Abstract
CD28 is a receptor expressed on T cells that regulates their differentiation after antigen stimulation to long-term-survival memory T cells. CD28 enhances T-cell receptor signals and reduces expression of CBL ubiquitin ligases, which negatively control T-cell activation. In the absence of CD28 ligation during the primary stimulation, CBL levels remain high and T cells fail to mount an efficient secondary response. CD28 associates with p85α, one of the regulatory subunits of phosphoinositide-3-kinase (PI3K), but the relevance of this interaction is debated. We examined here the contribution of the other ubiquitous PI3K regulatory subunit, p85β, in CD28 function. We describe that p85β bound to CD28 and to CBL with greater affinity than p85α. Moreover, deletion of p85β impaired CD28-induced intracellular events, including c-CBL and CBL-b down-regulation as well as PI3K pathway activation. This resulted in defective differentiation of activated T cells, which failed to exhibit an efficient secondary immune response. Considering that p85β-deficient T cells fail in recall responses and that p85β binds to and regulates CD28 signals, the presented observations suggest the involvement of p85β in CD28-mediated activation and differentiation of antigen-stimulated T cells.
Introduction
T-cell activation involves cooperation of signals triggered by the T-cell receptor (TCR) and costimulatory molecules such as CD28.1 TCR engagement induces a cascade of early signals including activation of Src family tyrosine kinases (TyrKs) pp56lck and pp59fyn, which in turn phosphorylate the TCRζ chain and activate the Syk family TyrK ZAP-70. ZAP-70 phosphorylates and associates numerous signaling molecules such as LAT and SLP-76, which propagate the activation signal.2,3 These cascades elicit expression of genes required for activation of hematopoietic cells, and for T-cell expansion, effector function, and differentiation.4
CD28 binding contributes to T-cell activation by stabilizing the immunologic synapse, by enhancing the magnitude and duration of TCR-induced signaling cascades,5 and by inducing long-term survival.6 CD28 is therefore necessary for optimal activation of T cells, which become anergic (nonresponsive) in the absence of CD28-derived signals.7,8 Although CD28 coreceptor function was initially described for CD4+ T cells,9 it is also required for optimal primary CD8+ T-cell responses to most pathogens as well as for their recall responses.10-13 CD28 can evade the anergy program by inducing down-regulation of Casitas B-lineage lymphoma b (CBL-b).14-17 CBL-b ubiquitinates the p85α regulatory subunit of PI3K; this reduces p85α binding to CD28 and TCRζ, resulting in diminished PI3K activation.18,19 CBL-b down-regulation, thus, is required for optimal PI3K pathway activation, and in turn for Rac-induced actin polymerization and immunologic synapse stability.20
c-CBL is another CBL family member that limits T-lymphocyte activation.21 c-CBL exerts this action by down-regulating ZAP-70 TyrK, although c-CBL enhances PLCγ activation.21 c-CBL–deficient mice show increased positive selection of T cells in thymus as well as peripheral lymphoid hyperplasia.22-24 Whereas changes in c-CBL expression after T-cell activation have not been reported, this process causes an early reduction in CBL-b levels, an event required for optimal T-cell activation.16,25,26 CBL-b levels recover at later time points, contributing to trigger TCR trafficking to the lysosomal compartment.27
Class I PI3K enzymes are formed by a p110 catalytic subunit and a regulatory subunit; the p110 subunit catalyzes formation of phosphatidylinositol (3,4)P2 and phosphatidylinositol (3,4,5)P3 after receptor stimulation. PI3Ks are classified as TyrK-controlled class IA enzymes (p110α, p110β, and p110δ) that associate p85-like regulatory subunits, and G protein–regulated class IB PI3K, p110γ.28 In the case of class IA PI3K, 3 genes (pik3r1, pik3r2, and pik3r3) and their alternative splice forms give rise to 5 regulatory subunits.28 Whereas p85α and p85β expression is ubiquitous, p55γ, p55α, and p50α expression is restricted to certain tissues.28 PI3K binds to the TCR and to CD28 through p85, thereby contributing to trigger T-cell activation, survival, and division28-35 ; nonetheless, the contribution of PI3K to CD28-mediated signaling remains incompletely understood.
We previously described that p85β-deficient T cells exhibit a moderately enhanced primary immune response.33 In the course of these studies, we noticed that secondary responses in these mice were defective. Considering that CD28 binds to p85 subunits,36-40 and that CD28 regulates recall T-cell responses,5-15 we examined whether p85β contributes to mediate CD28 signals. We describe that p85β associates with CD28, controls CD28-mediated signals, and is required for efficient secondary immune responses.
Methods
Mice and cDNA
p85β−/− mice, reported previously,33 were maintained in heterozygosis. F5TCR (Vβ11Vα4) transgenic (Tg) mice41 were provided by D. Kioussis (Medical Research Council [MRC], London, United Kingdom). The CNB ethics committee approved all animal studies. F5TCRTg mice were crossed with p85β−/− mice; offspring were analyzed by polymerase chain reaction (PCR) and flow cytometry to confirm the TCR transgene and correct MHC expression. Mice were bred and maintained in specific pathogen-free conditions in our animal facility; the CNB ethics committee approved all studies. Recombinant hemagglutinin p85α (rHAp85α) and rHAp85β were donated by L. Williams (Chiron, Emeryville, CA) and J. Janssen (Heidelberg University, Heidelberg, Germany); we cloned both into the pEF-BOS vector. Mutant forms of human p85β were cloned by PCR using primers encoding HA peptide, including the appropriate coding sequence encompassing the SH3 domain (pEF-BOS HA-SH3β) or SH2-SH2β region (pEF-BOS HA-SH2-SH2β). pSG5 p65βPI3K, similar to the previously described p65αPI3K,42 was prepared by introducing a stop codon at residue 567 by point mutation.
Antibodies and reagents
Antibodies used for T-cell activation were hamster antimouse CD3ϵ (145-2C11) and CD28 (37.51), mouse anti–human CD3ϵ (UCHT1) and CD28 (CD28.2), mouse anti–Armenian and -Syrian hamster IgG1 (G94-56), mouse anti–Armenian hamster IgG (G192-1; all from BD Pharmingen, San Jose, CA), and rabbit anti–mouse IgG (Fcγ-fragment specific; Jackson Immunoresearch, West Grove, PA). For immunoprecipitation, protein A Sepharose beads and the following antibodies were used: anti–mouse CD28 (37.51 [BD Biosciences, San Jose, CA] and M-20 [Santa Cruz Biotechnology, Santa Cruz, CA]), anti–human CD28 (152-2E10; Biosource, Camarillo, CA), anti-PI3K pan-p85 (Upstate Biotechnology, Lake Placid, NY), anti-SH3 PI3K p85α (Upstate Biotechnology), anti–c-CBL (BD Biosciences), anti–CBL-b (Santa Cruz Biotechnology), and anti-HA (Covalence, Franklin, MA). We prepared an anti-p85β Ab using His-tagged full-length murine p85β; the Ab recognizes murine and human p85β and not p85α (I.C. and A.C.C., in preparation). For Western blot, we used the appropriate antibodies and developed them with enhanced chemiluminescence (ECL; GE Healthcare, Little Chalfont, United Kingdom). We measured activity of PKC (protein kinase C) using anti–phospho PKC (γThr514); PKB (protein kinase B; also termed Akt) using anti–phospho-Akt (Ser473) and phospho-Akt (Thr308); p44/42 mitogen-activated protein kinase (MAPK) using anti–phospho-p44/42MAPK (Thr202/Tyr204); and p38 MAPK using anti–phospho-p38MAPK (Thr180/Tyr182; all from Cell Signaling Technology, Beverly, MA).
