Abstract
To evaluate the hypothesis that host germ line variation in immune genes is associated with overall survival in diffuse large B-cell lymphoma (DLBCL), we genotyped 73 single nucleotide polymorphisms (SNPs) from 44 candidate genes in 365 DLBCL patients diagnosed from 1998 to 2000. We estimated hazard ratios (HRs) and 95% confidence intervals (CIs) for the association of SNPs with survival after adjusting for clinical factors. During follow-up, 96 (26%) patients died, and the median follow-up was 57 months for surviving patients. The observed survival of this cohort was consistent with population-based estimates conditioned on surviving 12 months. An IL10 haplotype (global P = .03) and SNPs in IL8RB (rs1126580; HRAG/GG = 2.11; CI, 1.28-3.50), IL1A (rs1800587; HRCT/TT = 1.90; CI, 1.26-2.87), TNF (rs1800629; HRAG/GG = 1.44; CI, 0.95-2.18), and IL4R (rs2107356; HRCC/CT = 1.97; CI, 1.01-3.83) were the strongest predictors of overall survival. A risk score that combined the latter 4 SNPs with clinical factors was strongly associated with survival in a Cox model (P = 6.0 × 10−11). Kaplan-Meier 5-year survival estimates for low, intermediate-low, intermediate-high, and high-risk patients were 94%, 79%, 60%, and 48%, respectively. These data support a role for germ line variation in immune genes, particularly genes associated with a proinflammatory state, as predictors of late survival in DLBCL.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most commonly diagnosed subtype of non-Hodgkin lymphoma (NHL) in Western countries.1,2 DLBCL is potentially curable, but the course of the disease is variable.3-5 Established adverse prognostic factors in DLBCL include older age, higher stage, poor performance score, and above normal lactic dehydrogenase, and these factors have been validated as part of the International Prognostic Factor Index (IPI). However, the IPI predicts outcome incompletely. Molecular features of the tumor offer significant promise in providing additional information on prognosis.6,7 For example, gene expression profiling of DLBCL has defined 2 major subgroups, one with a gene expression profile similar to normal germinal center B cells (60% 5-year survival) and the other mimicking activated peripheral B cells (30% 5-year survival).8-10 Besides clinical and tumor molecular characteristics, there is a growing appreciation that the tumor microenvironment,7 and more broadly the host genetic background,11 may be an additional critical factor in cancer progression and outcome and therefore may be useful as a prognostic marker.12
Cytokines and related immune factors have been hypothesized to play an important role in lymphomagenesis,13 as they appear to influence proliferation, differentiation, and movement of both tumor and stromal cells, regulate communication between tumor and stroma, and regulate tumor interactions with the extracellular matrix.14 Immunologic function is in part influenced by host genetics, and germ line genetic variation in cytokine and related immune genes have been associated with risk of developing DLBCL15-17 and disease-free and overall survival after DLBCL diagnosis.18-20 To test the hypothesis that inherited variation in cytokine and related immune genes impact DLBCL survival more comprehensively, we evaluated the role of 73 single nucleotide polymorphisms (SNPs) from 44 candidate immune genes (Table 1) and overall survival in DLBCL, using cases that were recruited as part of a population-based case-control study. We have previously reported the association of immune SNPs with risk of developing DLBCL in this study population.16 Here we present the risks of dying from DLBCL according to those SNPs. By measuring the effects of the same markers on both etiology and survival, we examine a larger area of influence of genes on this immune system malignancy.
Methods
Study population
This study was reviewed and approved by human subjects review boards at the National Cancer Institute and each of the participating study centers, and written, informed consent was obtained from all participants in accordance with the Declaration of Helsinki. Methods for this molecular epidemiology study have been described previously.16 Briefly, we enrolled 1321 patients (20-74 years of age) with newly diagnosed, histologically confirmed NHL in 4 Surveillance, Epidemiology, and End Results (SEER) cancer registries (Detroit, MI, metropolitan area; northwestern Washington state; the state of Iowa; and Los Angeles County, CA) from July 1998 through June 2000. Known HIV-positive cases were excluded. All participants completed an in-person interview, and 1172 (89%) provided either a venous blood sample (n = 773) or mouthwash buccal cell sample (n = 399). This analysis is restricted to the 365 cases of DLBCL in the case group, as defined by SEER coding and using the InterLymph classification system,21 who had a DNA sample available for genotyping. Of the eligible DLBCL cases for this study, 21% died before we could contact them, 9% had a physician refusal or could not be located, 17% were approached but refused participation (many were too ill), and 53% participated.
