FITC), phycoerythrin (PE), PE-Cy5 or allophycocyanin (APC), AlexaFluor405, or AlexaFluor700. APC-Cy7-anti-CD3 (S4.1), PECy5.5-anti-CD56 (MEM-188) from Invitrogen (Carlsbad, CA) and PE-Cy5-anti-CD19 (HIB19) (BD Biosciences) were combined to identify CD56dim NK cells. Cell-surface staining was combined with antibodies against interferon-γ (IFN-γ) and CD107 to assess NK subset function. Dead cells were excluded from analyses by gating out nonviable cells (Dead-cell discrimination kit; Miltenyi Biotec, Auburn, CA). Cells were analyzed by 11-parameter flow cytometry using FACSAria (BD Biosciences, 407, 488, and 633 nm excitation), maintained by the Stanford Shared FACS Facility. Data analyses used FlowJo 6.4.7 software (TreeStar, Ashland, OR) and followed proposed guidelines.33
Assessment of missing-self response and class I–mediated inhibition
Cytokine response was assessed by intracellular staining for IFN-γ, and degranulation was measured using the CD107 mobilization assay. PBMCs at 106 per well were cocultured with either HLA class I–deficient 721.221 cells, allogeneic phytohemagglutinin (PHA) blasts from unrelated donors or without target cells as control at an optimized effector-target ratio of 5:1 in RPMI1640 medium, supplemented with 10% heat-inactivated bovine calf serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2500 units IL-2 (gift from the National Cancer Institute, Frederick, MD). To generate allogeneic PHA blasts, PBMCs were cultured in 0.5 μg/mL PHA (Sigma-Aldrich, St Louis, MO) and IL-2 (100 U/mL) for 4 days. Before assays, PHA blasts were stained with PE-Cy5-anti-CD56 (NCAM; BD Biosciences) to distinguish the CD56+ cells within the blasts from the effector NK cells. FITC-anti-CD107a and -b mAbs (H4A3, H4B4; BD Biosciences) were added for the degranulation assay. Total incubation time was 6 hours at 37°C in 5% CO2. Brefeldin A (GolgiPlug) and monensin (GolgiStop, both BD Biosciences) were added for the last 5 hours of coincubation. Cells were washed with 1% bovine calf serum and incubated with 10% human serum to reduce nonspecific immunofluorescent staining. After cell-surface staining, cells were fixed and permeabilized with Cytofix/Cytoperm solution (BD Biosciences), and intracellular IFN-γ production was assessed using PE-Cy7-anti–IFN-γ mAb (4S.B3; BD Biosciences). 721.221-HLA-B*5801 and HLA-Cw*1202 transfectants were prepared as described previously.34 Inhibition by HLA class I engagement was assessed by measuring reduction in the IFN-γ response of NK cells against 221-B*5801 or -Cw*1202 transfectant compared with parental 721.221.35
Simulation of NK subset frequencies under the “product rule”
NK subset frequency was calculated using overall frequencies of each receptor in the CD3−CD56dim NK-cell population. Given the expression frequency of each receptor in the CD3−CD56dim NK pool as: %2DL1 = x1, %2DL3 = x2, %3DL1 = x3, %3DL2 = x4, %NKG2A = x5, %LILBR1 = x6; and if receptor coexpression were entirely stochastic, subset frequencies would be calculated as the product of receptor frequencies, ie, frequency of the receptor-null subset: (1-x1)*(1-x2)*(1-x3)*(1-x4)*(1-x5)*(1-x6), or frequency of the 2DL1+2DL3+ receptor subset: x1*x2*(1-x3)*(1-x4)*(1-x5)*(1-x6).
Statistics
Nonparametric tests of significance were used in the analyses (2-tailed Mann-Whitney U test). Tests used SPSS version 8 software (SPSS, Chicago, IL).
Results
NK cells express all combinations of inhibitory MHC class I receptors
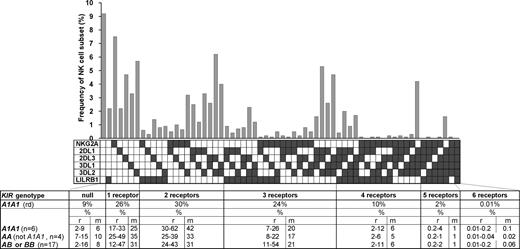
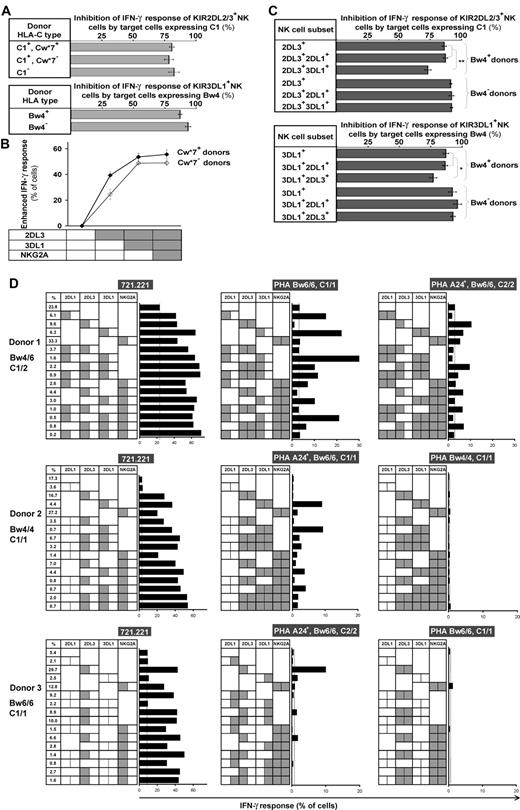
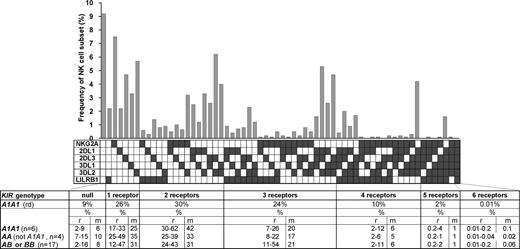
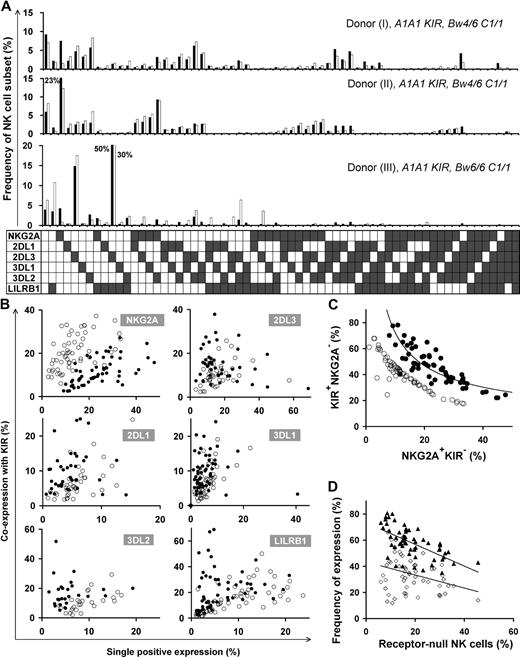
The variegated expression of inhibitory MHC class I receptors by NK cells was determined by multidimensional flow cytometry. Peripheral CD56dim NK cells from a panel of 27 KIR- and HLA-typed (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) human donors were simultaneously stained with mAbs against KIR2DL1, 2DL3, 3DL1, 3DL2, NKG2A, and LILRB1. In the 10 donors homozygous for group A KIR haplotypes, these antibodies react only with inhibitory receptors. NK subsets corresponding to all 64 possible combinations of the 6 inhibitory receptors were defined for all 10 AA donors. For the A1A1 homozygous donor shown in Figure 1, 26% of the NK cells express one receptor and 66% coexpress 2 or more receptors. Cells lacking all 6 receptors comprised 9% of the repertoire, whereas cells expressing all 6 receptors comprised 0.01%. These results demonstrate that for AA KIR donors no combination of inhibitory MHC class I receptors is disallowed or deleted.