Anti-PKCα (C-20; Santa Cruz Biotechnology), -Akt1/PKBα (Upstate Biotechnology), -p42/44 MAPK, and -p38 MAPK (Cell Signaling) antibodies were used to control loading. We analyzed the profile of tyrosine-phosphorylated proteins using anti-PTyr antibody (4G10; Upstate Biotechnology). We also used anti–CBL-b (G-1, C-20; Santa Cruz Biotechnology), anti–c-CBL (clone 17) and anti–mouse Nedd4 (clone 15; both from BD Transduction Laboratories), anti–human Nedd4 (Upstate Biotechnology), anti-βactin (Sigma-Aldrich, St Louis, MO), anti-HA (Covalence), and anti–pan-p85 antibody (Upstate Biotechnology). Ly294002 was from Calbiochem (San Diego, CA).
Cell lines and transfections
Jurkat T cells were maintained in complete Dulbecco modified Eagle medium (DMEM; BioWhittaker, Walkersville, MD) with 10% fetal bovine serum (FBS; Sigma-Aldrich), 2 mM glutamine, 50 μM 2-mercaptoethanol, and 100 U/mL penicillin/streptomycin. Cells (1.5 × 107) in 400 μL complete medium were transfected by electroporation of 25 μg DNA using a Gene Pulser (270 V, 950 μF; Bio-Rad, Hercules, CA). Cells were immediately transferred to 10 mL complete medium containing 50 μg/mL Z-VAD (benzyloxycarbonyl-Val-Ala-Asp(OMe)-fluoromethylketone) and assayed after 36 hours, when fluorescence-activated cell sorting (FACS) or Western blot analyses showed maximum expression of the transfected protein.
Spleen and lymph node cell suspensions were T-cell enriched by depletion of B cells using mouse pan B (B220) Dynabeads (Dynal, Oslo, Norway) followed by 2-hour incubation on plastic plates at 37°C to deplete adherent cells, or using mouse T cell–negative isolation kits (Invitrogen, Carlsbad, CA); T cells were more than 95% CD3+ by FACS analysis (Epics XL-MCL; Beckman Coulter, Hialeah, FL). For CD8+ cells, T cell–enriched suspensions were depleted of CD4+ cells using Dynabeads. Primary T and B cells were maintained in RPMI 1640 (BioWhittaker). Antigen-presenting cells (APCs) were prepared by depleting T cells from spleen suspensions with mouse pan-T (Thy1.2) Dynabeads.
Two-dimensional gel electrophoresis
Before activation, purified mouse or Jurkat T cells were incubated (2 hours) in DMEM with 0.1% BSA (fraction V low endotoxin; Sigma-Aldrich), after which cells were washed and resuspended in serum-free medium. For activation, 5 to 10 × 106 cells were incubated with soluble anti-CD3+anti-CD28 Ab and cross-linked with secondary antibody as for in vivo T-cell activation. Cells were lysed in CHAPS buffer (5% CHAPS, 20 mM Tris-HCl, pH 8, 1 mM MgCl2, 1 mM CaCl2, 10% glycerol, 137 mM NaCl, 5 mM NaF, 1 mM sodium orthovanadate, 1 mM PMSF, 1 mM EDTA, 1 μg/mL each aprotinin and leupeptin, and 2 nM okadaic acid; 4°C, 90 minutes). Extracts were immunoprecipitated with anti-p85 (1 μg/mL) or CD28 (1 μg/mL; 4°C, 16 hours). Two-dimensional electrophoresis was performed following standard procedures.
Statistical analyses
Statistical analyses were performed using StatView 512+ (Calabasas, CA). Gel bands were quantitated with ImageJ software (National Institutes of Health [NIH], Bethesda, MD).
Online supplemental material
T-cell activation protocols, coupling of peptides to Actigel resin, in vitro transcription translation, PI3K and PKB kinase assays, in vitro binding of proteins to peptide columns, immunoprecipitation and Western blot assays, TCR down-regulation analyses, flow cytometry assays, quantitative reverse-transcription (RT)–PCR, and proliferation assays are available in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Results
CD28 preferentially associates with p85β
CD28 binds directly to the p85 regulatory subunit of PI3K.36-40 T cells express both of the ubiquitous p85 isoforms: α and β;33 however, the relative contribution of these subunits to CD28-mediated signals has not been elucidated. We first examined the relative abundance of p85α and p85β in Jurkat cells and wild-type (WT) murine T cells. Because p85α and p85β have to be recognized by distinct Abs, for comparison rather than Western blot (WB), we performed quantitative RT-PCR (QRT-PCR) as p85 mRNA levels do correlate with p85 protein levels.43 Whereas the ratio of p85α/p85β was close to 1 in Jurkat cells, in murine T cells p85β levels were approximately 15% those of p85α (Figure 1A).
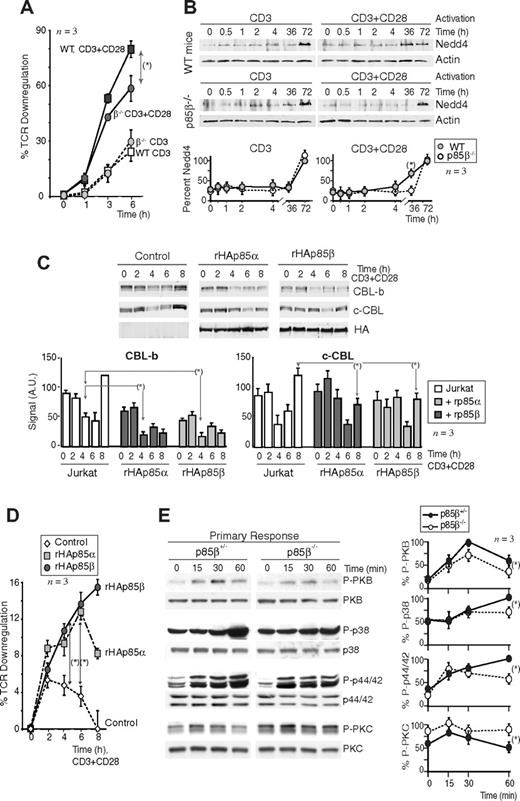
CD28 preferentially associates with p85β. (A) Ratio of p85α/p85β molecules per nanogram of total mRNA determined by QRT-PCR. (B) Jurkat cells transfected with control, rHAp85α, or rHAp85β vectors (36 hours), were activated via CD3+CD28 (15 minutes). WB examined protein expression in total extracts, extracts were also immunoprecipitated with anti-CD28 Ab, and associated p85 was analyzed in Western blot (WB) using anti-HA Ab (long and short exposure). (C) Number of p85α or p85β molecules per nanogram of total mRNA (from p85β−/− and p85β+/+ T cells) by QRT-PCR; each dot represents a mouse. Western blot analysis of p85 in extracts of a representative mouse (vertical line indicate a repositioned gel lane). (D) T cells from p85β−/− and p85β+/+ mice and Jurkat cells were activated as in panel A. Total extracts or CD28 immunoprecipitates were resolved in 2-dimensional electrophoresis and analyzed in WB using anti–pan-p85 antibody. β indicates the p85β spot (absent in p85β−/− mice). The isoelectric point (pI) of mp85β was 5.74, and the pIs were 5.96, 5.9, and 5.8 for mp85α and its phosphorylated forms; the pI for hp85β was 6.03, and the pIs were 5.84, 5.77, and 5.71 for hp85α and its phosphorylated forms. Graphs show the mean plus or minus SD (n = 3) of the β spot signal relative to the combined signal for all spots (100%). (E) Jurkat cells were activated as in panel B; WB shows the amount of p85 present in CD3 or CD28 immunoprecipitates. Graphs show the mean signal plus or minus SD (AU indicates arbitrary unit) of p85 bound to CD3 or CD28. (F) cDNA encoding human (h) or mouse rp85α or rp85β was transcribed/translated in vitro (rhp85α was fused to GFP) in the presence of 35S-Met. Purified proteins were examined by SDS-PAGE and autoradiography (top left). Sequence of the hCD28 peptides; the dots represent the nonconserved amino acids in mCD28 (H > P and P > A). In the bottom, p85 bound to the different CD28 peptide columns analyzed by SDS-PAGE and autoradiography. PDGFR (P) peptide was used as a positive control; column with no peptide and mp85α was used as the negative control. The histograms show the absolute binding of each rp85 form to CD28 motifs (mean signal ± SD in arbitrary units [AU]) on the right, and rp85 relative binding to M peptides and to P column (n = 3). *t test, P < .05.