Genotyping
The strategy for candidate gene selection focused on genes involved in key immune pathways, particularly those related to cytokine regulation and function. Priority in gene selection was given to those genes with data suggesting functional and biologic significance, an association with NHL etiology or prognosis, an association with other immune diseases, and a minor allele frequency (MAF) of more than 5% in the white population. Full details on candidate gene selection have been previously published.16
Details on DNA extraction and genotyping have been published previously.16 All genotyping was conducted at the National Cancer Institute Core Genotyping Facility using the Taqman or EPOCH platforms (http://cgf.nci.nih.gov), and sequence data and assay conditions are provided at http://snp500cancer.nci.nih.gov/home_1.cfm?CFID=2106952&cftoken=53492492.22 Genotyping was conducted first on blood-based DNA samples (n = 215 DLBCL patients) and then was expanded to patients with only buccal samples (n = 150 DLBCL patients) for 52 of the 73 candidate SNPs. The decision to genotype buccal samples was based on a risk association in the subset of participants with blood-based DNA samples in the parent case-control study.16 For the 21 SNPs that we genotyped on DLBCL patients with blood samples (N = 215), the call rates ranged from 91.2% to 100% (median call rate, 93.0%); for the 52 SNPs that we genotyped on DLBCL patients with blood or buccal (n = 365), the call rates ranged from 89.6% to 99.5% (median, 95.9%). Although we evaluated all SNPs with survival (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), only those 52 SNPs that were also genotyped in patients with buccal samples were eligible for multi-SNP models.
Positive and negative controls and 140 replicate samples were interspersed for all genotyping assays and blinded from the laboratory. Agreement for quality control replicates and duplicates was more than 99% for all assays. Only 1 SNP (rs1801157) in black controls was not in Hardy-Weinberg equilibrium (P < .01); reviewing of all genotyping data (including quality control samples) confirmed the accuracy of this assay.
Prognosis study
Full details on the prognosis study using cases from the case-control study have been previously published.12 Briefly, age, sex, race, Hispanic ethnicity, and education level were derived from patient interviews as part of the case-control study. Date of diagnosis, histology, stage, presence of B symptoms, first course of therapy, date of last follow-up, and vital status were derived from linkage to individual SEER registry databases in early 2005. Data on first course of therapy include use of single-agent or multiagent chemotherapy, radiation, and other therapies exclusive of chemotherapy and/or radiation. Individual agents and doses, as well as indications for use of nonstandard therapy, were not available. The SEER registries collect date and cause of death but do not collect data on treatment response or disease recurrence or progression.
Data analysis
Evaluation of single SNPs and haplotypes.
Our overall data analysis approach has been previously published.12 We first used Cox proportional hazards regression23 to estimate hazard ratios (HRs) and 95% confidence intervals (95% CIs) for the association of each individual genotype with overall survival, adjusting for age and clinical and demographic factors (Table S1). The primary test of association for each SNP with survival used a codominant coding of the alleles (ie, 0, 1, and 2 variant alleles). This statistic was chosen because it has good power to detect genotype associations under a range of genetic models.24 Age was modeled according to the standard IPI score as less than 60 versus 60 or more years.25 Clinical and demographic factors were each modeled as 2 separate risk scores, analogous to a propensity score for logistic regression.26 The clinical risk score was a linear combination of stage (local, regional, distant, missing), presence of B symptoms (no, yes, missing), and type of initial therapy (chemotherapy + radiation, chemotherapy + other therapy, radiation only, all other, or missing therapy). The demographic risk score was a linear combination of sex, race (white, all other), study center (Detroit, Iowa, Los Angeles, Seattle), and years of education (< 12, 12-15, 16+ years). For multi-SNP models, the demographic and clinical risk scores were combined into a single score, with values of 0 to 2 (low to high risk) as previously described.12
We were able to construct haplotypes for 4 genes: TNF/LTA, IL8, IL8RB, and IL10. Haplotype frequencies for selected genes were estimated by an expectation-maximization algorithm,27 and the posterior probabilities for each haplotype were included in a Cox model to assess the association with survival.
To address concerns about multiple testing, we computed the tail strength of all 73 SNPs initially evaluated for their role in survival (Table S1). This measure28 is closely related to the false discovery rate29 and assesses the relative strength of the collection of observed P values from an analysis of a large number of markers.
Selection of the best multi-SNP risk score.