All combinations of inhibitory MHC class I receptors are represented in human NK-cell repertoires. In the upper part of the figure, the gray bars give the frequencies for CD56dim NK cells having the receptor combination denoted by the black, filled boxes below each bar; shown are data for a representative donor (rd). The lower part of the figure gives mean (m) and range (r) frequencies for NK subsets expressing different numbers of inhibitory MHC class I receptors, both for the representative donor alone and all 27 donors analyzed. The donors are grouped by KIR haplotype combination. A and B refer to KIR haplotypes defined at the low resolution of gene content16 ; A1 is defined at the high resolution of allele content. The representative donor has the A1A1 genotype. See Figure S1 for donor KIR and HLA class I genotypes.
All combinations of inhibitory MHC class I receptors are represented in human NK-cell repertoires. In the upper part of the figure, the gray bars give the frequencies for CD56dim NK cells having the receptor combination denoted by the black, filled boxes below each bar; shown are data for a representative donor (rd). The lower part of the figure gives mean (m) and range (r) frequencies for NK subsets expressing different numbers of inhibitory MHC class I receptors, both for the representative donor alone and all 27 donors analyzed. The donors are grouped by KIR haplotype combination. A and B refer to KIR haplotypes defined at the low resolution of gene content16 ; A1 is defined at the high resolution of allele content. The representative donor has the A1A1 genotype. See Figure S1 for donor KIR and HLA class I genotypes.
Similar analysis was performed for 17 donors who had either one or 2 group B haplotypes and for whom the anti-KIR2DL1 and/or the anti-KIR2DL2/3 antibodies could also react with the related activating receptors: KIR2DS1 and KIR2DS2, respectively. Overall the results obtained for these donors were comparable with those obtained with the AA donors (Figure 1), suggesting that similar principles govern formation of the NK-cell repertoires in donors of different KIR genotype.
Parameters that characterize variation in human NK repertoires
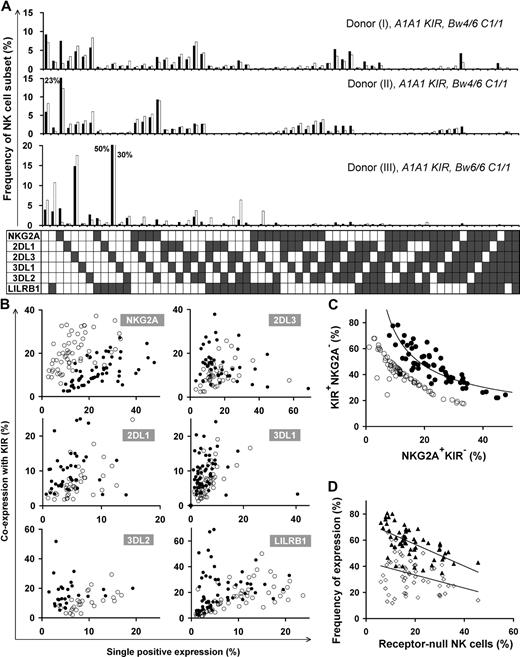
Different HLA types in donors resulted in disparate repertoires, as depicted in the comparison between repertoires of 3 donors with identical KIR genotype in the top 2 panels (Bw4+) versus the bottom panel (Bw4−) in Figure 2A. We also found substantial variation in repertoire between donors with the same KIR genotype and the same HLA class I epitopes that bind KIR (Figure 2A top 2 panels). Here, the main difference is the degree of coexpression between KIR and NKG2A; coexpression among KIR is more prominent than KIR coexpression with NKG2A in donor I, whereas KIRs are coexpressed more with NKG2A in donor II.
Diverse NK repertoires are characterized by the degree of receptor coexpression, balance of NKG2A/KIR, and receptor-null cell content. (A) Shown are NK-cell repertoires for 3 KIR A1A1 haplotype donors. Each observed repertoire (■) is compared with a simulated repertoire (□), which is the repertoire expected by the product rule. Donors I and II have Bw4 and C1; donor III has only C1. (B) Comparison of the expression frequencies for the 6 MHC class I receptors expressed independently (single-positive cells) and in combination with KIR. ● represent the frequencies observed for the 58 donors; ○ represent frequencies predicted by the product rule. (C) The inverse correlation between the frequencies of KIR+NKG2A− and KIR−NKG2A+ NK cells in 58 donors with varied KIR and HLA types. The observed frequencies (●) are compared with those expected from the product rule (○). (D) For each donor, the frequency of receptor-null NK cells is plotted against the frequency of cells expressing one or more KIR (▴) and the frequency of cells expressing NKG2A (◇).
Diverse NK repertoires are characterized by the degree of receptor coexpression, balance of NKG2A/KIR, and receptor-null cell content. (A) Shown are NK-cell repertoires for 3 KIR A1A1 haplotype donors. Each observed repertoire (■) is compared with a simulated repertoire (□), which is the repertoire expected by the product rule. Donors I and II have Bw4 and C1; donor III has only C1. (B) Comparison of the expression frequencies for the 6 MHC class I receptors expressed independently (single-positive cells) and in combination with KIR. ● represent the frequencies observed for the 58 donors; ○ represent frequencies predicted by the product rule. (C) The inverse correlation between the frequencies of KIR+NKG2A− and KIR−NKG2A+ NK cells in 58 donors with varied KIR and HLA types. The observed frequencies (●) are compared with those expected from the product rule (○). (D) For each donor, the frequency of receptor-null NK cells is plotted against the frequency of cells expressing one or more KIR (▴) and the frequency of cells expressing NKG2A (◇).
Receptor coexpression among the 6 inhibitory NK receptors was studied, comparing NK-cell repertoires expected under stochastic expression against that observed for 58 donors (Figure 2B). This revealed bias in the extent to which the 6 inhibitory MHC class I receptors are coexpressed. NKG2A+KIR− expression frequency always exceeds that predicted by the product rule, whereas NKG2A-KIR coexpression is less than predicted. KIR2DL1/2DL3/3DL1 are coexpressed with other KIR in a majority of donors, whereas, in some cases, KIR single-positive cells are dominant. KIR3DL2 and LILRB1 coexpress with other KIRs more frequently than expected by stochastic expression. The degree of receptor coexpression, distinct to each donor, is a primary factor that characterizes human repertoire structure.
A second factor characterizing NK-cell repertoires is the balance between KIR and NKG2A content. Our studies, for the first time, detail the dynamics in this balance where KIR and NKG2A expression are inversely correlated in the 58 donors, following a tight curve (Figure 2C). This correlation establishes a continuum in which persons vary greatly in their proportions of NKG2A+KIR− and NKG2A−KIR+ cells.
We find that receptor-null cells are a third factor that affects repertoire structure. These cells are an essential component of the human NK-cell repertoire, present in all donors, and in frequencies unique to each repertoire. We observed an inverse correlation between the frequency of receptor-null cells and that of KIR, rather than NKG2A (Figure 2D).
