Abstract
Thrombin-activatable fibrinolysis inhibitor (TAFI) is a pro-metallocarboxypeptidase that can be proteolytically activated (TAFIa). TAFIa is unique among carboxypeptidases in that it spontaneously inactivates with a short half-life, a property that is crucial for its role in controlling blood clot lysis. We studied the intrinsic instability of TAFIa by solving crystal structures of TAFI, a TAFI inhibitor (GEMSA) complex and a quadruple TAFI mutant (70-fold more stable active enzyme). The crystal structures show that TAFIa stability is directly related to the dynamics of a 55-residue segment (residues 296-350) that includes residues of the active site wall. Dynamics of this flap are markedly reduced by the inhibitor GEMSA, a known stabilizer of TAFIa, and stabilizing mutations. Our data provide the structural basis for a model of TAFI auto-regulation: in zymogen TAFI the dynamic flap is stabilized by interactions with the activation peptide. Release of the activation peptide increases dynamic flap mobility and in time this leads to conformational changes that disrupt the catalytic site and expose a cryptic thrombin-cleavage site present at Arg302. This represents a novel mechanism of enzyme control that enables TAFI to regulate its activity in plasma in the absence of specific inhibitors.
Introduction
TAFI1,2 is a pro-metallocarboxypeptidase that links the coagulation and fibrinolytic systems. TAFI is activated by thrombin, the thrombin-thrombomodulin complex or plasmin.3 Activated TAFI (TAFIa) inhibits plasmin-mediated blood clot lysis by removing C-terminal lysine residues from partially degraded fibrin that are required for positive feedback in tissue plasminogen-activator dependent plasmin generation. In addition, TAFIa has been implicated in modulation of the inflammatory response by inactivating bradykinin and the anaphylatoxins C3a and C5a.4,5 Although it is a powerful antifibrinolytic agent, there are no known physiologic inhibitors of TAFIa. Instead, the half-life of TAFIa activity is regulated by its intrinsic instability. The inactivation rate, 5 to 10 minutes at 37°C, is highly temperature-dependent, suggesting that inactivation involves a large conformational change.6 This is also suggested by the susceptibility of the inactive enzyme, TAFIai to proteolytic cleavage by thrombin at Arg302, a site that is cryptic in TAFI and TAFIa.6,7 The stability of TAFIa is an important determinant for its antifibrinolytic potential because TAFIa inhibits fibrinolysis through a threshold-dependent mechanism.8-10
Full-length TAFI consists of 401 amino acids divided into 2 domains: the first 92 amino acids form the activation peptide; the next 309 amino acids form the catalytic domain. The activation peptide restricts substrate access to the catalytic cleft in the zymogen. TAFI is activated through cleavage at Arg92, which releases the activation peptide.
TAFI is highly homologous to the pancreatic procarboxypeptidases with 42% sequence identity to human procarboxypeptidase B (proCPB) and 38% to the various procarboxypeptidase A forms (proCPA1, proCPA2, and proCPA4).11-13 Unlike TAFIa, the pancreatic carboxypeptidases have stable activities. A model of TAFI built on the basis of crystal structures of CPA and CPB was unable to explain the differences in thermal stability between these enzymes.13,14 Thermal stability of TAFIa is increased by binding of small peptide substrates, like hippuryl-arginine,15 or carboxypeptidase inhibitors such as 2-guanidino-ethyl-mercaptosuccinic acid (GEMSA).16,17 In addition, extensive mutagenesis studies have identified 5 mutations (S305C, T325I, T329I, H333Y, and H335Q) which, in combination, stabilize TAFIa activity 180-fold.18-20
In this paper we present crystal structures of wild-type TAFI, a TAFI-inhibitor complex, and a TAFI mutant with a 70-fold more stable active enzyme. These structures explain the observed stabilization of TAFIa activity by specific mutations and inhibitors and provide the basis for a model for TAFI autoregulation by local protein dynamics.