CD28 preferentially associates with p85β. (A) Ratio of p85α/p85β molecules per nanogram of total mRNA determined by QRT-PCR. (B) Jurkat cells transfected with control, rHAp85α, or rHAp85β vectors (36 hours), were activated via CD3+CD28 (15 minutes). WB examined protein expression in total extracts, extracts were also immunoprecipitated with anti-CD28 Ab, and associated p85 was analyzed in Western blot (WB) using anti-HA Ab (long and short exposure). (C) Number of p85α or p85β molecules per nanogram of total mRNA (from p85β−/− and p85β+/+ T cells) by QRT-PCR; each dot represents a mouse. Western blot analysis of p85 in extracts of a representative mouse (vertical line indicate a repositioned gel lane). (D) T cells from p85β−/− and p85β+/+ mice and Jurkat cells were activated as in panel A. Total extracts or CD28 immunoprecipitates were resolved in 2-dimensional electrophoresis and analyzed in WB using anti–pan-p85 antibody. β indicates the p85β spot (absent in p85β−/− mice). The isoelectric point (pI) of mp85β was 5.74, and the pIs were 5.96, 5.9, and 5.8 for mp85α and its phosphorylated forms; the pI for hp85β was 6.03, and the pIs were 5.84, 5.77, and 5.71 for hp85α and its phosphorylated forms. Graphs show the mean plus or minus SD (n = 3) of the β spot signal relative to the combined signal for all spots (100%). (E) Jurkat cells were activated as in panel B; WB shows the amount of p85 present in CD3 or CD28 immunoprecipitates. Graphs show the mean signal plus or minus SD (AU indicates arbitrary unit) of p85 bound to CD3 or CD28. (F) cDNA encoding human (h) or mouse rp85α or rp85β was transcribed/translated in vitro (rhp85α was fused to GFP) in the presence of 35S-Met. Purified proteins were examined by SDS-PAGE and autoradiography (top left). Sequence of the hCD28 peptides; the dots represent the nonconserved amino acids in mCD28 (H > P and P > A). In the bottom, p85 bound to the different CD28 peptide columns analyzed by SDS-PAGE and autoradiography. PDGFR (P) peptide was used as a positive control; column with no peptide and mp85α was used as the negative control. The histograms show the absolute binding of each rp85 form to CD28 motifs (mean signal ± SD in arbitrary units [AU]) on the right, and rp85 relative binding to M peptides and to P column (n = 3). *t test, P < .05.
We then examined p85-CD28 complex formation in Jurkat T cells expressing similar levels of recombinant HA-tagged p85α or p85β (rHAp85α or rHAp85β), which were approximately double that of endogenous proteins (Figure 1B). We immunoprecipitated CD28, and examined CD28-p85 association by WB detection with anti-HA antibody. Despite similar abundance of rHAp85α and rHAp85β, CD28 bound a larger amount of p85β than p85α (Figure 1B).
We also examined endogenous CD28-p85 complex formation in mouse T cells. As mentioned in Figure 1A, p85β mRNA levels were roughly one sixth those of p85α; a moderate increase in p85α mRNA levels was found in approximately two-thirds of p85β−/− mice (Figure 1C, WB of a p85β−/− representative mouse with enhanced p85α levels). We analyzed total WT T-cell extracts as well as CD28 immunoprecipitates obtained from CD3+CD28-activated T cells in 2-dimensional gel electrophoresis. Whereas in WT cells the p85β spot (the most acidic spot) represented approximately 15% of the total p85, in CD28 immunoprecipitates the p85β proportion was double, showing that relative to its abundance p85β exhibits a greater affinity for CD28 (Figure 1D). Similarly, in Jurkat cells, the human p85β right-most spot (the more basic spot) was approximately 50% of the total signal in whole extracts, and approximately 80% in CD28 immunoprecipitates (Figure 1D).
We also compared the amount of endogenous p85β bound to CD3 or CD28 receptors after T-cell activation by CD3, CD28, or both. CD3 activation did not induce a significant association of p85β to either CD3 or CD28 (Figure 1E). In contrast, CD28 activation induced the binding of p85β to CD28 but triggered as well p85β binding to CD3 (Figure 1E), suggesting that CD28 stimulation is required to bring p85β into complex with CD3. Finally, after CD3+CD28 stimulation, a larger amount of p85β was found in CD28 immunoprecipitates, suggesting a greater affinity of p85β for CD28 than for CD3 (Figure 1E). A similar analysis performed with p85α showed that CD3 stimulation triggered association of p85α to CD3, whereas CD28 activation favored p85α association to CD28; a similar amount of p85α was found in CD3 and CD28 receptors after CD3+CD28 stimulation (Figure 1E). Therefore, whereas p85α associates similarly with both CD3 and CD28, p85β association to CD28 is greater than to CD3, moreover, CD28 activation is required for association of p85β to CD28 and to CD3. The analysis of CD28-bound p85 using a pan-p85 Ab resembled the pattern of p85β, supporting that CD28 binds greater levels of p85β than p85α (Figure 1E).