Diverse human NK-cell repertoires are classified into 5 types
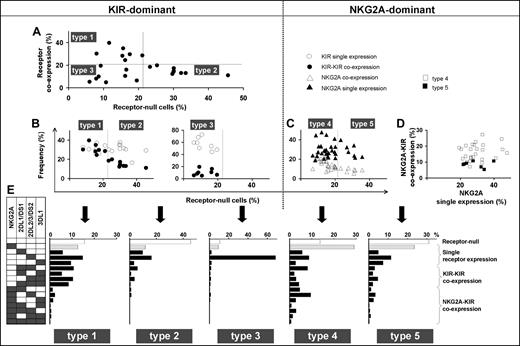
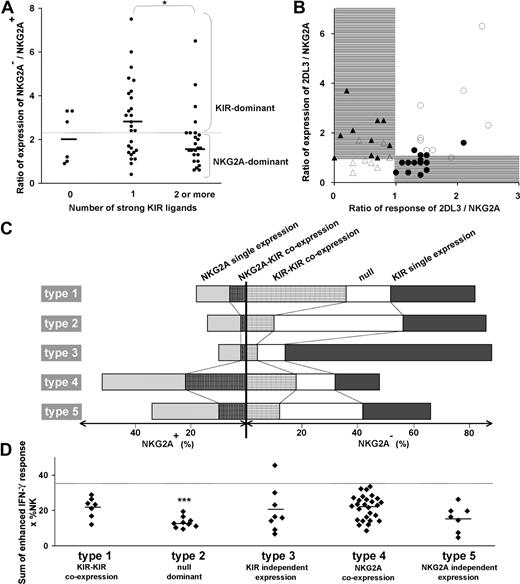
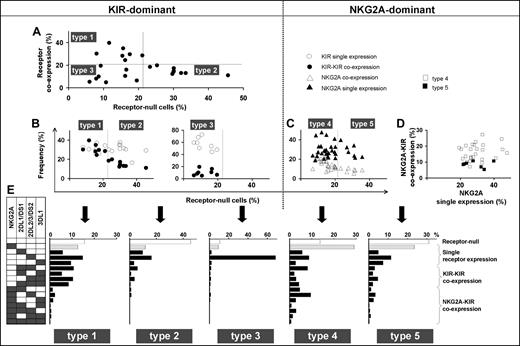
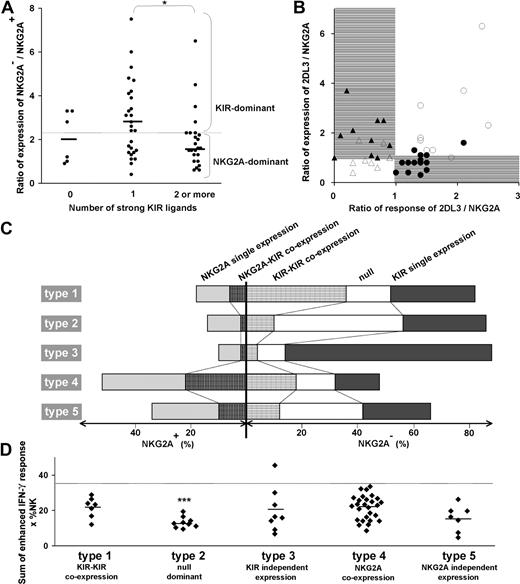
These 3 parameters combine to divide the spectrum of 58 human NK-cell repertoires into 5 broad types (Table 1; Figure 3). In the type 1, 2, and 3 repertoires, KIR expression is dominant, whereas in the type 4 and 5 repertoires, NKG2A expression dominates. The type 1, 2, and 3 repertoires are distinguished by the frequency of receptor-null NK cells and the frequency of KIR-KIR coexpression (Figure 3A,B). Type 1 repertoires have fewer null cells and higher KIR coexpression. Type 2 repertoires have more null cells, less coexpression, and most closely resemble the repertoires predicted by the product rule (Figure S2). Type 3 repertoires are dominated by a single KIR, for which the skewed expansion of the single-positive subset expressing this KIR is associated with reduced KIR coexpression and fewer null cells (Figure 3B).
Human NK-cell repertoires form 5 broad groups according to inhibitory receptor phenotypes. NK-cell repertoires divide into KIR-dominant (left half of figure) and NKG2A-dominant (right half of figure) repertoires. (A) KIR-dominant repertoires further divide into 3, based on the frequencies of receptor-null cells and KIR-KIR coexpression (●). (B) Whereas frequencies of KIR single expression (○) and KIR-KIR coexpression (●) are similar in type 1 repertoires, KIR coexpression is less frequent in type 2 repertoires and even more so in type 3 repertoires. (C,D) NKG2A-dominant repertoires are further divided according to the frequency of receptor-null cells (C). The difference between type 4 (□) and type 5 (■) repertoires is apparent in panel D where the ratio of NKG2A coexpression with KIR against NKG2A single expression is higher in type 4 than type 5 repertoires. (E) Representative profiles of the 5 repertoire types.
Human NK-cell repertoires form 5 broad groups according to inhibitory receptor phenotypes. NK-cell repertoires divide into KIR-dominant (left half of figure) and NKG2A-dominant (right half of figure) repertoires. (A) KIR-dominant repertoires further divide into 3, based on the frequencies of receptor-null cells and KIR-KIR coexpression (●). (B) Whereas frequencies of KIR single expression (○) and KIR-KIR coexpression (●) are similar in type 1 repertoires, KIR coexpression is less frequent in type 2 repertoires and even more so in type 3 repertoires. (C,D) NKG2A-dominant repertoires are further divided according to the frequency of receptor-null cells (C). The difference between type 4 (□) and type 5 (■) repertoires is apparent in panel D where the ratio of NKG2A coexpression with KIR against NKG2A single expression is higher in type 4 than type 5 repertoires. (E) Representative profiles of the 5 repertoire types.
Among NKG2A-dominant repertoires, type 4 repertoires have more NKG2A-KIR coexpression, whereas type 5 repertoires have more cells expressing NKG2A independently as the dominant receptor (Figure 3C,D). The increase in NKG2A-KIR coexpression in type 4 repertoires is associated with reduced numbers of receptor-null cells (Figure 3C,D). We find also that NKG2A-dominant repertoires tend to be more common in people with group B KIR haplotypes than in group A homozygotes (Figure S3).
Strength of missing-self response distinguishes NK subsets
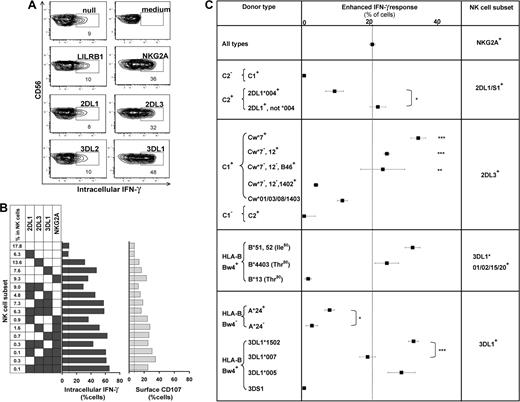
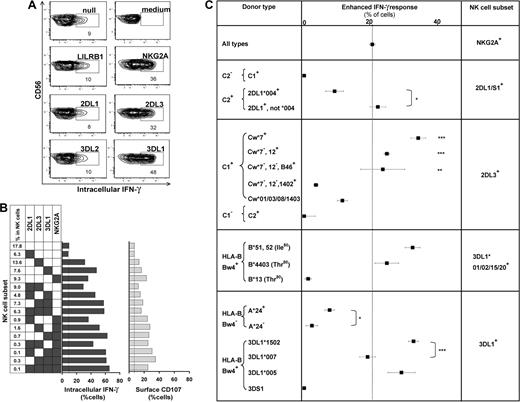
The missing-self response of NK subsets against class I–deficient 721.221 cells was compared by measuring cytokine production (intracellular IFN-γ) and degranulation (CD107 mobilization). Figure 4 shows results for the AA KIR donor of Figure 1, who has the HLA-C1 ligand for KIR2DL3, the HLA-Bw4 ligand for KIR3DL1, and the ubiquitous ligands for NKG2A and LILRB1.7,16,36 KIR2DL3, 3DL1, or NKG2A-single-positive subsets gave enhanced responses, whereas the “receptor-null” subset lacking all 6 receptors exhibited a low, basal response as well as KIR3DL2, LILRB1, and KIR2DL1 single-positive cells (Figure 4A). Thus, enhanced responses correlated with the expression of one or more receptors that engage a self-MHC class I ligand (Figure 4B). Enhancement of degranulation was greater in NK cells expressing NKG2A. Similar analysis for 58 donors with various KIR and HLA genotypes demonstrated that KIR2DL1, 2DL3, and 3DL1 single-positive cells can exhibit enhanced responses to missing-self, but only if the donor expresses a cognate HLA class I ligand for the inhibitory KIR. The NKG2A-positive subsets displayed enhanced responses in all donors. Analysis of KIR2DL2 alone was not possible because it is always present in donors with KIR2DS2.