Methods
Expression and purification
Cloning of wild-type TAFI isoform Thr147 was described previously.7 For cloning purposes, BglII and NotI cleavage sites were introduced at either end of the TAFI coding sequence and the internal BglII site was removed. The T325I, T329I, H333Y, and H335Q mutations were introduced using the Quikchange site-directed mutagenesis protocol (Stratagene, La Jolla, CA). The wild-type and mutant coding sequences were cloned into pABC345, which introduces the cystatin S secretion signal21 followed by a TEV-cleavable hexa-histidine tag. Constructs were transiently expressed in a modified HEK293S cell line. HEK293S cells22 are deficient in N-acetylglucosaminyltransferase I and therefore produce N-linked glycans of the (Man)5(GlcNAc)2-type. As has been done previously for HEK293 cells,23 we stably transfected HEK293S cells with the EBNA1 protein of the Epstein-Barr virus, which, in combination with the OriP replication origin on the pABC345 expression plasmid, increases protein production. Details of the pABC345 expression vector and HEK293ES cell line will be published elsewhere (R.A.R., P.F.M., T.P., T.H.C.B., J.C.M.M., P. Gros, E.G.H., W.H.).
Six days after transfection, medium was collected and concentrated 10-fold using a Quixstand hollow fiber system (GE Healthcare, Diegem, Belgium) followed by diafiltration against 25 mM Tris-Cl, 500 mM NaCl (pH 8.2). TAFI was purified using nickel-Sepharose (GE Healthcare) followed by antibody affinity chromatography on a Nik9H10-Sepharose column as described previously.7 L-arginine and L-glutamate were added to 5 mM, the pooled protein fractions were concentrated using a UFV5BCC25, 0.5-mL filter (Millipore, Amsterdam, The Netherlands) and dialyzed against 7 mM MES, 50 mM L-arginine, 50 mM L-glutamate, 10 mM glycine (pH 6.0).
Crystallization, data collection, and structure determination
Crystals of wild-type TAFI and TAFI-IIYQ were grown using either hanging drop or sitting drop vapor diffusion at 4°C (in 16%-18% PEG-3k + 0.18-0.22 mM Na/K-tartrate) in the presence of 50 mM L-glutamate and 50 mM L-arginine. Protein concentration before addition to the crystallization drop was approximately 4 mg/mL. Crystals appeared after approximately 1 week. For cryoprotection, crystals were incubated in the crystallization solution supplemented with stepwise increments of glycerol to 20% (vol/vol) final concentration. TAFI crystals were soaked by transferring a single TAFI crystal to a drop of mother liquor supplemented with 2 mM 2-guanidino-ethyl-mercaptosuccinic acid (GEMSA) and 20% (vol/vol) glycerol. Crystals were left to soak for 3.5 days before freezing. Diffraction data were collected at 100 K at beamline ID-29 (TAFI) and beamline ID23-1 (TAFI-IIYQ and TAFI-GEMSA) at the European Synchrotron Radiation Facility (Grenoble, France) and processed with XDS software.24
The TAFI structure was solved by molecular replacement with the catalytic domain of the homologous human pancreatic carboxypeptidase B (PDB entry 1KWM) as a search model using the program PHASER.25 The activation peptide was built in part using RESOLVE26 and completed by manual building. The TAFI-IIYQ and TAFI-GEMSA structures were solved using molecular replacement with the TAFI structure. Refine-ment was performed using the program REFMAC5 from the CCP4 package27 and manual building using Coot.28 During refinement, positional noncrystallographic symmetry (NCS) constraints were applied, except in those few regions where a molecule appeared to be significantly different. B-factors were not restrained between the NCS-related molecules. Stereochemistry of the final model was checked using the program PROCHECK,29 which indicated that 83.4%, 14.7%, 0.9%, and 1.0% of TAFI residues were in the core, allowed, generously allowed, and disallowed regions of the Ramachandran plot, respectively. For TAFI-GEMSA these values were 81.5%, 16.5%, 1.6%, and 0.4%, respectively, and for TAFI-IIYQ they were 84.3%, 13.9%, 1.0%, and 0.8%. The final model contains 3 copies of TAFI protein, 9 GlcNAc moieties for TAFI and 11 GlcNAc moieties for TAFI-GEMSA and TAFI-IIYQ (4 on molecules A and B, and 1 or 3 on molecule C) and 3 zinc ions. An arginine molecule in TAFI-IIYQ and a GEMSA molecule in TAFI-GEMSA were modeled in the catalytic site S1′ pocket. Data collection and refinement statistics are listed in Tables 1 and 2. Accessibility of the catalytic pocket was assessed using the program CAVER.30 Molecular graphics were generated using PyMOL (DeLano Scientific, Palo Alto, CA).