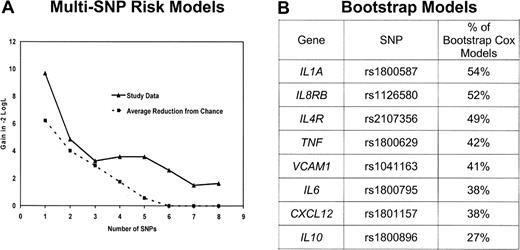
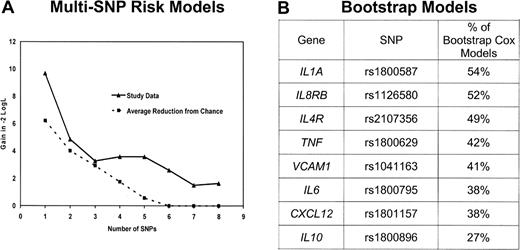
Because many of these genes have overlapping functions and are part of complex networks, it is useful to identify a parsimonious multivariable prediction model. To achieve this, we first brought forward 17 SNPs (from 13 genes) with a P less than or equal to .15 based on the recoded results in Table 2. To preserve power, we eliminated from further consideration for multivariable modeling the 5 SNPs with an MAF less than 0.05 or more than 10% missing data (missing data resulted mainly from buccal samples not being genotyped in the parent study). After removing these SNPs, remaining SNPs with missing genotype were then assigned to the low-risk genotype. For the remaining SNPs in high linkage disequilibrium, we selected a single SNP based on the highest MAF and the strongest HR. After this data reduction process, there were 8 SNPs from 8 genes. To evaluate each of the 255 possible combinations of these 8 SNPs, we created an SNP score variable by summing the number of deleterious genotypes (as categorized in Table 2) in each particular combination.12,30 Each multi-SNP score variable was fit in a Cox model adjusting for age and clinical and demographic risk scores. The models were grouped by number of SNPs included in the SNP score variable (ie, 1 SNP, 2 SNPs, … 8 SNPs) and were ranked by likelihood. A comparison of the likelihood for the best 1 SNP, 2 SNP, … 8 SNP models according to number of SNPs included in the model is shown in Figure 1A. In parallel, we ran 1000 bootstrap stepwise selection Cox models (adjusting for age and clinical and demographic risk scores) using the 8 SNPs and calculated the percentage of models that included each SNP (Figure 1B). The plot of the likelihood suggested a model with 5 or fewer SNPs, as the reduction in likelihood was marginal after 5 SNPs (Figure 1A). The 4 highest ranked SNPs from the bootstrap analyses coincided with the most prognostic 4-SNP risk model; we therefore proceeded with a 4-SNP risk score as the final model.
Selection of the multi-SNP model. (A) Reduction in the −2 log likelihood comparing the best 1 SNP, 2 SNP, … 8 SNP model from the study data (—) as well as expected reduction resulting from chance ( ). (B) Ranking of the percentage of 1000 stepwise bootstrap Cox models that included each SNP. The top 4 SNPs in the bootstrap model are the same 4 SNPs in the best 4 SNP risk model.
). (B) Ranking of the percentage of 1000 stepwise bootstrap Cox models that included each SNP. The top 4 SNPs in the bootstrap model are the same 4 SNPs in the best 4 SNP risk model.
Selection of the multi-SNP model. (A) Reduction in the −2 log likelihood comparing the best 1 SNP, 2 SNP, … 8 SNP model from the study data (—) as well as expected reduction resulting from chance ( ). (B) Ranking of the percentage of 1000 stepwise bootstrap Cox models that included each SNP. The top 4 SNPs in the bootstrap model are the same 4 SNPs in the best 4 SNP risk model.
). (B) Ranking of the percentage of 1000 stepwise bootstrap Cox models that included each SNP. The top 4 SNPs in the bootstrap model are the same 4 SNPs in the best 4 SNP risk model.
Evaluation of the multi-SNP risk score.
We assessed the association of the 4-SNP risk score with overall survival using Kaplan-Meier curves, Cox proportional hazards models, and time-dependent receiver-operator characteristic (ROC) curves for censored data.31 We also developed a single risk score that combined the number of deleterious genotypes from the 4 SNPs (0-4) plus a combined clinical and demographic risk score (0-2) to create a single SNP and clinical risk score (0-6). To compare the significance of the SNP and clinical and demographic risk score observed in our data to what we would expect from chance, we performed a permutation analysis. The test statistic observed in our study data was then compared with the distribution of test statistics from 200 permutation iterations.
Results
Descriptive results
The median age at diagnosis of the 365 cases was 57 years (range, 20-74 years; 41% were age 60 or older). A majority of the patients (88%) were white, and 56% were male. Clinically, 41% had advanced stage disease and 27% had B symptoms. Based on cancer registry data, the most common initial therapy was a chemotherapy-based regimen (88%). During follow-up, 96 (26%) of the patients died, and 68% of the underlying causes of death were coded on the death certificate as lymphoma. The median follow-up of living patients was 57 months (range, 27-78 months). The age (age 60 years or older; HR = 1.80; 95% CI, 1.22-2.76), demographic (combination of sex, race, study center, and education; HR = 2.26; 95% CI, 1.36-3.76), and clinical (combination of stage, B symptoms, and type of treatment; HR = 2.66; 95% CI, 1.63-4.33) risk scores were associated with overall survival when included in the same Cox model.
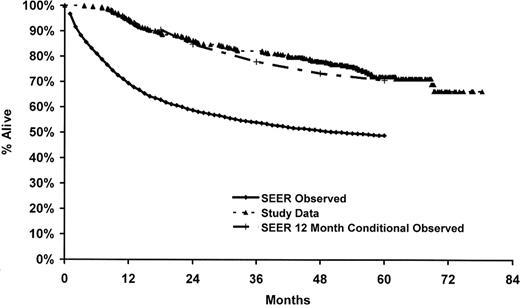
In the parent case-control study, only 53% of the eligible cases were enrolled in the study, and we did not have genotype and survival data on the nonparticipants. Therefore, to address the potential impact of nonresponse on our results, we compared our observed survival to survival reported in the SEER program from the same registries as our cases (ie, Detroit, Iowa, Los Angeles, and Seattle) for white DLBCL patients 20 to 74 years of age and diagnosed from 1995 to 2000.32 As shown in Figure 2, our observed survival was higher than that observed in the SEER data for DLBCL from the same time frame of this study but was very similar to SEER for 12-month conditional survival (ie, survival given that a patient survives 12 months). This is consistent with the enrollment pattern of these patients into our case-control study, whereby patients with early mortality were less likely to be enrolled into the study.