To map p85-binding regions in the CD28 cytoplasmic tail, and to quantify more accurately the amount of p85α and p85β bound to CD28, we analyzed direct binding of purified in vitro transcribed and translated p85 subunits to CD28 peptide columns. We selected 4 CD28 peptides including the YMNM motif (M1), the C-terminal YRS region (M2), and the Tyr-phosphorylated forms of these peptides (M1p, M2p; Figure 1F), regions involved in CD28 binding to the p85 subunit.40
Translated murine and human 35S-Met–labeled p85β and p85α (h-p85α fused to GFP) were resolved in SDS–polyacrylamide gel electrophoresis (PAGE) gels to measure p85 levels (Figure 1F), equal amounts of purified p85α and p85β were loaded onto CD28 peptide columns, and we examined the binding of each isoform to the columns. As negative controls we used columns with no-peptide, as positive controls we used PDGF receptor (R) peptide columns.42 p85 association to CD28 peptides was analyzed by SDS-PAGE and autoradiography. Both p85β and p85α bound slightly better to phosphorylated CD28 peptides, in addition, p85β bound slightly better to M1 peptides and markedly better to M2 and M2p CD28 peptides, compared with p85α (Figure 1F). We also compared p85 binding to CD28 peptides and to PDGFR peptide. p85α binding to PDGFR was within the range of p85α and p85β binding to M1p peptide; binding to M2p was one-third to one-half (human and mouse p85β, respectively) that observed using PDGFR peptide (Figure 1F, n = 3). Although in these assays both p85α and p85β bound better to M1p than to M2p columns, the scenario in vivo might be distinct; deletion of the last 10 residues of CD28 (in M2) reduces p85 binding and PI3K activity by 90%.40 The greater affinity of p85β for the M2p columns explains the greater binding of p85β to CD28 in vivo.
Lower PKB activation in p85β-deficient cells
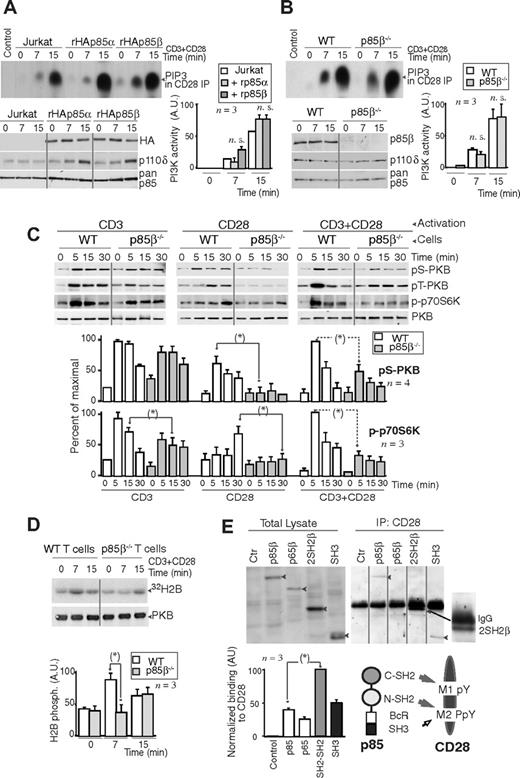
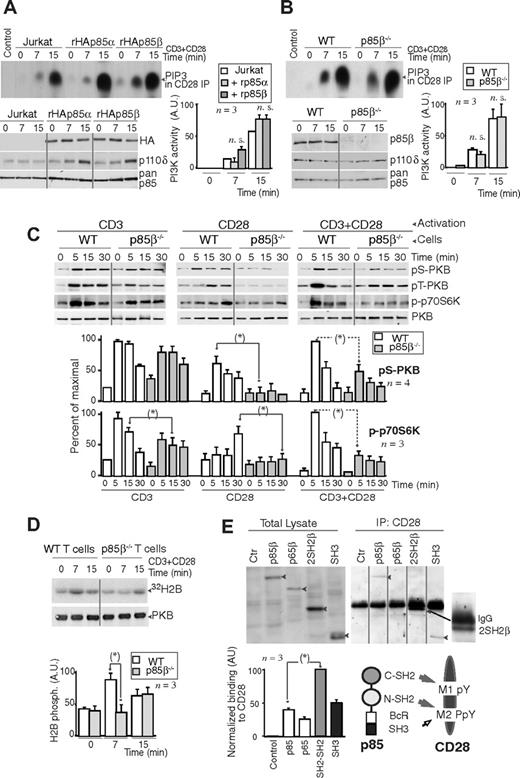
To determine whether the p85 regulatory subunits that bind to CD28 upon T-cell activation exhibit p110-associated PI3K activity, we transfected Jurkat cells with rHAp85α or rHAp85β in combination with p110δ. Cells were activated with CD3+CD28 and we measured CD28-associated PI3K activity in vitro. CD28 exhibited a similar associated PI3K activity in both cases, suggesting that CD28-bound p85α and p85β are in complex with p110 (Figure 2A). A faster PI3K activity association was observed in some cases in p85β cells, but the difference (n = 3) was not statistically significant. CD28 immunoprecipitates from CD3+CD28-activated murine WT or p85β−/− T cells also contained a similar associated PI3K activity (Figure 2B), suggesting that p110 recruitment to CD28 is observed in the presence or absence of p85β.
Lower PKB activation in p85β-deficient cells. (A,B) Jurkat cells were transfected with control, rHAp85α, or rHAp85β in combination with p110δ vectors and incubated for 36 hours (A). T cells were purified from Wild-type (WT) and p85β−/− and mice (B). Cells were activated with CD3 + CD28 antibodies (7 or 15 minutes) and collected. PI3K activity was assayed in CD28 immunoprecipitates in vitro. WB examined recombinant protein expression levels in cell extracts. The histogram represents the mean plus or minus SD (n = 3) of the PIP3 signal in AUs. (C,D) T cells were purified from p85β−/− and p85β+/− spleen and lymph node cell suspensions, then activated by CD3, CD28, or CD3+CD28 cross-linking (indicated). (C) Total cell lysates were examined in WB using anti–pSer473-PKB, anti–pThr308-PKB, anti–pThr389-p70S6K, or anti-PKB antibodies. Graphs show the percentage of p-PKB or p-p70S6K signal compared with maximal (with CD3+CD28 at 5 minutes). (D) PKB was immunoprecipitated from cell extracts and assayed in vitro using histone H2B as substrate. Graphs were as in panel A. (E) Jurkat cells were transfected with cDNA encoding the indicated p85β mutant forms, and 36 hours later cells were activated as in panel A, and total extracts or CD28 immunoprecipitates resolved by SDS-PAGE and examined by WB using anti-HA Ab. Vertical lines have been inserted to indicate a repositioned gel lane. The graph shows the mean signal plus or minus SD (AUs) of CD28-bound rp85β forms normalized for their expression levels. Scheme showing the potential interaction domains of p85 and CD28 and after activation. *P < .05.
Lower PKB activation in p85β-deficient cells. (A,B) Jurkat cells were transfected with control, rHAp85α, or rHAp85β in combination with p110δ vectors and incubated for 36 hours (A). T cells were purified from Wild-type (WT) and p85β−/− and mice (B). Cells were activated with CD3 + CD28 antibodies (7 or 15 minutes) and collected. PI3K activity was assayed in CD28 immunoprecipitates in vitro. WB examined recombinant protein expression levels in cell extracts. The histogram represents the mean plus or minus SD (n = 3) of the PIP3 signal in AUs. (C,D) T cells were purified from p85β−/− and p85β+/− spleen and lymph node cell suspensions, then activated by CD3, CD28, or CD3+CD28 cross-linking (indicated). (C) Total cell lysates were examined in WB using anti–pSer473-PKB, anti–pThr308-PKB, anti–pThr389-p70S6K, or anti-PKB antibodies. Graphs show the percentage of p-PKB or p-p70S6K signal compared with maximal (with CD3+CD28 at 5 minutes). (D) PKB was immunoprecipitated from cell extracts and assayed in vitro using histone H2B as substrate. Graphs were as in panel A. (E) Jurkat cells were transfected with cDNA encoding the indicated p85β mutant forms, and 36 hours later cells were activated as in panel A, and total extracts or CD28 immunoprecipitates resolved by SDS-PAGE and examined by WB using anti-HA Ab. Vertical lines have been inserted to indicate a repositioned gel lane. The graph shows the mean signal plus or minus SD (AUs) of CD28-bound rp85β forms normalized for their expression levels. Scheme showing the potential interaction domains of p85 and CD28 and after activation. *P < .05.