Coordinates and structure factors have been deposited at the Protein Data Bank (PDB; www.rcsb.org) under accession codes 3D66 for wt TAFI, 3D67 for TAFI-GEMSA, and 3D68 for TAFI-IIYQ.
Results
Crystallization and overall structure
The TAFI protein contains 5 putative N-glycosylation sites, which reportedly are all used.31 Crystallization of plasma and fully glycosylated recombinant TAFI resulted in nicely shaped but poorly diffracting crystals. Therefore, we produced recombinant TAFI in a HEK293ES cell line, which has a mutation in N-acetylglucosaminyltransferase-I, resulting in the production of homogeneous N-linked glycans of the (Man)5(GlcNAc)2 type. Using this approach we obtained wild-type TAFI crystals that diffracted to 3.1 Å (Table 1). The structure was solved by molecular replacement with the catalytic domain of human carboxypeptidase B13 (PDB entry 1KWM), yielding a solution consisting of 3 TAFI molecules per asymmetric unit. Although we observe no major differences in structure between the 3 TAFI molecules, 2 of the 3 molecules display partial disorder. We used the well-ordered molecule A for the overall structure description and comparison with other carboxypeptidase structures.
The TAFI structure (Figure 1A) consists of 2 domains. The catalytic domain, comprising residues 93-401, consists of a central 8-stranded mixed β-sheet flanked by 9 α-helices in a typical α/β-hydrolase fold. The N-terminal activation peptide is divided into 2 parts; the first 76 amino-acids form an open sandwich antiparallel α/β fold with 4 β-strands and 2 α-helices. The activation peptide is connected to the catalytic domain by the linker region comprising residues 77-92 which is partially α-helical. The activation peptide covers the catalytic pocket, which contains the catalytic zinc ion coordinated by His159, Glu162 and His288. We observe 4 N-linked glycans on the activation peptide at Asn22, Asn51, Asn63 and Asn86, which are disordered beyond the first N-acetyl-glucosamine moiety. The fifth reported N-glycosylation site at Asn21931 is completely buried within the catalytic domain and therefore glycosylation at this position is incompatible with the TAFI crystal structure
Overall structure of TAFI and comparison with pancreatic carboxypeptidases. (A) Ribbon drawing of TAFI with the activation peptide shown in blue, the catalytic domain in green, and dynamic flap residues 296-350 in orange. The catalytic zinc ion is shown as a magenta sphere and 4 N-linked glycans are shown in yellow stick representation. The cryptic thrombin cleavage site at Arg302 is indicated by an orange sphere. (B,C) TAFI molecule A superimposed on (B) human CPA2 (PDB entry 1AYE; yellow) and (C) human CPB (PDB entry 1KWM; yellow). Superimposition was done using catalytic domain residues only. The rmsd on Cα atoms between TAFI and CPA2 and CPB are 1.1 Å and 0.8 Å, respectively. (D) Radii of the access tunnels to the catalytic site zinc ion were calculated for TAFI (blue), CPA2 (red), and CPB (green).