Comparison of study data to SEER DLBCL survival. SEER data (observed and conditioned on surviving 12 months) are based on whites from Detroit, Iowa, Los Angeles, and Seattle sites, 20 to 74 years of age at diagnosis, and diagnosis dates 1995 to 2000.32
Comparison of study data to SEER DLBCL survival. SEER data (observed and conditioned on surviving 12 months) are based on whites from Detroit, Iowa, Los Angeles, and Seattle sites, 20 to 74 years of age at diagnosis, and diagnosis dates 1995 to 2000.32
Single SNP results
We identified 17 SNPs from 14 genes of potential interest based on our statistical criteria (P trend ≤ .15, Table S1). These SNPs were rescored so that all HRs were more than 1, and these are reported in Table 2. Most of the HRs were modest and in the range of 1.4 to 2.5. The smallest observed P value (< .001) was for an IL5 SNP (rs2069807, C-1551T, HRCT/TT = 4.56; 95% CI, 1.98-10.5), which was relatively rare (only 2.9% of patients carried a variant allele). The next smallest observed P value (.002) was for an IL8RB SNP (rs1126580; HRAG/GG = 2.11; 95% CI, 1.28-3.50). The only other P less than or equal to .01 was for an IL1A SNP (rs1800587; HRCT/TT = 1.90; 95% CI, 1.26-2.87). The tail strength of our set of 73 immune SNPs was 0.20 (95% CI, −0.03-0.43). A positive tail strength indicated that the observed P values were more significant than what would be expected resulting from chance; the tail strength of 0.20 in our study suggests that this set of SNPs displayed approximately 20% more signal than expected if all markers were null.
Haplotype results
There was no significant association with the risk of DLBCL (global P = .4) for the TNF/LTA haplotype constructed from 3 TNF SNPs (rs1799724, rs1800629, and rs361525) and 2 LTA SNPs, rs2239704 and rs909253 (Table S2), which has been previously reported to be associated with risk of developing DLBCL in this study population.16 However, using a haplotype limited to TNF G-308A (rs1800629) and LTA A252G (rs909253) reported to be associated with risk of DLBCL by the InterLymph Consortium,15 the AG versus GA haplotype was associated with a marginally significant higher risk of death (HR = 1.36; 95% CI, 0.96-1.94; Table 3). When the number of adverse alleles from TNF G-308A (A allele) and LTA A252G (G allele) were summed according to the approach of Warzocha et al,18 patients with 2 to 4 adverse alleles (38% of patients) had poorer survival compared with patients with 0 or 1 allele (HR = 1.27; 95% CI, 0.84-1.90), and this association was stronger for patients with 3 or 4 adverse alleles (10% of the patients) compared with patients with 0 to 2 alleles (HR = 1.72; 95% CI, 0.95-3.09).
An IL10 haplotype based on IL10 A-1082G (rs1800896) and IL10 T-3575A (rs1800890) showed a suggestive association with survival (global P = .07), and the GT haplotype was associated with poorer survival compared with the most common (AT) haplotype (HR = 1.82; 95% CI, 1.08-3.07; Table 3). Inclusion of 2 additional SNPs to the haplotype was even more strongly associated with survival (global P = .03), and 3 of the most common haplotypes were associated with poorer survival (Table 3). There were no associations of haplotypes in IL8 or IL8RB with survival (Table S2).
Multi-SNP risk score
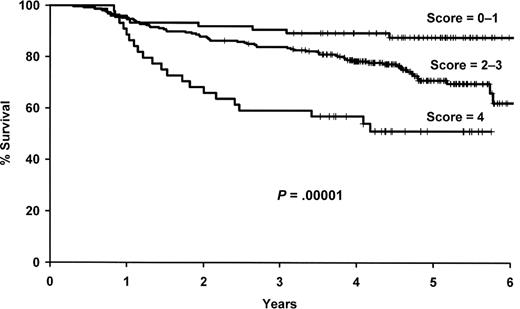
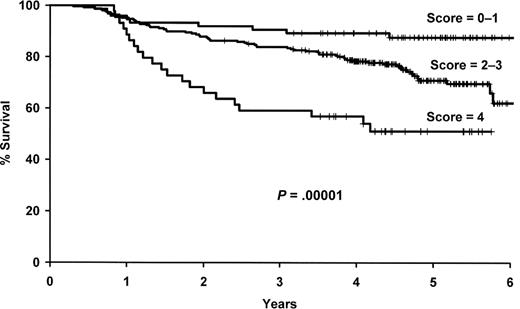
As outlined in “Evaluation of the multi-SNP risk score,” we selected a 4-SNP risk score for further evaluation, which included polymorphisms in IL1A (rs1800587), IL8RB (rs1126580), IL4R (rs2107356), and TNF (rs1800629). The number of deleterious genotypes was summed from these 4 SNPs (0-4), and this score was strongly associated with survival in both univariate (P = 2.8 × 10−5) and multivariate (P = 3.7 × 10−6) analyses (Figure 3, Table 4). Patients with 4 deleterious genotypes were more than 6 times more likely to die compare with patients with zero deleterious genotypes (95% CI, 3.05-15.0), and there was a gradient in risk with the number of deleterious SNP genotypes. Both the 4 SNP risk score and IL10 haplotype remained statistically significant when they were included in the same model along with the clinical and demographic variables (data not shown).