PI3K assay might underestimate quantitative differences in vivo; we thus examined the activation of the PI3K effectors PKB and p70S6K upon stimulation of WT and p85β−/− T cells via CD3, CD28, or both. p-p70S6K and p-PKB cell content was approximately one-third lower in p85β−/− T cells upon CD3 activation; in contrast, CD28 activation resulted in p-p70S6K and p-PKB levels that were approximately 60% lower in p85β−/− T cells than control cells (Figure 2C), showing that a greater signaling defect is observed upon stimulation of CD28. CD3+CD28 activation also yielded significantly lower levels of p-p70S6K and p-PKB in p85β−/− cells (Figure 2C); we confirmed the lower PKB activity in p85β−/− T cells in kinase assays (Figure 2D).
SH2 domains, and to lesser extent SH3 domains, are involved in p85 binding to CD28
The Tyr170 region (M1 peptide) is Tyr phosphorylated after T-cell activation; the C-terminal M2 region contains a phosphorylable Tyr and a Pro-rich motif. Because pTyr residues bind to SH2 domains and Pro-rich regions bind to SH3 domains, we tested the binding of p85β to these regions. Previous studies showed that the CD28 Tyr170 region binds to p85α C-SH2 domain in vitro, whereas the phosphorylated M2 region, containing Pro residues, binds both to the C-SH2 and the SH3 p85α domains.40 We examined, in extracts of CD3 + CD28-activated Jurkat cells, the binding of CD28 to p85β SH2-SH2 region, or to the SH3 region, or to a previously described p65PI3K mutant encompassing the SH3, Bcr, and the N-SH2 domains,42 in vivo. Cells were transfected with the different HA-tagged p85β constructs; 36 hours later, the cells were activated and the extracts immunoprecipitated with CD28; association to p85β constructs was determined in WB using HA Ab. When binding was normalized considering the expression levels, the binding of wild-type p85β was lower than that observed with p85 fragments, suggesting that p85 folding partially masks the interaction regions (Figure 2E). In the mutants, the greatest binding was observed with the SH2-SH2 region, whereas binding to p65PI3K mutant, which lacks the C-SH2 domain, was lower, suggesting that the C-SH2 domain (or else both SH2 domains) is required for binding to CD28 (Figure 2E). Moreover, the SH3 domain also bound CD28, showing that, although to a lesser extent, SH3 binding to Pro residues also contributes to CD28-p85β association (Figure 2E). Thus, p85β C-SH2 and SH3 domains participate in p85β binding to CD28.
c-CBL and CBL-b preferentially associate with p85β
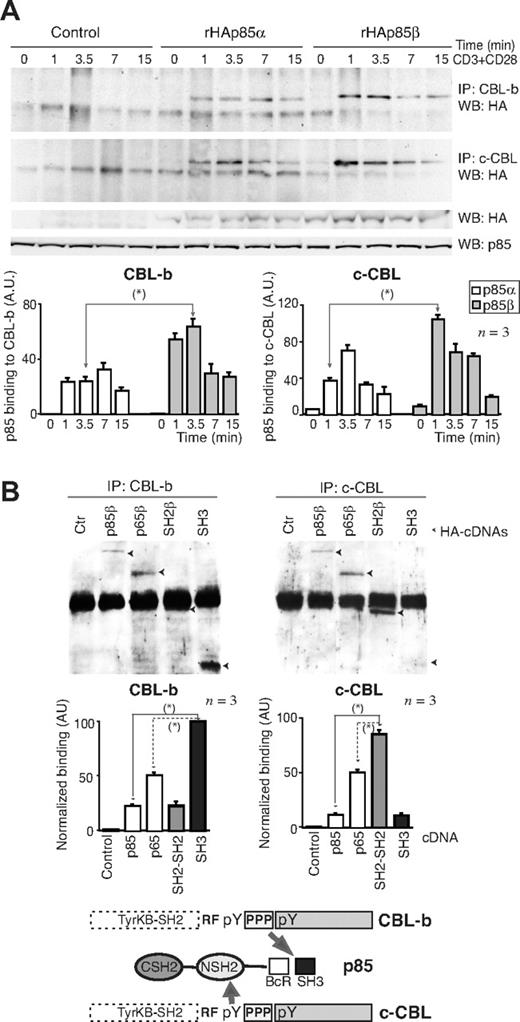
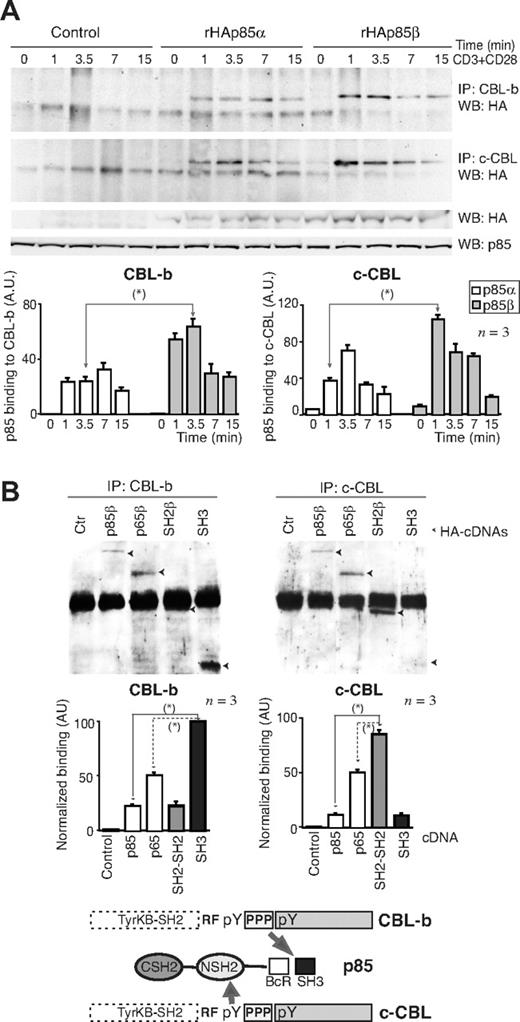
CD28 binds to p85,36-40 which in turn associates with CBL-b and c-CBL.18,44 Therefore, p85 might link CD28 and CBL. Because c-CBL associates better with p85β than p85α in vitro,44 we tested whether p85β might also associate better than p85α with CBL proteins in vivo. We measured CBL association with p85α and p85β in Jurkat T cells transfected with rHAp85α or rHAp85β. Although rHAp85α and rHAp85β were expressed to a similar extent, CBL-b and c-CBL immunoprecipitates contained more rHAp85β than rHAp85α (Figure 3A). p85β thus not only binds CD28 preferentially, but also shows greater affinity for CBL-b and c-CBL, potentially acting as a bridge between CD28 and the CBL. CBL contains Tyr-phosphorylated residues and a Pro-rich domain24 (Figure 3B bottom) that might bind SH2 and SH3 regions, respectively. To determine the p85 region involved in association with CBL-b and c-CBL, cells were transfected with the different HA-tagged p85β constructs; we immunoprecipitated the CBL and determined association to different p85β constructs in WB using HA Ab. Expression levels were as in Figure 2D. As in p85β/CD28 association, p85β fragments showed greater binding than full-length p85 to both CBL ligases (Figure 3B). In addition, whereas CBL-b bound preferentially to the SH3 domain and to the p65PI3K mutant (which includes the SH3 domain), c-CBL bound preferentially to SH2-SH2 and to p65PI3K (which includes the NSH2 domain), showing that the SH3 and NSH2 domains are likely the main regions of p85β binding to CBL-b and c-CBL, respectively.