Overall structure of TAFI and comparison with pancreatic carboxypeptidases. (A) Ribbon drawing of TAFI with the activation peptide shown in blue, the catalytic domain in green, and dynamic flap residues 296-350 in orange. The catalytic zinc ion is shown as a magenta sphere and 4 N-linked glycans are shown in yellow stick representation. The cryptic thrombin cleavage site at Arg302 is indicated by an orange sphere. (B,C) TAFI molecule A superimposed on (B) human CPA2 (PDB entry 1AYE; yellow) and (C) human CPB (PDB entry 1KWM; yellow). Superimposition was done using catalytic domain residues only. The rmsd on Cα atoms between TAFI and CPA2 and CPB are 1.1 Å and 0.8 Å, respectively. (D) Radii of the access tunnels to the catalytic site zinc ion were calculated for TAFI (blue), CPA2 (red), and CPB (green).
Comparison with other pro-carboxypeptidases
TAFI is homologous to human zinc-metalloprocarboxypeptidases (CPs) with highest similarity between the catalytic domains (44% and 48% for A-type and B-type CPs, respectively) and less conservation between the activation peptides (approximately 20%). Comparison of the TAFI structure with human pro-carboxypeptidaseA2 and B (PDB entries 1AYE and 1KWM, respectively) reveals few differences in overall fold. Like proCPA2 and in contrast to proCPB, the catalytic site residues of TAFI are in the “active” conformation,13 which is consistent with the reported activity of TAFI for small peptide substrates.32 The main difference between the 3 structures is a rotation of the activation peptide of TAFI away from the catalytic domain by 16° and 12° compared with proCPA2 and proCPB, respectively (Figure 1B,C). In addition there is a difference in the length of the α-helix in the connecting linker, which is longer in pro-carboxypeptidases of the A type compared with CPB's, whereas in TAFI this helix is of an intermediate length. The rotation of the activation peptide suggests that the catalytic site in zymogen TAFI could be more accessible to substrate compared with other CPs as was suggested by Valnickova et al.33 Calculation of the radius of the access tunnel to the catalytic site in TAFI, proCPA2 and proCPB with the program CAVER30 shows, however, that this is not the case (Figure 1D). As it turns out the rotation of the activation peptide in TAFI is compensated for by larger side chains, for example TAFI-Gln45 is a Thr in the corresponding position in CPB and CPA2. In conclusion, comparison of the overall TAFI structure with that of homologous peptidases does not reveal any obvious differences that could explain the thermal instability of TAFI.
A highly dynamic region in TAFI
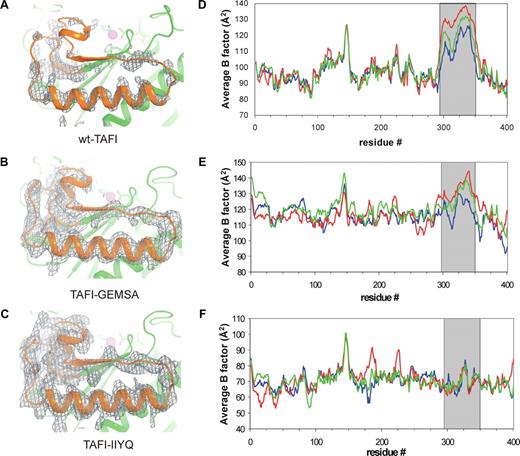
We observe striking differences in electron density levels for residues 296-350 among the 3 TAFI molecules in the asymmetric unit. In molecule A this region is well defined with electron density comparable to the remainder of the protein, in molecule C only backbone density is visible, whereas in molecule B density for residues 325-346 is completely lacking (Figure 2A). The observed reduction in electron density levels must be caused by mobility, which averages the electron density of these residues over a large volume. Protein mobility is also reflected by increased values of atomic B-factors, which describe atomic motions assuming an isotropic displacement of atoms around their average position. In molecule A, B-factors of residues 296-350 are well above the average for the whole protein (Figure 2D), whereas in molecules B and C the B-factors are even higher, emphasizing that this region of the protein is highly dynamic. The dynamic region can be described as a flap that starts with the loop after β-strand 10 near the catalytic site of the enzyme, then loops away from the catalytic site to a helix-loop-β-strand motif at the surface of the protein and terminates with residues 340-350 which form again part of the catalytic cleft wall (Figures 1A, 3A). The dynamic flap includes the cryptic thrombin cleavage site at Arg302. It also contains Asp348, which determines substrate specificity of TAFI and Tyr341 that is strictly conserved in all carboxypeptidases and is thought to stabilize the carboxylate of the terminal residue of the substrate.34 The dynamic flap interacts with the activation peptide through hydrophobic interactions between Tyr341 and residues Val35 plus Leu39 (Figure 3B). The observed differences in dynamics of the flap region in the 3 molecules in the asymmetric unit most likely result from crystal packing effects. As crystal packing tends to reduce protein mobility, we consider it likely that the highly dynamic molecule B most closely reflects TAFI in solution. The presence of the dynamic flap and in particular its contribution to the active site wall, led us to speculate that it could be directly involved in TAFIa inactivation.