Results for the 4 SNP risk score. Kaplan-Meier curves by the number of deleterious genotypes from the 4 SNP risk score based on IL1A (rs1800587), IL8RB (rs1126580), IL4R (rs2107356), and TNF (rs1800629).
Results for the 4 SNP risk score. Kaplan-Meier curves by the number of deleterious genotypes from the 4 SNP risk score based on IL1A (rs1800587), IL8RB (rs1126580), IL4R (rs2107356), and TNF (rs1800629).
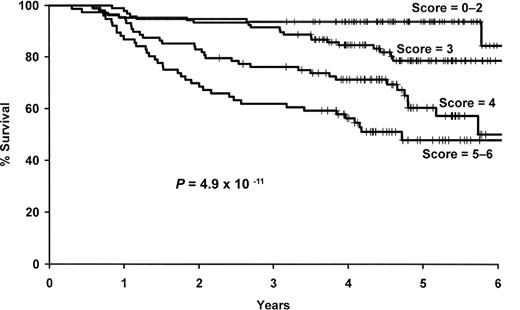
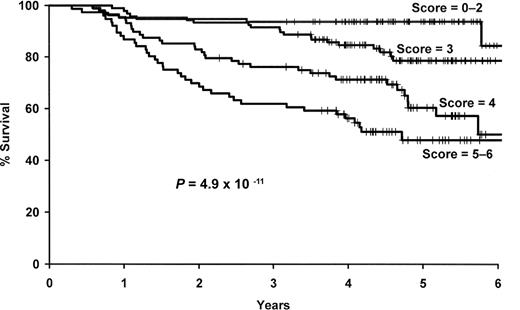
We next combined the number of deleterious genotypes (0-4) with the clinical and demographic risk score (0-2). This combined score was strongly associated with survival (P = 6 × 10−11), and patients with a score of 5 or 6 were more than 9 times more likely to die compared with those with a low (0-2) risk score (95% CI, 4.22-21.4; Figure 4, Table 5).
Results for the combined SNP and clinical and demographic risk score. Kaplan-Meier curves by level of the combined 4 SNP and clinical and demographic risk score.
Results for the combined SNP and clinical and demographic risk score. Kaplan-Meier curves by level of the combined 4 SNP and clinical and demographic risk score.
To further evaluate the predictive ability of our model, we conducted a time-dependent ROC analysis for censored data.31 This analysis uses sensitivity and specificity, both of which are time-dependent, to measure the prognostic capacity of the survival model as measured by the area under the curve (AUC). As shown in Figure 5, our clinical and demographic risk score (0-2) compared favorably in a time-dependent ROC results to the IPI from a previously published series.8 The time-dependent ROC analysis for the 4 SNP risk score showed a lower ability to predict outcome; but when it was combined with the clinical and demographic risk score, the AUC at 2 years was more than 0.70, and at 5 years the AUC was 0.75 (95% CI, 0.68-0.80). Figure 5 also includes the predictive ability of the germinal center phenotype versus all others from a previously published DLBCL survival dataset,8 and this characteristic predicted outcome at the same level as our 4 SNP risk score. Furthermore, when the germinal center phenotype was combined with the IPI, it predicted at the same level as our SNP plus clinical and demographic risk score.
Time-dependent ROC analysis. Time-dependent receiver-operator (ROC) analysis using the NCI-SEER dataset (clinical and demographic risk score, 4 SNP risk score, combined 4 SNP and clinical and demographic risk score) and using the Lymphoma/Leukemia Molecular Profiling Project (LLMPP) dataset8 (International Prognostic Index [IPI], Germinal-center B cell–like [GCB], and GCB + IPI).
Time-dependent ROC analysis. Time-dependent receiver-operator (ROC) analysis using the NCI-SEER dataset (clinical and demographic risk score, 4 SNP risk score, combined 4 SNP and clinical and demographic risk score) and using the Lymphoma/Leukemia Molecular Profiling Project (LLMPP) dataset8 (International Prognostic Index [IPI], Germinal-center B cell–like [GCB], and GCB + IPI).
To assess the robustness of our multi-SNP risk score, we repeated our model-building strategy (starting with selection of SNPs with a P ≤ .15 through the final multi-SNP risk model) with the datasets generated in a permutation analysis. Our observed results were more significant than 82% of the results from randomly generated datasets, which suggests that our multi-SNP risk score has some degree of significance given the intense model building approach performed. In addition, the likelihood plot (Figure 1A) of the best 1 SNP, 2 SNP, … 8 SNP models suggests that as many as 5 SNPs may add information to predicting survival beyond what would be expected resulting from chance, although we opted for 4 SNPs based on the combined results that included the bootstrap modeling in selecting the most robust and parsimonious model.