CBL-b and c-CBL associated preferentially with p85β. (A) Jurkat T cells were transfected with empty vector, or with vectors encoding rHAp85α and β. At 36 hours after transfection, cells were collected and activated with CD3+CD28 for different times. Total extracts were immunoprecipitated with CBL-b or c-CBL antibodies, and p85 binding was analyzed in WB using anti-HA antibody. Total p85 was used as equal protein concentration control. Histograms show relative signal plus or minus SD (n = 3) of rHAp85α or rHAp85β binding to CBL-b and c-CBL. (B) Extracts from the assay shown in Figure 2D were immunoprecipitated using anti–c-CBL or anti–CBL-b Ab, resolved by SDS-PAGE, and examined by WB using anti-HA Ab. The graphs are as in Figure 2D. Scheme showing the potential interaction domains of p85β to CD28 and CBL after activation. *P < .05.
CBL-b and c-CBL associated preferentially with p85β. (A) Jurkat T cells were transfected with empty vector, or with vectors encoding rHAp85α and β. At 36 hours after transfection, cells were collected and activated with CD3+CD28 for different times. Total extracts were immunoprecipitated with CBL-b or c-CBL antibodies, and p85 binding was analyzed in WB using anti-HA antibody. Total p85 was used as equal protein concentration control. Histograms show relative signal plus or minus SD (n = 3) of rHAp85α or rHAp85β binding to CBL-b and c-CBL. (B) Extracts from the assay shown in Figure 2D were immunoprecipitated using anti–c-CBL or anti–CBL-b Ab, resolved by SDS-PAGE, and examined by WB using anti-HA Ab. The graphs are as in Figure 2D. Scheme showing the potential interaction domains of p85β to CD28 and CBL after activation. *P < .05.
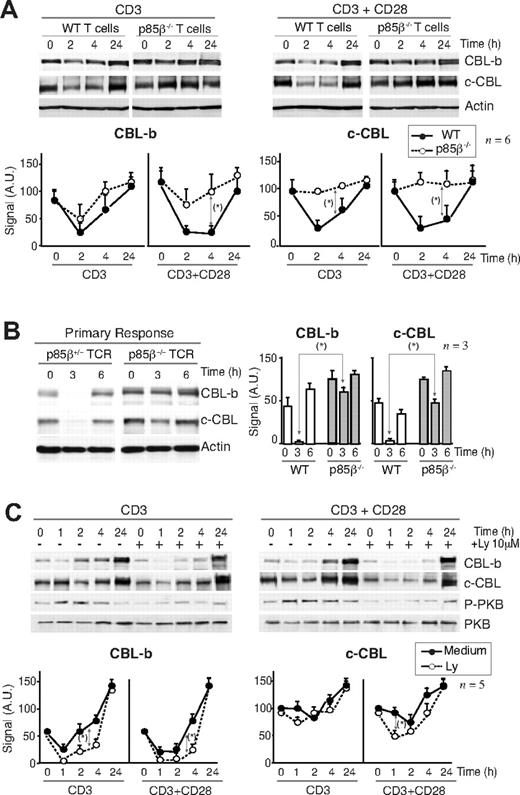
Defective CBL regulation in p85β-deficient T cells
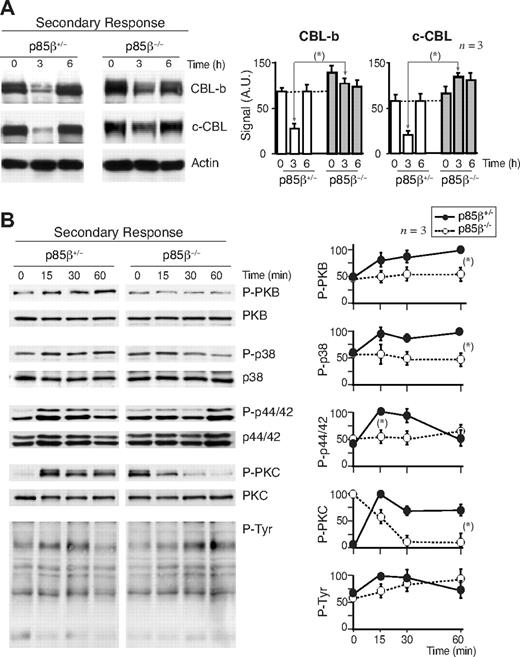
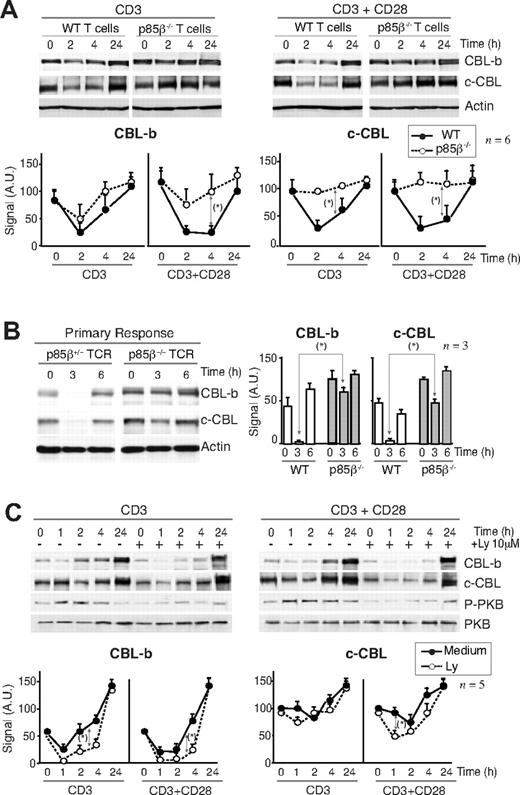
CBL limits T-cell activation; indeed, defects in CBL-b down-regulation yield T cells anergic.45-47 For this reason, one of the critical actions of CD28 is to induce CBL-b down-regulation after T-cell activation.15-20 Considering that p85β binds preferentially to CD28 and the CBL, we examined whether p85β deletion impaired CBL down-regulation. T cells from p85β+/− and p85β−/− mice were activated in vitro with anti-CD3 or with anti-CD3+anti-CD28 antibodies; in control cells, CD3 activation reduced CBL-b levels by 2 hours and reduction was more pronounced after CD3 + CD28 activation (Figure 4A). In contrast, CD3-induced CBL-b reduction was significantly less pronounced in p85β−/− T cells and was unaffected by the presence of CD28 (Figure 4A). Similarly, whereas in controls CD3 induced a moderate c-CBL down-regulation at approximately 2 hours upon stimulation that was enhanced in the presence of CD28Ab, neither CD3 nor CD3+CD28 reduced c-CBL levels in p85β−/− T cells (Figure 4A). Although we detect CBL down-regulation defects upon CD3 stimulation this might also reflect CD28 signaling defects because CD28−/− mice show CD3-induced CBL down-regulation defects,16 implying that activation via TcR also involves CD28.