Inhibitor-binding and mutations reduce dynamic-flap mobility. Panels A through C depict σA-weighted 2Fo-Fc electron density maps at 1.0 σ contour level for residues 296-350 in molecule B of TAFI (A), TAFI-GEMSA (B), and TAFI-IIYQ (C). Panels D through F express TAFI mobility as the average B-factor per residue for the 3 molecules in the asymmetric unit of the crystal. In wild-type TAFI (D), mobility of residues 296-350 is more than the average for the entire molecule, in particular for molecule B. In TAFI-GEMSA (E), B-factors in this region are reduced. In TAFI-IIYQ (F), B-factors in the flap are similar to the average for the molecule. Molecule A is shown in blue, molecule B in red, and molecule C in green. Gray area denotes residues of the dynamic flap.
Inhibitor-binding and mutations reduce dynamic-flap mobility. Panels A through C depict σA-weighted 2Fo-Fc electron density maps at 1.0 σ contour level for residues 296-350 in molecule B of TAFI (A), TAFI-GEMSA (B), and TAFI-IIYQ (C). Panels D through F express TAFI mobility as the average B-factor per residue for the 3 molecules in the asymmetric unit of the crystal. In wild-type TAFI (D), mobility of residues 296-350 is more than the average for the entire molecule, in particular for molecule B. In TAFI-GEMSA (E), B-factors in this region are reduced. In TAFI-IIYQ (F), B-factors in the flap are similar to the average for the molecule. Molecule A is shown in blue, molecule B in red, and molecule C in green. Gray area denotes residues of the dynamic flap.
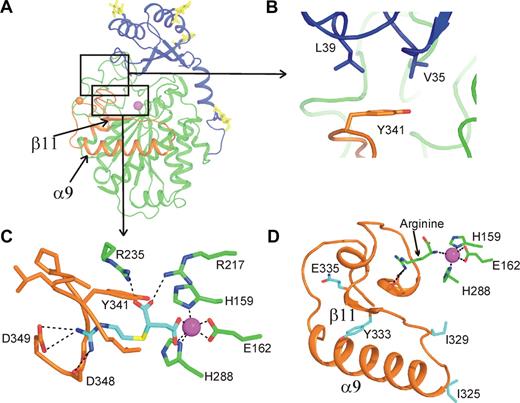
Interactions modulating flap dynamics. (A) Overview of TAFI indicating the interactions shown in panels B and C. (B) Interactions between the dynamic flap and the activation peptide. (C) GEMSA (cyan) bound in the active site of TAFI. (D) Ribbon diagram of the dynamic flap indicating the positions of stabilizing TAFI-IIYQ mutations (cyan), relative to the catalytic site indicated by the bound arginine molecule in the S1′ pocket (green). TAFI colors as in Figure 1A with oxygen (red), nitrogen (blue), and sulfur (yellow). H-bonds are indicated by black dashes.