Sensitivity analyses
All results were also similar when we excluded all nonwhites from the analysis (data not shown).
We fit the final 4-SNP plus clinical and demographic risk score model based on deaths resulting from lymphoma coded on the death certificate (n = 65 of the 96 deaths; other deaths censored) and found the HR for the continuous score increased slightly from 1.9 (Table 5) to 2.0 (95% CI, 1.6-2.5).
Another potential concern is that not all patients received standard of care. Although we did not have sufficient data to fully evaluate treatment decisions and standard of care for these patients, we were able to exclude patients who did not receive multiagent chemotherapy, the presumed standard of care at the time of enrollment (rituximab was unlikely to have been used in the community setting from 1998 to 2000). After excluding these patients (n = 73), results from Tables 4 and 5 were essentially unchanged (data not shown). Finally, there was no survival difference by enrollment year (P = .42) and no impact on the final results in Table 5 after adjusting for enrollment year (data not shown).
Discussion
Using a population-based sample of 365 DLBCL cases diagnosed from 1998 to 2000 and followed through early 2005, we identified 17 SNPs from 14 cytokine and related immune regulation genes and a haplotype from the IL10 gene that were all associated with overall survival from DLBCL independent of clinical and demographic factors. Furthermore, we observed a strong effect from the combination of 4 SNP markers: 3 SNP markers from 3 genes, namely, IL1A1, IL8RB, and IL4R, that had not been previously reported in NHL prognosis, and 1 SNP marker in TNF, which has been shown to have a deleterious effect on survival. The preliminary results are particularly encouraging when combined into a common carrier model that linearly summed the number of deleterious genotypes. Combining the SNP risk score with demographic and clinical factors increased the predictive ability of the model, with AUCs of more than 0.70 after 24 months of follow-up in the time-dependent ROC analysis, which is approaching the predictive range needed for clinically useful tests. Compared with gene expression profiling data from biopsy samples,8 host genetics demonstrated a similar prognostic ability. Overall, these results support the importance of germ line variation in immune genes as predictors of DLBCL prognosis. Although steps were taken to guard against overfitting the data, including a permutation analysis of our entire model building strategy to assess the significance of our results compared with chance, these results clearly require replication in independent populations. In addition, it will be important to evaluate a population of DLBCL patients treated with rituximab in combination with CHOP chemotherapy and evaluate these findings in conjunction with molecular subtypes of DLBCL.
Even with rapid reporting by population-based cancer registries, we were able to enroll only approximately 50% of eligible DLBCL patients; therefore, we systematically missed those patients with the most aggressive disease leading to early mortality. Indeed, the observed survival of our patient cohort was much better than SEER population-based estimates for DLBCL patients overall but was quite consistent with survival estimates that were conditioned on DLBCL patients who survived 12 months after diagnosis (Figure 2). Therefore, our results will not apply to early mortality, and this will need to be addressed in future studies. A majority of deaths during the first year after diagnosis resulted from disease; and in the SEER data from Figure 2, approximately 58% of the deaths that occurred in the first 5 years after diagnosis occurred during the first year, and 75% of those deaths resulted from disease. Although our data are not informative for early mortality, our data will be robust to patients who survive to 12 months after their diagnosis. Furthermore, in sensitivity analyses, we found that our results held for patients who died of their disease (68%).
A proinflammatory state may both contribute to lymphomagenesis and lead to overall poorer survival. Of the genes in our final multi-SNP model, TNF has been most extensively studied as a prognostic factor in DLBCL. Higher levels of TNF have been associated with poorer outcome in NHL.33 Warzocha et al18 reported that an extended haplotype in TNF (G allele) and LTA (A allele) was associated with higher TNF production, and DLBCL patients (n = 126) with 2 to 4 high-risk alleles (33% of patients) had lower progression-free (HR = 2.33; 95% CI, 1.17-4.64) and overall (HR = 1.92; 95% CI, 0.63-5.80) survival. These results are consistent with our findings that patients with 2 to 4 risk alleles (38% of patients) had a lower overall survival compared with patients with 0 or 1 risk alleles (HR = 1.27; 95% CI, 0.63-1.48), although our HR was weaker and not statistically significant. However, our HR increased to 1.72 when we compared patients with 3 or 4 versus 0, 1, or 2 risk alleles, suggesting a gradient in risk with the number of adverse alleles. In a follow-up study, Warzocha et al reported that of SNPs in LTA+252, TNF (TNF−376, TNF−308, TNF−238, TNF−163), and HLA DRB1*02, only the TNF −308A allele was associated with higher levels of TNF and its associated receptors p55 and p75. The TNF −308A allele was also an independent predictor of freedom from progression (relative risk (RR) = 1.63) and overall survival (RR = 1.51) in DLBCL,19 and there was no evidence of a TNF/LTA haplotype effect in this analysis. Of note, the TNF G-308A promoter polymorphism has been associated with the development of DLBCL in this study population16 and in the InterLymph consortium pooled dataset,15 suggesting a role for this SNP (or another in strong linkage disequilibrium with it) in both the etiology and prognosis of DLBCL.