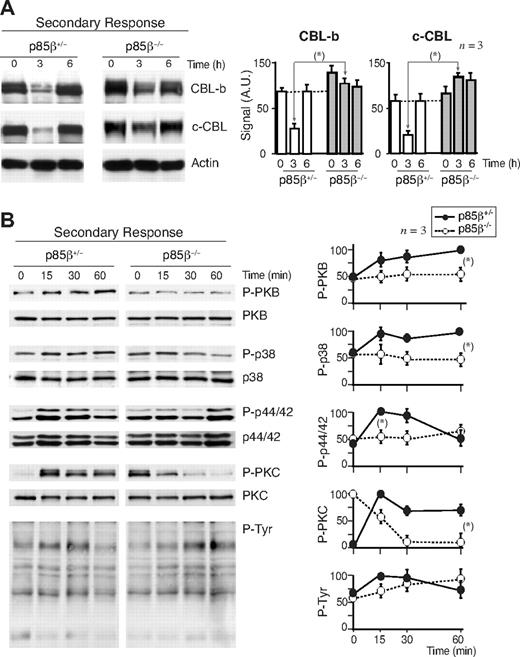
Defective CBL and TCR down-regulation in p85β-deficient cells. (A) Purified peripheral T cells from p85β+/− and p85β−/− mice were stimulated with CD3+CD28 Ab for the indicated times. CBL-b and c-CBL levels were analyzed in WB. Actin was used as loading control. (B) For primary responses, p85β+/− and p85β−/− F5TCRTg mice were injected with antigenic peptide. Mice were sacrificed at 0, 3, and 6 hours after injection and CBL levels were examined in extracts of purified T cells. (C) Jurkat T cells were activated with CD3 or CD3 + CD28 for the indicated times in the presence or absence of 10 μM Ly294002. We analyzed CBL-b and c-CBL levels in cell extracts by WB. Graphs (A-C) show CBL-b and c-CBL signal intensity in AUs, mean plus or minus SD (n = 5). *P < .05.
Defective CBL and TCR down-regulation in p85β-deficient cells. (A) Purified peripheral T cells from p85β+/− and p85β−/− mice were stimulated with CD3+CD28 Ab for the indicated times. CBL-b and c-CBL levels were analyzed in WB. Actin was used as loading control. (B) For primary responses, p85β+/− and p85β−/− F5TCRTg mice were injected with antigenic peptide. Mice were sacrificed at 0, 3, and 6 hours after injection and CBL levels were examined in extracts of purified T cells. (C) Jurkat T cells were activated with CD3 or CD3 + CD28 for the indicated times in the presence or absence of 10 μM Ly294002. We analyzed CBL-b and c-CBL levels in cell extracts by WB. Graphs (A-C) show CBL-b and c-CBL signal intensity in AUs, mean plus or minus SD (n = 5). *P < .05.
CBL down-regulation was also observed in vivo (Figure 4B). p85β+/− and p85β−/− F5TCRTg mice were injected with antigenic peptide. Antigenic peptide–loaded APCs activate T cells through TcR and CD28.5 Primary antigen stimulation in vivo induced a pronounced, transient reduction in CBL-b and c-CBL levels in control mice T cells; p85β−/− T cells, however, showed a significantly lower CBL-b and c-CBL down-regulation (Figure 4B).
Considering that PI3K/PKB pathway activation was lower in p85β−/− T cells, we examined whether PI3K activity is required to trigger CBL down-regulation. Jurkat T cells were activated in the presence or absence of the PI3K inhibitors wortmannin (not shown) or Ly294002 (Figure 4C). CD3 stimulation induced and CD3+CD28 enhanced CBL down-regulation in Jurkat cells; however, PI3K inhibition did not block but moderately enhanced CBL down-regulation, more clearly that of CBL-b (Figure 4C). Because p85β deletion impairs CBL down-regulation despite reduction of PI3K/PKB pathway activity, these observations suggest that p85β controls CBL down-regulation in a PI3K activity–independent manner.
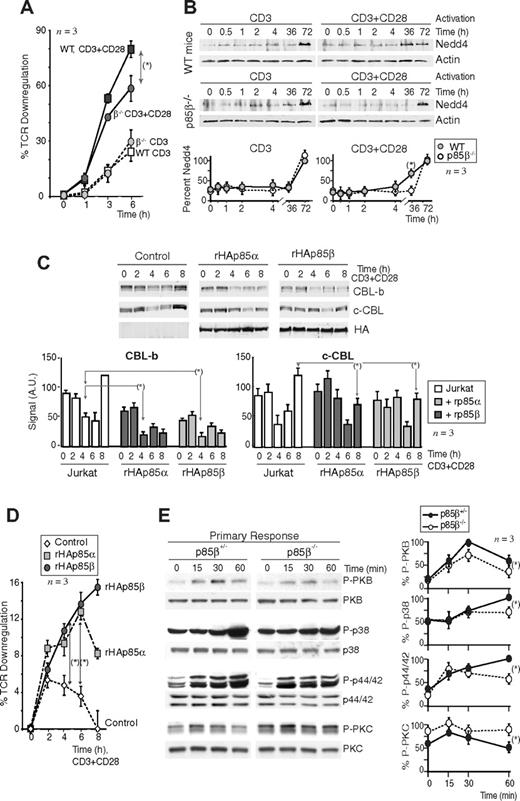
c-CBL and CBL-b regulate TCR down-regulation at late time points.25,27,48 Considering that p85β links CD28 and the CBL, it was possible that p85β-deficient T cells exhibited defects in activation-induced TCR down-regulation. Indeed, whereas CD3 cross-linking induced a similar TCR down-regulation in WT and p85β−/− purified T cells, CD3+CD28–triggered TCR down-regulation was significantly lower in p85β−/− T cells than in WT cells (Figure 5A), confirming the signaling defects of CD28 in p85β−/− T cells.
p85β overexpression enhances CBL-b and TCR down-regulation. (A) TCR down-modulation was measured by flow cytometry in purified peripheral CD8+ T cells from p85β−/− and p85β+/− F5TCRTg mice stimulated in vitro with CD3 or CD3+CD28 for the times indicated. TCR down-regulation was estimated as the reduction in the mean fluorescence index of TCRβ expression on stimulated CD8+ versus nonstimulated CD8+ T cells (mean ± SD, n = 3). (B) Cells were activated as in panel A, and Nedd4 levels were examined in WB. The graph quantitates the mean plus or minus SD (n = 3) of Nedd4 signal relative to maximal levels (100%). (C,D) Jurkat T cells were transiently transfected with empty vector or rHAp85α or β. Cells were activated as in panel A using CD3+CD28 36 hours after transfection and then collected. We analyzed CBL-b and c-CBL levels in cell extracts by WB (C) or TCR down-modulation as in panel A (D); MFI of TCRβ expression on stimulated CD4+ versus gated nonstimulated CD4+ T cells (mean ± SD, n = 4; D). Graphs (C) show CBL signal intensity in AUs (mean ± SD, n = 3). (E) Purified peripheral CD8+ T cells from p85β−/− and p85β+/− F5TCRTg mice were stimulated in vitro with peptide-pulsed APCs. Cell extracts were examined in WB (indicated). The graphs represent the mean plus or minus SD (n = 3) of the signal for each of the proteins in 3 different assays, compared with the maximal signal (100%) in WT cells at 15 minutes (pPKC), 30 minutes (pPKB), or 1 hour (p-p38, p-p44/42) and normalized in comparison with loading controls (n = 3). P values are shown for the time points of maximal signal in WT cells (*P < .05).