Interactions modulating flap dynamics. (A) Overview of TAFI indicating the interactions shown in panels B and C. (B) Interactions between the dynamic flap and the activation peptide. (C) GEMSA (cyan) bound in the active site of TAFI. (D) Ribbon diagram of the dynamic flap indicating the positions of stabilizing TAFI-IIYQ mutations (cyan), relative to the catalytic site indicated by the bound arginine molecule in the S1′ pocket (green). TAFI colors as in Figure 1A with oxygen (red), nitrogen (blue), and sulfur (yellow). H-bonds are indicated by black dashes.
Inhibitor binding reduces flap dynamics
Carboxypeptidase inhibitors are known stabilizers of TAFIa. The stabilizing potency of different carboxypeptidase inhibitors depends on their Ki, as it appears that only the free enzyme inactivates.16 We decided to soak TAFI crystals with the inhibitor GEMSA, which has a Ki of 11 μM. The TAFI-GEMSA complex diffracted to 3.4 Å. GEMSA was found bound in the catalytic cleft S1′ pocket where the carboxy-terminal arginine or lysine residue of the substrate would bind. The observed binding mode is consistent with that of GEMSA inhibited carboxypeptidase D35 (PDB entry 1H8L). One carboxylate group of GEMSA coordinates the catalytic zinc ion, and the second carboxylate is coordinated by catalytic site residues Arg217 and Arg235. Additional hydrogen bonds are formed with Asp348 and Asp349, and hydrophobic interactions are formed with residues 299 and 340-349 that are all part of the dynamic flap of TAFI (Figure 3C). GEMSA binding caused a marked improvement in electron density of residues in the flap, with the expected concurrent decrease in B-factors (Figure 2B,E). Surprisingly, the reduction of flap dynamics is not restricted to the active site region but occurs throughout the flap. The stabilizing effect of GEMSA on the flap region supports the idea that flap dynamics and the instability of TAFIa are directly linked.
Stabilizing mutations have long-range effects
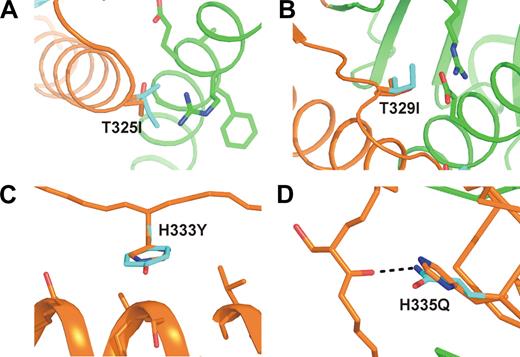
A direct link between flap dynamics and TAFIa inactivation is also suggested by the fact that all known stabilizing mutations are located in the flap region. To investigate the effect of stabilizing mutations on TAFI structure we crystallized TAFI-IIYQ, a mutant of TAFI that harbors the T325I, T329I, H333Y, and H335Q mutations, which result in a 70-fold more stable active enzyme. The mutant protein crystallized under the same conditions as the wild-type protein and crystals diffracted to 2.8 Å. As expected, the structure of the mutant protein is highly similar to the wild-type enzyme. However, unlike the wild-type protein, electron density in the flap region is well-defined for all 3 molecules in the asymmetric unit, including molecule B (Figure 2C) that is highly disordered in wild-type TAFI (Figure 2A). Furthermore, B-factors of residues 296-350 are similar to the average B-factor of the 3 TAFI-IIYQ molecules (Figure 2F). The TAFI-IIYQ mutations are located distal from the catalytic site in the loop between helix α-9 and strand β-11 (T325I and T329I) and in and just after strand β-11 (H333Y and H335Q; Figure 3D). Although side-chain conformations for these residues cannot be assigned unequivocally, it is clear that the 2 threonine-to-isoleucine mutations increase interactions between the dynamic region and the stable core of the catalytic domain (Figure 4A,B). The Thr329-to-Ile substitution enables more extensive hydrophobic interactions with the side chain of Arg365, whereas Ile325 fits nicely into a hydrophobic pocket formed by the side chains of Arg384, Glu385, and Ala388 on helix α-12. The 2 histidine mutations increase interactions within the dynamic segment; the H333Y mutation enables more extensive hydrophobic interactions with residues in helix α-9, whereas the replacement of His335 with Gln results in the formation of an additional hydrogen bond with the backbone oxygen of Ser303 (Figure 4C,D).