The other SNPs in our multi-SNP model (IL1A, IL8RB, and IL4R) have not been previously evaluated as DLBCL prognostic factors. The IL1A (rs1800587) −889T allele has been associated with higher interleukin-1 (IL1) production34 and an elevated erythrocyte sedimentation rate,35 but does not appear to be associated with risk of developing DLBCL.15,36 IL8RB encodes for the receptor of the chemokine IL8, a potent neutrophil chemoattractant whose expression is greatly enhanced by IL1 and TNF.37 We observed an association with the IL8RB SNP rs1126580, which is located in the 3′ untranslated region of the gene, but we did not observe any associations with the other IL8RB or IL8 SNPs or with haplotypes in these genes. Although we observed lower survival with the IL8RB rs1126580 AG/GG genotype, this genotype was associated with lower risk of developing DLBCL in this study population,16 although there was no association between this SNP and DLBCL risk in another study.36 The common allele (C) for the IL4R SNP rs2107356 was associated with poorer survival in our study; in etiology studies, this allele has been associated with a lower risk of developing DLBCL in this study population16 but not in 2 other studies.17,36 IL4 is central to B cells switching to IgE antibody production and maturation of helper T cells to a Th2 phenotype, and the IL4 receptor is crucial for binding and signal transduction of both IL4 and IL13.38 Several genes that correlate with DLBCL survival (eg, BCL6, HGAL)7 are IL4-specific target genes,39,40 although IL4 may use different signaling pathways in the germinal center B cell–like versus the activated B cell–like subtypes of DLBCL,41 and future studies should consider these subtypes.
We found a suggestive positive association of the IL10 A-1082G allele with DLBCL survival (HRAG/GG = 1.48; 95% CI, 0.91-2.38); and although this SNP did not make it into our final multi-SNP risk score, it did enter 27% of the multigene bootstrap models (overall ranked 8th). In contrast, Lech-Maranda et al20 found that this allele was inversely associated with overall survival in 199 DLBCL patients (RR = 0.78, P = .001), although 2 other studies reported no association.42,43 We observed no association of IL10 rs1800871 (C-819T), rs1800872 (C-592A), and rs1800890 (T-3575A) with DLBCL survival; the results of the former 2 SNPs are consistent with other studies,20,43 whereas the latter SNP has not been previously evaluated for DLBCL survival. Two microsatellite loci44 and 4 SNPs (−819C, −592C, −1082G, and −3575T)20,45,46 in the IL10 promoter have been associated with greater IL10 production, and higher IL10 levels have been associated with poorer prognosis in DLBCL in some,20,47 but not all,48-50 studies. We found that IL10 haplotypes that included alleles with putative greater IL10 production were also the alleles most strongly associated with lower overall survival. With respect to etiology, the IL10 −1082G and −3575T alleles have been associated with risk of developing DLBCL.15,17,36
A major strength of this study was the population-based ascertainment of incident cases of DLBCL. Whereas lack of standardization in treatment and clinical follow-up is a limitation of observational studies relative to clinical trials, clinical trials are often conducted in highly selected patient populations and therefore may not be representative of patients in the community. Furthermore, protection from confounding by the clinical trial design is less compelling in this context, where genotype is unlikely to confound treatment choice. Our observations, if validated, could be considered for general application to community-based patients. This study is the largest study of immune candidate SNPs in relation to survival conducted to date in DLBCL. The genes and SNPs were selected based on either functional data or their association with cancer or other immune-related diseases, and extensive quality controls were used to ensure high-quality genotyping. Our statistical analyses were comprehensive, and we have been cautious to evaluate the robustness of our results to both false positives and false negatives. Nevertheless, this analysis must be acknowledged as a first step, as other SNPs or haplotypes for these genes, or other immune genes we did not assess, may be of greater prognostic relevance.
A limitation of this study was the lack of detailed data on prognostic factors or treatment. However, we did have age, stage, B symptoms, and treatment class, and these variables predicted survival with a level of predictive ability similar to the IPI for a large study of DLBCL patients.8 Pathology classification was based on the cancer registry report without central review. Our sample was 20 to 74 years of age and may not generalize to patients 75 years of age and older. As discussed in “Descriptive results,” the study design did not capture patients with aggressive disease who died shortly after diagnosis. Finally, all of these patients were initially treated before 2000, before the widespread use of rituximab in the treatment of DLBCL.
In conclusion, host genetic variation in the cytokine and chemokine genes IL1A, IL8RB, IL4R, IL10, and TNF, individually and particularly in combination, were associated with late survival (> 12 months) in DLBCL after accounting for clinical and demographic factors. Our results suggest that patients with a greater propensity to produce TNF-α, IL-10, and IL-1 (and thus a proinflammatory state) may promote lymphomagenesis, decrease the ability of the host to eradicate lymphoma, or perhaps impair therapeutic efficacy, leading to poorer overall survival. These same TNF and IL10 SNPs and haplotypes also appear to increase risk of developing DLBCL, supporting a shared mechanism in the etiology and prognosis of DLBCL. The association with IL8RB supports a role for the tumor microenvironment in the biologic and clinical behavior of DLBCL. Thus, immunogenetics represents a promising class of prognostic factors that warrants further evaluation in DLBCL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Peter Hui and Cristine Allmer for programming assistance and Sondra Buehler for assistance in manuscript preparation.