NK subsets display defined levels of enhanced response to “missing-self” that are determined by the combination of KIR and HLA polymorphisms in the donor. (A) NK subsets defined by single inhibitory MHC class I receptors respond differently to 721.221 target cells. For this donor (HLA-C1, Bw4; KIR A1A1), the NK subsets bearing only LILRB1, KIR2DL1, or 3DL2 gave weak responses, such as the receptor-null subset, whereas NK cells bearing NKG2A, KIR2DL3, or 3DL1 gave strong responses. (B) NK subsets, from the same donor, expressing different combinations of KIR3DL1, 2DL3, 2DL1, and NKG2A were analyzed for their cytokine and cytotoxic response to 221 cells. The vertical line gives the response level of the receptor-null subset. (C) How different combinations of HLA class I ligands and NK-cell receptors enhance the missing-self response of NK subsets to 221 cells. In all 58 donors, the enhanced response of NKG2A single-positive cells was constant (dotted vertical line), whereas the effects on KIR single-positive cells varied with donor HLA and KIR genotype. NKG2A, n = 58; 2DL1/S1+ C2−, n = 42; C2+2DL1*004+, n = 4; C2+2DL1*004−, n = 11; 2DL3+ Cw*7+, n = 16; Cw*7−Cw*12+, n = 7; Cw*7−Cw*12−B*46+, n = 4; Cw*7−Cw*12− Cw*1402+, n = 4; Cw*01/03/08/1403+, n = 10; 2DL2/3+ C2/2, n = 5; 3DL1*001/002/15/20+ B*51/52+, n = 13; B4403+, n = 5; B*13+, n = 2; 3DL1+Bw4− A*24+, n = 13; A*24−, n = 9; 3DL1*1502+, n = 22; 3DL1*007+, n = 5; 3DL1*005+, n = 8; 3DS1+, n = 1. *P < .05, ** P < .005, *** P < .001. Comparison of the effect by C1 was significant by ANOVA. P < .05 was considered statistically significant.
NK subsets display defined levels of enhanced response to “missing-self” that are determined by the combination of KIR and HLA polymorphisms in the donor. (A) NK subsets defined by single inhibitory MHC class I receptors respond differently to 721.221 target cells. For this donor (HLA-C1, Bw4; KIR A1A1), the NK subsets bearing only LILRB1, KIR2DL1, or 3DL2 gave weak responses, such as the receptor-null subset, whereas NK cells bearing NKG2A, KIR2DL3, or 3DL1 gave strong responses. (B) NK subsets, from the same donor, expressing different combinations of KIR3DL1, 2DL3, 2DL1, and NKG2A were analyzed for their cytokine and cytotoxic response to 221 cells. The vertical line gives the response level of the receptor-null subset. (C) How different combinations of HLA class I ligands and NK-cell receptors enhance the missing-self response of NK subsets to 221 cells. In all 58 donors, the enhanced response of NKG2A single-positive cells was constant (dotted vertical line), whereas the effects on KIR single-positive cells varied with donor HLA and KIR genotype. NKG2A, n = 58; 2DL1/S1+ C2−, n = 42; C2+2DL1*004+, n = 4; C2+2DL1*004−, n = 11; 2DL3+ Cw*7+, n = 16; Cw*7−Cw*12+, n = 7; Cw*7−Cw*12−B*46+, n = 4; Cw*7−Cw*12− Cw*1402+, n = 4; Cw*01/03/08/1403+, n = 10; 2DL2/3+ C2/2, n = 5; 3DL1*001/002/15/20+ B*51/52+, n = 13; B4403+, n = 5; B*13+, n = 2; 3DL1+Bw4− A*24+, n = 13; A*24−, n = 9; 3DL1*1502+, n = 22; 3DL1*007+, n = 5; 3DL1*005+, n = 8; 3DS1+, n = 1. *P < .05, ** P < .005, *** P < .001. Comparison of the effect by C1 was significant by ANOVA. P < .05 was considered statistically significant.
Not all HLA-specific inhibitory receptors can produce an enhanced response as demonstrated by the hyporesponsiveness of KIR3DL2 single-positive cells in donors with the HLA-A*03/11 ligand, or LILRB1. We find also that the basal level of response in the receptor-null NK subset differs between persons (Figure S4A). The heterogeneity in the basal level could reflect variation in other factors that contribute to NK-cell recognition of 221 cells. To facilitate more meaningful comparison of the enhancement in different donors, in subsequent analyses we subtracted for each donor the basal level given by receptor-null cells from the responses obtained with the receptor-positive subsets (Figure S4B).
KIR and HLA class I polymorphisms define levels of enhancement in individual NK subsets
We find that the missing-self responses conferred by inhibitory KIR vary according to the donors' KIR and HLA polymorphisms, whereas that conferred by NKG2A is remarkably constant between donors (Figure 4C). These effects demonstrate a new, functional consequence of HLA class I allotype diversity.
In this analysis, substantial differences were seen for the C1 allotypes examined, whereas various C2 allotypes (HLA-Cw*04/05/06/15) gave no distinct difference. KIR2DL3-mediated enhancement by C1 was strongest for HLA-Cw*07, followed by Cw*12 and B*46, an unusual B allotype having the C1 epitope.37 Several C1 allotypes (Cw*01/03/08/1403) enhanced to a lesser extent, and the C1 epitope of Cw*1402 had no enhancing effect. Notably, the enhancement mediated by C2 was less for KIR2DL1*004 than other 2DL1 allotypes. No significant difference in the response of EB6+ cells was observed between 2DS1+ and 2DS1− donors.
The enhancement mediated by KIR3DL1 varied according to the HLA-A or -B allotype providing the Bw4 epitope. High DX9-binding 3DL1 allotypes (3DL1*01/02/15/20)+ NK cells were strongly enhanced by HLA-B*51, B*52, and by B*4403. In contrast, the Bw4 epitope from the HLA-B*13 allotype had no enhancing effect. Epidemiologic studies have correlated different effects, with Bw4 epitopes having isoleucine (Ile80) or threonine (Thr80) at position 80.38 Here we see that allotypes having Ile80 (B*51/B*52) or Thr80 (B*4403) can both enhance, whereas polymorphism outside the Bw4 motif can abrogate the effect, as seen for the Thr80-containing B*13. HLA-A*24, an HLA-A allotype with a Bw4 epitope, is a known ligand for KIR3DL1.39 We found that HLA-A*24 also enhances the response of 3DL1+ NK cells, but the effect is weak and detectable only in donors lacking any Bw4+ HLA-B. KIR3DL1 polymorphism also influences enhancement, the effect being significantly stronger for 3DL1*1502 than 3DL1*007, which corresponds to the hierarchy in inhibition conferred by these 3DL1 allotypes.27 No enhancing effect of 3DS1 was detected.