Stabilizing effects of TAFI-IIYQ mutations. Mutations T325I (A) and T329I (B) form additional interactions with residues from the catalytic domain outside the dynamic flap. Mutations H333Q (C) and H335Y (D) stabilize the dynamic flap through interactions within the flap. Because the side chain conformation of Ile325 and Ile329 could not be determined unequivocally from the electron density, details of these interactions remain subject to speculation. Wild-type residues are shown in orange and mutated residues in cyan. H-bonds are shown as black dashes.
Stabilizing effects of TAFI-IIYQ mutations. Mutations T325I (A) and T329I (B) form additional interactions with residues from the catalytic domain outside the dynamic flap. Mutations H333Q (C) and H335Y (D) stabilize the dynamic flap through interactions within the flap. Because the side chain conformation of Ile325 and Ile329 could not be determined unequivocally from the electron density, details of these interactions remain subject to speculation. Wild-type residues are shown in orange and mutated residues in cyan. H-bonds are shown as black dashes.
Surprisingly, the catalytic site of TAFI-IIYQ contained residual electron density in the S1′ pocket, which we interpreted as a bound arginine molecule. The arginine carboxylate interacts with catalytic site residues Asn234, Arg235, and Tyr341, and the amino group coordinates the catalytic zinc ion. The arginine side chain is hydrogen-bonded by Ser399 and Asp348 from the dynamic flap. Arginine was present during preparation of both wild-type and mutant protein, and some residual density observed in the active-site pocket of wild-type TAFI likely also originates from the presence of arginine. However, in the wild-type protein the arginine is apparently disordered, which supports the link between active site disorder and flap dynamics.
The structural data on mutant and inhibitor-bound TAFI demonstrate that changes in flap dynamics occur in a concerted manner and can be initiated far from the active site by stabilizing mutations and from within the active-site pocket by inhibitor binding.
Discussion
We presented the crystal structures of wild-type TAFI, a TAFI-GEMSA complex, and a TAFI mutant whose active form is stabilized 70-fold. A striking finding is the differential mobility of residues 296-350, which are highly mobile in wild-type TAFI but become ordered upon inhibitor binding and in the presence of specific mutations. Because the factors that reduce mobility of this region also increase the stability of TAFIa, we consider it justifiable to assume a direct correlation between protein mobility observed in zymogen TAFI and TAFIa stability.
Comparison of the 3 TAFI structures suggests the following mechanism for TAFI autoregulation (Figure 5): In zymogen TAFI, the flap region consisting of residues 296-350 already displays considerable mobility, but is prevented from irreversible unfolding by interactions with the activation peptide. Loss of these stabilizing interactions upon TAFI activation and dissociation of the activation peptide increases flap mobility even further. Flap mobility can be reduced by binding of inhibitor or substrate. Ultimately, the flap suffers an irreversible conformational change that destroys TAFIa activity due to the displacement of critical residues Tyr341 and Asp348 and exposes the cryptic thrombin cleavage site at Arg302. Thus the dynamic flap acts as an “activity timer” that limits the lifetime of the activated protein by exploiting local protein dynamics.
Model of TAFIa inactivation. In zymogen TAFI, the dynamic flap (orange) is stabilized by the activation peptide (blue), which also hinders substrate access to the active site. Cleavage by thrombin releases the activation peptide, the substrate can enter, and mobility in the dynamic flap is increased. In time an irreversible conformational change occurs in the dynamic flap. The catalytic site is disrupted and the cryptic thrombin cleavage site at Arg302 (indicated by x) becomes accessible.