This work was supported by National Institutes of Health (Bethesda, MD) grants R01 CA96704 and P50 CA97274; National Cancer Institute Intramural Program; and SEER contracts N01-PC-67 010, N01-PC-67 008, N01-PC-67 009, N01-PC-65 064, and N02-PC-71 105.
National Institutes of Health
Authorship
Contribution: J.R.C. and P.H. designed the study; J.R.C., P.H., N.R., and S.J.C. obtained funding; J.R.C., W.C., S.D., P.H., C.F.L., and R.K.S. obtained clinical data; S.J.C., S.S.W., and N.R. obtained genetic data; M.J.M. and S.M.G. performed statistical analysis; T.M.H. and J.R.C. drafted the manuscript; and all authors revised the manuscript and reviewed and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: James R. Cerhan, Division of Epidemiology, Mayo Clinic College of Medicine, 200 1st Street SW, Rochester, MN 55905; e-mail: cerhan.james@mayo.edu.

![Figure 5. Time-dependent ROC analysis. Time-dependent receiver-operator (ROC) analysis using the NCI-SEER dataset (clinical and demographic risk score, 4 SNP risk score, combined 4 SNP and clinical and demographic risk score) and using the Lymphoma/Leukemia Molecular Profiling Project (LLMPP) dataset8 (International Prognostic Index [IPI], Germinal-center B cell–like [GCB], and GCB + IPI).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/7/10.1182_blood-2007-09-111658/6/m_zh80190824660005.jpeg?Expires=1765910102&Signature=Rpq2W9W8mMDVa-UIRXYL1tfr2tBYmNg7M9C2U~RWCyNSwh93DVxp4zDajH1JMQM-z3pFJ~qE5QUEYzAY2i5q4GbnESKxAISuoyJUnFekMMXsN0p13bVCyLTGvy30fmIObCc2C-9tCH7TJWeTcprRjl2kcrQWKOm7XUbber-Ka8pGW2WQDIZLN62bh6C6OPCRuZaOq2YSihZNtyJ-LuSbnEYiyPIZnRIn1uzhqd9VH-R0Wwv77Vs99Fm4Q6rykHuZn53tTou-lHghEqHAWTPE0IXBFxAI0HXMeAgR4YplMcNGdefFYdxpfQ6oLgGIDRA6VD9LJiKJMau7pww3JE0j4w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

 ). (B) Ranking of the percentage of 1000 stepwise bootstrap Cox models that included each SNP. The top 4 SNPs in the bootstrap model are the same 4 SNPs in the best 4 SNP risk model.
). (B) Ranking of the percentage of 1000 stepwise bootstrap Cox models that included each SNP. The top 4 SNPs in the bootstrap model are the same 4 SNPs in the best 4 SNP risk model.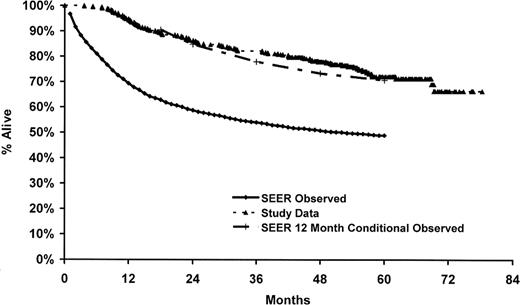
![Figure 5. Time-dependent ROC analysis. Time-dependent receiver-operator (ROC) analysis using the NCI-SEER dataset (clinical and demographic risk score, 4 SNP risk score, combined 4 SNP and clinical and demographic risk score) and using the Lymphoma/Leukemia Molecular Profiling Project (LLMPP) dataset8 (International Prognostic Index [IPI], Germinal-center B cell–like [GCB], and GCB + IPI).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/7/10.1182_blood-2007-09-111658/6/m_zh80190824660005.jpeg?Expires=1765914109&Signature=xpyL6Epr6MNKNdJv33H1sg8SHzVLl~SQrN3Q6RFBI3y9ZYktgta5cqJYhtEpmmRYRtTYYi3sRTH4S~HEOAYBP5POAQ8OUbNhf2J7WPuEwmnWE1I4KnPbT4yWZldP5-cDWyzMunyfIN2KOw4TwMQMKHVvn8Gt2p4mNNCph~ozwbvxR27wYE0kqqwFGoWbwL~S2vldffySd8eeXcuMICz13lP69wNoQfunKcAxb24x87U9LNGihk4h1KkKn2YGOaVVl9fl44ys3gl40XpiS53~uvSIklqgveJv8utiF2JdETzUcFLm1BZVJLEMlOhjcnNLWjd-M0MYG0ZtplI~dwwh9Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)