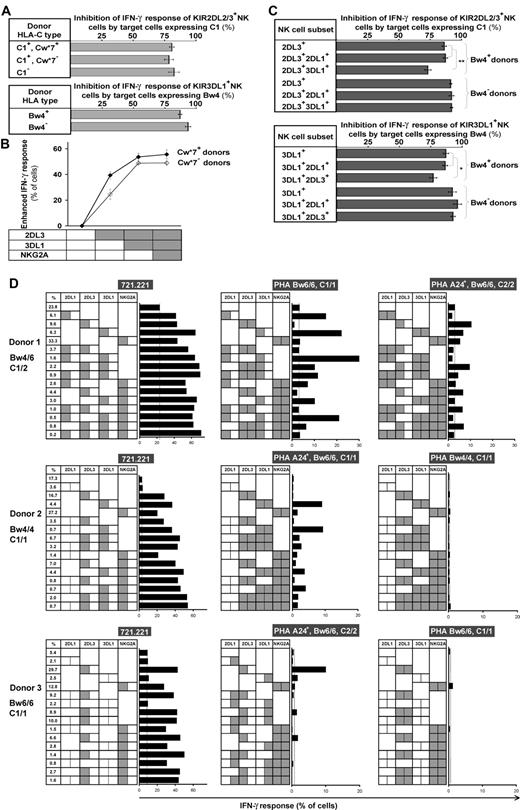
The differential enhancing effects that HLA class I ligands have on the missing-self response are not directly reflected in their capacity to inhibit NK-cell effector function (Figure 5A). Cytokine production by NK cells expressing KIR2DL3 is equivalently inhibited by HLA-C1, irrespective of the HLA-C type of the NK- cell donors, and likewise for 3DL1*1502-mediated inhibition in Bw4+ versus Bw4− donors.
The enhanced levels of subset response conferred by receptor coexpression are dampened to various degrees by the combinations of HLA allotypes on target cells. (A) The capacity of 221 transfectant expressing C1 (top subpanel) and Bw4 (bottom subpanel) to inhibit NK cells bearing their cognate KIR2DL2/3 and 3DL1*1502 receptors did not differ between NK cells from donors who have or lack the cognate ligand. The C1 inhibitor was Cw*1202, and the Bw4 inhibitor was B*5801. The results are the means obtained from the following donors: KIR2DL2/3+ Cw*07+, n = 13; Cw*07−, n = 19; C1−, n = 6; 3DL1*1502+ Bw4+, n = 15; Bw4−, n = 5. (B) Coexpression of inhibitory MHC class I receptors increases the enhanced IFN-γ response to missing-self, but the effect is attenuated in cells with multiple receptors. The response of NK cells expressing different combinations of receptors (2DL3, 3DL1, and NKG2A) was compared among donors who have the identical A1A1 KIR genotype and both the Bw4 and C1 ligands. Donors having Cw*07 (♦, n = 3) are distinguished from donors lacking Cw*07 (♢, n = 4). (C) NK cells expressing inhibitory receptors for 2 different HLA class I ligands are not fully inhibited by either of the ligands alone. NK cells expressing KIR2DL3 and 3DL1 from Bw4− donors are fully inhibited by C1, whereas 2DL3+ 3DL1+ cells from Bw4+ donors are incompletely inhibited by C1 (top subpanel). Similarly, NK cells expressing KIR2DL3 and 3DL1 from Bw4− donors are fully inhibited by Bw4, whereas 2DL3+3DL1+ cells from Bw4+ donors are incompletely inhibited by Bw4 (bottom subpanel). The C1 inhibitor was Cw*1202, the Bw4 inhibitor was Bw*5801, and the 3DL1 allele was 1502. 5 Bw4+ donors and 2 Bw4− donors with the A1A1 KIR genotype were studied. Plots show mean values with the range of variation indicated by the SEM. *P < .05, **P < .005. (D) NK subset responses against allogeneic PHA blasts and 721.221. KIR ligands of 3 group A homozygote donors are shown on the left, and HLA genotypes of PHA blasts are shown at the top of the each panel. Receptor combinations of KIR2DL1, 2DL3, 3DL1, or NKG2A on each NK subset are indicated by the presence of a dotted line in the center of each box; gray shading in the left half of each box indicates presence of a ligand conferring enhancement of missing-self response to a subset; gray shading in the right half of each box indicates presence of the ligand on the target cell that induces inhibition. Length of bar indicates missing-self response as assessed by IFN-γ production. Subset frequencies for each donor are indicated in the far left column. Donor HLA allotypes are: donor 1, HLA-B*07/57, Cw*06/07; donor 2, HLA-B*52/58, Cw*03/12; donor 3, HLA-B*39/40, Cw*07/07.
The enhanced levels of subset response conferred by receptor coexpression are dampened to various degrees by the combinations of HLA allotypes on target cells. (A) The capacity of 221 transfectant expressing C1 (top subpanel) and Bw4 (bottom subpanel) to inhibit NK cells bearing their cognate KIR2DL2/3 and 3DL1*1502 receptors did not differ between NK cells from donors who have or lack the cognate ligand. The C1 inhibitor was Cw*1202, and the Bw4 inhibitor was B*5801. The results are the means obtained from the following donors: KIR2DL2/3+ Cw*07+, n = 13; Cw*07−, n = 19; C1−, n = 6; 3DL1*1502+ Bw4+, n = 15; Bw4−, n = 5. (B) Coexpression of inhibitory MHC class I receptors increases the enhanced IFN-γ response to missing-self, but the effect is attenuated in cells with multiple receptors. The response of NK cells expressing different combinations of receptors (2DL3, 3DL1, and NKG2A) was compared among donors who have the identical A1A1 KIR genotype and both the Bw4 and C1 ligands. Donors having Cw*07 (♦, n = 3) are distinguished from donors lacking Cw*07 (♢, n = 4). (C) NK cells expressing inhibitory receptors for 2 different HLA class I ligands are not fully inhibited by either of the ligands alone. NK cells expressing KIR2DL3 and 3DL1 from Bw4− donors are fully inhibited by C1, whereas 2DL3+ 3DL1+ cells from Bw4+ donors are incompletely inhibited by C1 (top subpanel). Similarly, NK cells expressing KIR2DL3 and 3DL1 from Bw4− donors are fully inhibited by Bw4, whereas 2DL3+3DL1+ cells from Bw4+ donors are incompletely inhibited by Bw4 (bottom subpanel). The C1 inhibitor was Cw*1202, the Bw4 inhibitor was Bw*5801, and the 3DL1 allele was 1502. 5 Bw4+ donors and 2 Bw4− donors with the A1A1 KIR genotype were studied. Plots show mean values with the range of variation indicated by the SEM. *P < .05, **P < .005. (D) NK subset responses against allogeneic PHA blasts and 721.221. KIR ligands of 3 group A homozygote donors are shown on the left, and HLA genotypes of PHA blasts are shown at the top of the each panel. Receptor combinations of KIR2DL1, 2DL3, 3DL1, or NKG2A on each NK subset are indicated by the presence of a dotted line in the center of each box; gray shading in the left half of each box indicates presence of a ligand conferring enhancement of missing-self response to a subset; gray shading in the right half of each box indicates presence of the ligand on the target cell that induces inhibition. Length of bar indicates missing-self response as assessed by IFN-γ production. Subset frequencies for each donor are indicated in the far left column. Donor HLA allotypes are: donor 1, HLA-B*07/57, Cw*06/07; donor 2, HLA-B*52/58, Cw*03/12; donor 3, HLA-B*39/40, Cw*07/07.
Functional consequences of inhibitory receptor coexpression
Analysis of NK cells expressing more than one inhibitory receptor showed that the enhancement was greater than with the individual receptors. In donors without the strong enhancement mediated by Cw*07, the effect of 2 receptors was additive, whereas in donors with Cw*07, the enhancement obtained by their combination was less than the sum of the parts (Figure 5B).
NK cells that coexpress inhibitory receptors for 2 self-MHC class I ligands are not fully inhibited by only one of those ligands. Whereas 2DL3+3DL1+ NK cells from C1+Bw4+ donors were incompletely inhibited by either C1 or Bw4, the same NK subset from C1+Bw4− donors was inhibited completely by either of these ligands (Figure 5C). These results indicate that inhibition by one cognate ligand is insufficient to suppress fully those NK subsets for which multiple KIR-HLA combinations enhance the missing-self response.