Model of TAFIa inactivation. In zymogen TAFI, the dynamic flap (orange) is stabilized by the activation peptide (blue), which also hinders substrate access to the active site. Cleavage by thrombin releases the activation peptide, the substrate can enter, and mobility in the dynamic flap is increased. In time an irreversible conformational change occurs in the dynamic flap. The catalytic site is disrupted and the cryptic thrombin cleavage site at Arg302 (indicated by x) becomes accessible.
The activation peptide must provide critical interactions that stabilize the dynamic flap in the zymogen. Extensive interactions exist between the activation peptide and the catalytic domain. However, direct contacts involving the dynamic flap are limited, involving only activation peptide residues Val35 and Leu39 and dynamic flap residue Tyr341 (Figure 3B). We cannot exclude that the N-glycans on the propeptide stabilize the dynamic flap. In particular, a complex N-glycan attached to Asn22 could come sufficiently close to the dynamic flap to establish direct contacts. Future mutagenesis studies will have to address this issue.
The proposed model of TAFIa inactivation implies considerable mobility of active site residues that are part of the dynamic flap. Catalysis would therefore require formation of a properly formed active site by induced fit. This may proceed in a fashion similar to the observed induction of order in the active site of GEMSA-bound TAFI. Substrate binding through an induced-fit mechanism involving the equivalent of Tyr341 in pancreatic carboxypeptidases has previously been suggested.34 Apparently, in TAFI the region involved in the induced-fit mechanism has been extended to encompass a much larger region of the protein.
TAFIa inactivation is accompanied by a biphasic decrease in Trp fluorescence.6,36 Consistent with this observation, unfolding and/or dislocation of the dynamic flap would significantly change the environment and, consequently, fluorescence of Trp350 and most likely Trp153, which is completely buried within the core of the catalytic domain. Therefore, although we cannot exclude the possibility that other parts of TAFI are involved in the inactivation process, we propose that a dramatic conformational change of residues 296-350 would be sufficient to explain the observations related to TAFIa inactivation.
The use by TAFI of a dynamic flap as activity timer for its autoregulation is a fundamentally new discovery and extends the limited range of regulatory mechanisms known to operate in extracellular space. Moreover, understanding the mechanism of inactivation of TAFIa may have important implications for therapeutic strategies for bleeding and thrombosis. Because the activity of activated TAFI is determined by a threshold mechanism,8-10 the effect of stabilizing or destabilizing agents on the function of TAFI is much larger than the effect of modulating TAFI concentration. Our findings provide structural information that opens the path to the development of new drugs that target TAFI to influence blood clot stability.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank P. Gros (Crystal and Structural Chemistry, Utrecht University) for critically reading the manuscript. We thank the European Synchrotron Radiation Facility (ESRF), Grenoble, France for providing data collection facilities and the ESRF beamline scientists at ID-29 and ID23-1 for their help with data collection.
The research was supported in part by a VENI grant from the Netherlands Organization for Scientific Research, Den Haag, The Netherlands (NWO grant no. 916.36.104 to P.F.M.) and research grants from the Netherlands Heart Foundation, Den Haag, The Netherlands (NHS2003B245 to J.C.M.M. and NHS2005B238 to E.G.H.).
Authorship
Contribution: P.F.M. and J.C.M.M. designed research. P.F.M. and T.P. carried out mutagenesis and biochemical research R.A.R. and W.H. established the HEK293ES cell line and protein expression. T.H.C.B. performed crystallographic research and analyzed data. E.G.H directed design of crystallographic studies and data analysis. T.H.C.B. and E.G.H wrote the manuscript with input from all other authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: E. G. Huizinga, Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Department of Chemistry, Faculty of Science, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands; e-mail: e.g.huizinga@uu.nl; or J. C. M. Meijers, Department of Experimental Vascular Medicine, Academic Medical Center, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands; e-mail: j.c.meijers@amc.uva.nl.
References
Author notes
* P.F.M. and T.H.C.B. contributed equally to this work.