In an allogeneic situation, the enhanced response of NK subsets is dampened to various degrees by the inhibition mediated by HLA allotypes expressed on target cells. In Figure 5D, NK subset responses in 3 donors with disparate HLA types are quantified for their reaction against allogeneic PHA blasts. The donor carrying multiple KIR ligands (donor I) has a better chance of ligand mismatch and higher NK-cell reactivity against allogeneic targets than donors with fewer KIR ligands (donors II and III). The residual alloresponse of the KIR3DL1-expressing subsets in donor II demonstrates that HLA-A*24 is both a weak inhibitor and a weak enhancer as its inhibition is insufficient to fully dampen the strong enhancement conferred by HLA-B*52/58 in this donor (Figure 5D center panel). Also of practical relevance are the active responses of multiple receptor-expressing subsets that are only partially inhibited in donor I (the KIR2DL3+3DL1+, 2DL1+NKG2A+, 3DL1+NKG2A+, 2DL1+2DL3+3DL1+, 2DL1+2DL3+NKG2A+ subsets, Figure 5D top center panel).
The number of strong KIR ligands sets the balance between KIR- and NKG2A-dominance in the repertoire
The source of variability between subsets and donors was investigated and linked to the variability in effector function and repertoire phenotype. A correlation was observed between the number of strong ligands possessed by a donor and the ratio between the frequencies of NK cells lacking and expressing NKG2A (Figure 6A). Persons with one strong KIR ligand have an increased frequency of cells that lack NKG2A and express KIR. In persons who have 2 or more strong ligands, the trend is reversed and the ratio of NKG2A− to NKG2A+ cells becomes lower than for persons with no strong ligand.
The balance in KIR/NKG2A expression is determined by the number of strong KIR-HLA interactions and results in equilibrium of total NK-cell repertoire response. (A) For the donor panel, the ratio in the frequency of NK cells that lack NKG2A (sum of the null subset and the NKG2A−KIR+ subset) to NK cells that have NKG2A is plotted against the number of strong ligands. All C2, Cw*07, Cw*12, B*46, and all Bw4+ HLA-B allotypes, with the exception of B*13 were considered strong ligands, but to NK cells that have NKG2A in donors having the cognate inhibitory KIR. The mean ratio (2.4) for the panel is given by the horizontal dotted line. Ratios greater than the mean represent repertoires dominated by KIR (KIR-dominant) and ratios below the mean represent repertoires dominated by NKG2A (NKG2A-dominant). The correlation of KIR-dominant repertoires with one ligand and of NKG2A-dominant repertoires with 2 or more ligands was significant. *P < .05. (B) Comparison of the ratio of the expression frequencies of KIR2DL3 and NKG2A with the ratio of their enhanced missing-self responses. Closed symbols represent donors having strong KIR3DL1 and/or 2DL1 responses; open symbols represent donors with weak 3DL1 and 2DL1 responses. Between receptor expression and response, 4 patterns were discerned. On the right side of the panel are donors with a stronger response of 2DL3 over NKG2A and either strong (●) or weak (○) KIR3DL1 and/or 2DL1 responses. On the left side are donors with stronger response of NKG2A over 2DL3 and either strong (▴) or weak (Δ) 3DL1 and 2DL1 responses. (C) The relative proportions of 5 NK subsets in 5 donors (Figure 3E) representing the 5 types of NK-cell repertoire. The type 1 repertoire is characterized by a larger subset of NK cells coexpressing inhibitory KIR, type 2 by receptor-null cells, type 3 by cells expressing single inhibitory KIR, type 4 by cells coexpressing KIR and NKG2A, and type 5 by a higher proportion of single-positive NKG2A cells compared with coexpression of NKG2A with KIR. (D) Overall levels of enhancement in missing-self response of 58 NK-cell repertoires. The donors are grouped according to the 5 repertoire types and plotted according to the sum of the enhanced response for all receptors having a cognate ligand. The type 2 null-dominant repertoires have a significantly lower enhanced response with a mean of 13 compared with 22, 20, 22, and 16 for the type 1, 3, 4, and 5 repertoires, respectively (P < .005). The summed enhanced response for the donors was less than 35 (dotted horizontal line) with one exception. *P < .05. ***P < .005.
The balance in KIR/NKG2A expression is determined by the number of strong KIR-HLA interactions and results in equilibrium of total NK-cell repertoire response. (A) For the donor panel, the ratio in the frequency of NK cells that lack NKG2A (sum of the null subset and the NKG2A−KIR+ subset) to NK cells that have NKG2A is plotted against the number of strong ligands. All C2, Cw*07, Cw*12, B*46, and all Bw4+ HLA-B allotypes, with the exception of B*13 were considered strong ligands, but to NK cells that have NKG2A in donors having the cognate inhibitory KIR. The mean ratio (2.4) for the panel is given by the horizontal dotted line. Ratios greater than the mean represent repertoires dominated by KIR (KIR-dominant) and ratios below the mean represent repertoires dominated by NKG2A (NKG2A-dominant). The correlation of KIR-dominant repertoires with one ligand and of NKG2A-dominant repertoires with 2 or more ligands was significant. *P < .05. (B) Comparison of the ratio of the expression frequencies of KIR2DL3 and NKG2A with the ratio of their enhanced missing-self responses. Closed symbols represent donors having strong KIR3DL1 and/or 2DL1 responses; open symbols represent donors with weak 3DL1 and 2DL1 responses. Between receptor expression and response, 4 patterns were discerned. On the right side of the panel are donors with a stronger response of 2DL3 over NKG2A and either strong (●) or weak (○) KIR3DL1 and/or 2DL1 responses. On the left side are donors with stronger response of NKG2A over 2DL3 and either strong (▴) or weak (Δ) 3DL1 and 2DL1 responses. (C) The relative proportions of 5 NK subsets in 5 donors (Figure 3E) representing the 5 types of NK-cell repertoire. The type 1 repertoire is characterized by a larger subset of NK cells coexpressing inhibitory KIR, type 2 by receptor-null cells, type 3 by cells expressing single inhibitory KIR, type 4 by cells coexpressing KIR and NKG2A, and type 5 by a higher proportion of single-positive NKG2A cells compared with coexpression of NKG2A with KIR. (D) Overall levels of enhancement in missing-self response of 58 NK-cell repertoires. The donors are grouped according to the 5 repertoire types and plotted according to the sum of the enhanced response for all receptors having a cognate ligand. The type 2 null-dominant repertoires have a significantly lower enhanced response with a mean of 13 compared with 22, 20, 22, and 16 for the type 1, 3, 4, and 5 repertoires, respectively (P < .005). The summed enhanced response for the donors was less than 35 (dotted horizontal line) with one exception. *P < .05. ***P < .005.
These results infer that the frequency of KIR expression is reduced, not only in donors carrying several ligand-receptor pairs with potential to induce overly strong NK-cell responses, but also in donors who lack strong ligand-receptor pairs altogether. This relationship was investigated further by plotting the ratio of expression in KIR2DL3 and NKG2A and plotting it against the ratio of the response mediated by KIR2DL3 and NKG2A (Figure 6B). The comparison of 2DL3 to NKG2A is particularly informative because 2DL3 is the most frequent KIR expressed by NK cells.27 The graph reveals that KIR2DL3 is expressed at higher frequencies only when one of 3 receptors, KIR2DL3, 3DL1, or 2DL1, confers a strong response, whereas NKG2A frequencies are higher when KIR2DL3 and 2DL1/3DL1 both confer either strong responses or weak responses. These correlations infer a balance between frequencies of the KIR2DL3+ and NKG2A+ subsets that prevents formation of NK-cell repertoires with responses that are either too strong or too weak. In this system, the reduction in KIR frequencies is complemented by an increase in NKG2A frequency, which underlies the inverse correlation seen in Figure 2C. NKG2A is particularly suited for this role because the response it confers remains constant regardless of donor HLA type.
Human NK-cell repertoires are structured toward a balance of missing-self response
The proportions of the 5 major groups of NK subset vary considerably in the 5 types of repertoires (Figure 6C). To assess the overall enhancement of the missing-self response for each donor, we summed the response for all NK subsets, weighting them according to their frequency. For 57 of the 58 donors, these values range between 5 and 35 (Figure 6D). The ranges and mean values are comparable for the type 1, 3, 4, and 5 repertoires, whereas the type 2 repertoire has a narrower range and a significantly lower mean. The results point to regulation during NK-cell development that alters subset frequencies according to the combined KIR and HLA genotype and produces diverse repertoires with a limited range of response. They also emphasize the significantly reduced response of type 2 repertoires that contain more receptor-null NK cells.
The amount of deviation from the product rule is unique to each person and signifies the independent nature of KIR and NKG2A receptor expression
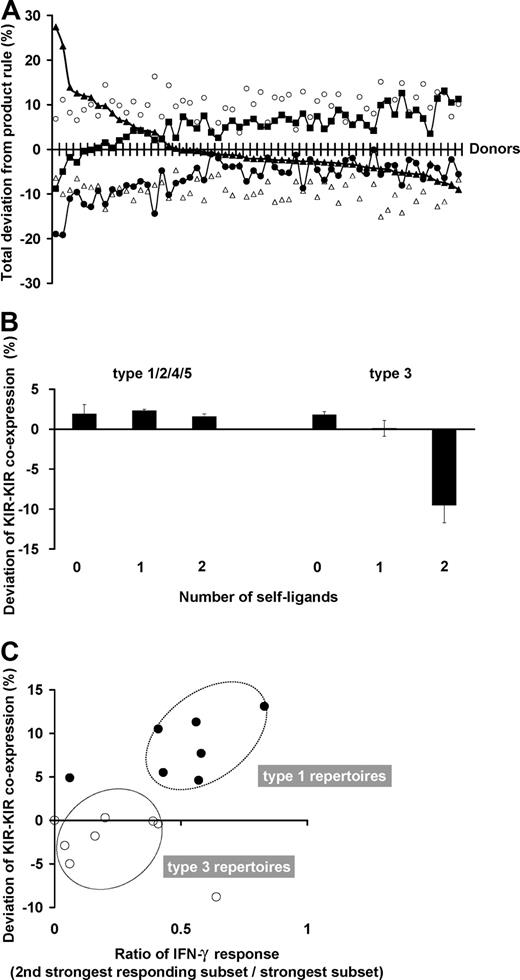
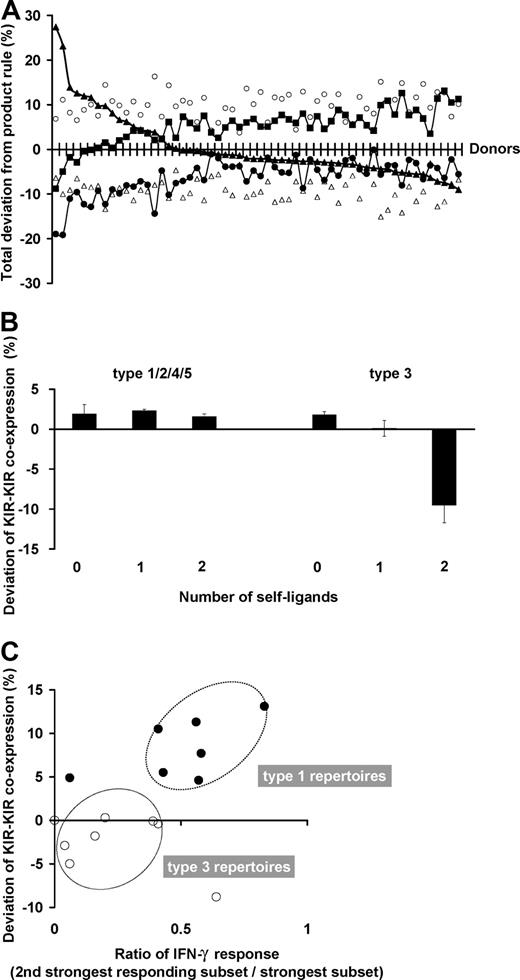
The degree to which subset frequencies conform to the stochastic model varies with the donor. For each donor, 5 parameters of the NK-cell repertoire were assessed for their deviation from stochastic expression: the frequencies of KIR single-positive cells, NKG2A single-positive cells, NKG2A+KIR+ cells, receptor-null cells, and of KIR-KIR coexpression. The deviation between the observed and expected values for these 5 parameters is shown on a donor-by-donor basis in Figure 7A. Relatively constant characteristics within the donor panel are the extent to which the frequencies of NKG2A single-positive cells are greater and KIR-NKG2A coexpression is less than expected. More variable deviation is observed in receptor-null cell frequencies and cells with single- or multiple-KIR.
NK subset frequencies in human NK-cell repertoires vary in their amount of departure from the product rule. (A) For each donor, the deviation of the observed NK-cell repertoire from the product rule was assessed for 5 parameters: single KIR expression (▴), KIR-KIR coexpression (■), NKG2A expression alone (○), NKG2A-KIR coexpression (▵), and no receptor expression (●). The donors are ordered from left to right according to decreasing deviation in single KIR expression. (B) KIR coexpression is higher than expectation under the product rule regardless of ligand presence. The amount of deviation from expectations under the product rule is shown for NK subsets coexpressing 2 KIR (2DL1-2DL3, 2DL3-3DL1, 3DL1-2DL1). This amount is shown separately by the number of cognate ligands (0-2) for the 2 KIR. The deviations are all positive in these subsets from donors with type 1, 2, 4, and 5 repertoires. The deviations display a different pattern for cells from donors with type 3 repertoires, where KIR coexpression is drastically reduced from frequencies predicted under stochastic coexpression, but only in those subsets that have developed in the presence of autologous cognate ligands. All donors are group A homozygotes. Donors for type 1, 2, 4, and 5 repertoires: n = 24; those for type 3 repertoires: n = 5. (C) KIR coexpression is enhanced when multiple KIR confer similar levels of enhanced missing-self response. The ratio of missing-self response between KIR with the strongest response and that from the second strongest are compared in donors with type 1 (●, n = 7) and type 3 (○, n = 8) repertoires. The plots display an association with the amount of deviation in KIR-KIR coexpression.
NK subset frequencies in human NK-cell repertoires vary in their amount of departure from the product rule. (A) For each donor, the deviation of the observed NK-cell repertoire from the product rule was assessed for 5 parameters: single KIR expression (▴), KIR-KIR coexpression (■), NKG2A expression alone (○), NKG2A-KIR coexpression (▵), and no receptor expression (●). The donors are ordered from left to right according to decreasing deviation in single KIR expression. (B) KIR coexpression is higher than expectation under the product rule regardless of ligand presence. The amount of deviation from expectations under the product rule is shown for NK subsets coexpressing 2 KIR (2DL1-2DL3, 2DL3-3DL1, 3DL1-2DL1). This amount is shown separately by the number of cognate ligands (0-2) for the 2 KIR. The deviations are all positive in these subsets from donors with type 1, 2, 4, and 5 repertoires. The deviations display a different pattern for cells from donors with type 3 repertoires, where KIR coexpression is drastically reduced from frequencies predicted under stochastic coexpression, but only in those subsets that have developed in the presence of autologous cognate ligands. All donors are group A homozygotes. Donors for type 1, 2, 4, and 5 repertoires: n = 24; those for type 3 repertoires: n = 5. (C) KIR coexpression is enhanced when multiple KIR confer similar levels of enhanced missing-self response. The ratio of missing-self response between KIR with the strongest response and that from the second strongest are compared in donors with type 1 (●, n = 7) and type 3 (○, n = 8) repertoires. The plots display an association with the amount of deviation in KIR-KIR coexpression.
Although the pattern in deviation is unique to each donor, the sum of the deviations in NKG2A+ NK cells and NKG2A− NK cells cancel out almost completely in every donor (Figure S5). This extends previous reports suggesting that NKG2A is acquired before KIR in NK-cell development17-20 ; our findings indicate it is a 2-stage process in which the stages are regulated separately. Consequently, this implies that the frequencies of KIR+ cells and receptor-null cells are in a compensatory relationship, which is independent of the NKG2A+ cells.
“Weaker” levels of ligand-receptor interactions affect the degree of receptor coexpression
In the great majority of donors, the presence of cognate ligands does not affect the degree of coexpression among KIRs. This is exemplified in Figure 7B, where for the type 1, 2, 4, and 5 repertoires, coexpression among KIRs is always more than expected by the product rule regardless of whether or not their cognate ligands are present, and KIR-KIR coexpression is increased the most in type 1 repertoires. Type 3 repertoires are different, where coexpression among KIRs is less than expected under the product rule but only when the ligand is present. This reduction is enhanced further by the presence of a second receptor-ligand interaction.
We find that the “weaker” KIR-HLA combinations have a role in distinguishing the amount of KIR-KIR coexpression (Figure 7C). Donors who combine one strong KIR-HLA combination with weaker ligand-receptor combinations have type 1 repertoires, which are characterized by higher proportions of cells that coexpress 2 or more inhibitory KIRs. In contrast, donors who have a single strong combination without weaker KIR-HLA combinations have type 3 repertoires, in which single-positive NK cells carrying the strong inhibitory KIR predominate.
Discussion
The human NK-cell population is a complex mixture of cells with highly variable levels of missing-self response and differing specificity to inhibitory ligands. Our high-resolution analyses on a number of diverse, primary NK-cell populations show that all possible receptor combinations are present and that there is no wholesale deletion of any NK subset. This retainment of all possible NK subsets necessitates a unique mechanism to adjust cell frequencies and effectively compensate for the wide range of variation in NK subset response that is hardwired to the genetic polymorphisms of KIR and HLA brought to match in a person. As a result of this “repertoire calibration,” human NK-cell repertoires have similar levels of overall response.
Donors having one “strong” KIR-HLA combination generate KIR-dominant repertoires, whereas donors having multiple strong combinations or only weak combinations generate NKG2A-dominant repertoires. By this means, the NKG2A receptor, which is of medium strength, buffers the effects of KIR to balance the overall strength of the missing-self response within a limited range. For donors who have only weak KIR-HLA combinations, NKG2A provides a stronger receptor, whereas for donors with strong KIR-HLA combinations, it provides a weaker receptor to be used instead of KIR.
Our high-dimension cytometric analyses have provided new insight into receptor coexpression. Quantification of receptor coexpression in the repertoires has shown that a substantial portion of the NK-cell population coexpress multiple receptors, resulting in dual consequences on NK subset function: enhancement of missing-self response and broader ligand specificity for inhibition.
Of the 6 receptors examined, NKG2A, KIR2DL1, 2DL3, and 3DL1 enhance the missing-self response in donors having a cognate ligand. The lack of enhancement in missing-self response by KIR3DL2 corresponds to the lack of binding to HLA-A*03/11 except when particular virally derived peptides are loaded.40 The lack of enhancement conferred by LILRB1 is probably because of its low affinity for HLA-B and -C.36,41 In all donors, NKG2A enhanced to the same degree, emphasizing its role as a constant and conserved inhibitory MHC class I receptor. In contrast, the enhancement mediated by KIR2DL1, 2DL3, and 3DL1 was highly variable and depended on the particular allotypes of KIR and HLA that each donor combined. Of note, HLA-Cw*07, which comprises a common and distinctive lineage of HLA-C, is a particularly strong enhancer.
The lack of enhancement by HLA-B*13 infers a uniqueness of this allotype and underscores the importance of investigating the effects of HLA polymorphisms in NK-cell biology. HLA-B*13 has been considered unique in that Bw4-specific allo-antibodies produced in women carrying HLA-B*13 do not react with their HLA-B*13-expressing cells (Thomas Fuller, University of Utah, written correspondence, May 12, 2008). Notably, Foley et al have recently shown that, among 13 HLA-Bw4 alleles investigated, only HLA-B*13 lacks the protective effect from NK-cell lysis.42
The KIR and HLA alleles represented in this panel are limited, and this report is by no means a comprehensive study of the interactions between all combinations of HLA and KIR allotypes. However, the donors studied here have revealed the effects for the most common KIR allotypes and gene content KIR haplotypes known to date. Of clinical importance, our experiments enable quantification of NK-cell alloreactivity, which is determined by the balance between levels of missing-self response and inhibition conferred by the combined effects KIR and NKG2A allotypes (Figure 5D).
Inhibitory receptor acquisition is an orderly process during NK-cell development, where CD94:NKG2A is first expressed and followed by acquisition of KIR in human and Ly49 in mouse.13,18-20 This hierarchy is reflected in patients recovering from a hematopoietic stem cell transplantation, whose NK cells are initially NKG2A-positive and KIR-negative, and only later begin to express KIR.17,43 Recent reports from other laboratories have shown that receptor-null CD56dim NK cells can express KIR on activation.43,44 These observations lead to the question as to whether KIR-expressing NK cells arise primarily from the receptor-null compartment. Our analyses support this perspective, as demonstrated by the independent deviations within NKG2A+ and NKG2A− (KIR+ and receptor-null) compartments and the inverse correlation in KIR+ frequencies against receptor-null subset frequencies in CD56dim NK cells (Figure 2). The complementary deviation between NKG2A+KIR+ and NKG2A+KIR− subsets (Figure 7A) implies that the former subsets are probably derived from the cells that have acquired NKG2A before KIR in NK-cell development, a view supported by experiments in mouse demonstrating that NKG2A phenotypes remain stable after their acquisition.43,45
From this analysis of 58 donors, human NK-cell repertoires are seen to be highly variable and to depend on the combinations of KIR and HLA class I alleles a donor inherits. However, we find that receptor-null subset frequency is a variable that also affects repertoire structure and is a factor not predicted by KIR and HLA type. The frequency of receptor-null cells is inversely correlated with the amount of deviation from the product rule, where type 1, 3, and 4 repertoires contain less receptor-null cells and more deviation in subset frequencies contrasting to the subset frequencies in type 2 repertoires, which best follow predictions of the product rule. Such differences could be produced by critical, non-KIR/HLA-related factors, such as the amount of IL-15 available for expansion of the NK-cell population at the stages before the MHC-specific inhibitory receptors express.46-49 Other extrinsic factors might influence the repertoire of MHC class I-specific inhibitory NK receptors, such as the modulation of NKG2C and NKp46 activating receptors by human cytomegalovirus.50
Thus, this is a system that guarantees an array of effector cells with various levels of sensitivity and strength of response. From our analyses, NKG2A is seen to be the common foundation on which all NK-cell repertoires are built. However, by itself, NKG2A can generate only modest diversity. In stark contrast is the KIR system, which is highly sensitive to allotypic differences in KIR and MHC class I, as well as to the bound peptides. This report reveals how these 2 contrasting and complementary systems of MHC class I–specific inhibitory receptors cooperate to produce highly diverse NK-cell repertoires with a similarly measured capacity to respond to changes in MHC class I occurring during malignancy, infection, pregnancy, and allogeneic transplantation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all blood donors and the staff of the Stanford Shared FACS Facility and Mutsuhiko Minami for support in sample processing.
This work was supported by the National Institutes of Health (grants AI-22039, AI-17892, and AI-31168) (P.P.) and a grant from the Leukemia Research Foundation (Evanston, IL; M.Y.).
National Institutes of Health
Authorship
Contribution: M.Y. and N.Y. designed the project, performed experiments, and wrote the manuscript; F.P. and A.-M.L. performed HLA typing; M.D. contributed technical expertise; and P.P. contributed intellectually and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Makoto Yawata, Stanford University, School of Medicine, Department of Structural Biology, 299 Campus Drive West, Stanford, CA 94305-5126; e-mail: myawata@stanford.edu.
References
Author notes
*M.Y. and N.Y. contributed equally to this study.