p85β overexpression enhances CBL-b and TCR down-regulation. (A) TCR down-modulation was measured by flow cytometry in purified peripheral CD8+ T cells from p85β−/− and p85β+/− F5TCRTg mice stimulated in vitro with CD3 or CD3+CD28 for the times indicated. TCR down-regulation was estimated as the reduction in the mean fluorescence index of TCRβ expression on stimulated CD8+ versus nonstimulated CD8+ T cells (mean ± SD, n = 3). (B) Cells were activated as in panel A, and Nedd4 levels were examined in WB. The graph quantitates the mean plus or minus SD (n = 3) of Nedd4 signal relative to maximal levels (100%). (C,D) Jurkat T cells were transiently transfected with empty vector or rHAp85α or β. Cells were activated as in panel A using CD3+CD28 36 hours after transfection and then collected. We analyzed CBL-b and c-CBL levels in cell extracts by WB (C) or TCR down-modulation as in panel A (D); MFI of TCRβ expression on stimulated CD4+ versus gated nonstimulated CD4+ T cells (mean ± SD, n = 4; D). Graphs (C) show CBL signal intensity in AUs (mean ± SD, n = 3). (E) Purified peripheral CD8+ T cells from p85β−/− and p85β+/− F5TCRTg mice were stimulated in vitro with peptide-pulsed APCs. Cell extracts were examined in WB (indicated). The graphs represent the mean plus or minus SD (n = 3) of the signal for each of the proteins in 3 different assays, compared with the maximal signal (100%) in WT cells at 15 minutes (pPKC), 30 minutes (pPKB), or 1 hour (p-p38, p-p44/42) and normalized in comparison with loading controls (n = 3). P values are shown for the time points of maximal signal in WT cells (*P < .05).
Nedd4 is not responsible for the CBL down-regulation defects of p85β−/− mice
The CBL regulation defects of p85β-deficient T cells might result from defects in Nedd4 ubiquitin ligase, which targets c-CBL and CBL-b for degradation.49 Anti-CD3 stimulation triggered Nedd4 expression at 72 hours in both p85β+/− and p85β−/− cells; moreover, CD3 + CD28 activation accelerated Nedd4 expression in control cells but not in p85β−/− cells (Figure 5B). These observations support the CD28 signaling defects in p85β−/− T cells, because whereas CD3 induced Nedd4 expression similarly in control and p85β−/− T cells, CD28 failed to enhance Nedd4 expression in p85β−/− cells. In addition, these data show that defective Nedd4 expression is not the cause of CBL down-regulation defects because CBL levels decrease at 2 to 6 hours and Nedd4 increases at approximately 36 hours after T-cell activation.
Increased expression of p85β enhances CBL and TCR down-regulation
Because p85β deletion impaired CBL and TCR down-modulation, we analyzed whether p85β overexpression might enhance these processes. We expressed rHAp85α or rHAp85β in Jurkat cells. Whereas in Jurkat cells transfected with empty vector, CD3+CD28 stimulation triggered a reduction in CBL-b and c-CBL levels at 4 to 6 hours (Figure 5C), both rHAp85α and rHAp85β expression resulted in a greater and more sustained CBL down-regulation, more markedly in the case of p85β. Moreover, in agreement with the idea that p85 links CD28 to CBL, increased p85α expression, and even more markedly that of p85β, enhanced TCR down-regulation compared with controls (Figure 5D). These observations confirm the role of p85 as a molecule regulating TCR-CD28–induced CBL down-regulation with a greater action of p85β, which might cooperate with p85α for this function.
CD28 defective signaling in primary activation of p85β−/− T cells
APC-antigenic T-cell stimulation activates T cells in the context of CD28 costimulation, which stabilizes the immunologic synapse and prolongs TCR-induced signaling cascades.5-15 Because CD28 binds preferentially to p85β, we analyzed the effect of p85β deletion on the signaling cascades triggered by APC-induced T-cell activation in vivo using phosphoantibodies to analyze TyrK, PKC, PKB, p38MAPK, and p44/42MAPK activation.
In agreement with the lower PKB activation in CD3 + CD28–stimulated p85β−/− T cells, primary antigenic stimulation in p85β−/− F5TCRTg T cells induced a reduced pPKB content compared with controls (Figure 5E). In addition, whereas at 15 to 30 minutes after stimulation, p44/42MAPK and p38MAPK activation occurred similarly in p85β−/− and p85β+/− F5TCRTg cells, activation at 60 minutes was lower in p85β−/− T cells (Figure 5E). As CD28 prologues signals induced by the TCR,5 these observations concur with the idea that CD28 signaling is defective in p85β−/− cells. A similar defect was detected in TyrK stimulation (not shown). PKC activation was greater in p85β−/− F5TCRTg T cells than in controls (Figure 5E), in agreement with the higher stability of c-CBL in these cells, as c-CBL enhances PLCγ/PKC activation.24 Thus, a higher PKC activation, a less sustained MAPK activation, and lower pPKB content were observed in primary-stimulated p85β−/− T cells.
Reduced secondary immune response in p85β-deficient mice
CD28-deficient T cells undergo an incomplete primary activation resulting in a defective differentiation of activated T cells, which become unresponsive on a secondary Ag challenge.7,8 As p85β binds to CD28 and regulates CD28 downstream signaling, we examined whether p85β deficiency impaired secondary immune responses. We first compared the primary and secondary immune responses in p85β−/− and p85β+/− mice using as antigen Candida albicans, which triggers mainly a CD4+ cell response.50,51 The primary response to C albicans was slightly higher in p85β−/− mice than in controls; in contrast, p85β−/− mice showed a lower CD4+ spleen and lymph node secondary immune response (Figure 6A).
Reduced secondary responses in p85β-deficient mice. (A) p85β−/− and p85β+/− mice received an intraperitoneal injection of heat-inactivated C albicans to induce a primary response; 21 days later, some mice received an identical injection to induce the secondary response. Splenocytes or lymph nodes (LNs) were isolated and the cells analyzed by flow cytometry using appropriate antibodies. The figure shows CD4+ cell numbers at different times after injection (mean ± SD; n = 3). P values were calculated at day 5. (B) The NP366-374 influenza peptide was injected into p85β−/− and p85β+/− F5TCRTg mice for a primary response; injection was repeated after 11 days for the secondary response. The figure shows CD8+/Vβ11+ spleen cell numbers at different times after injection (mean ± SD, n = 3).  indicates peptide injection. (C) For primary in vitro proliferation assays, peripheral T cells from p85β−/− and p85β+/− F5TCRTg mice were activated with peptide-pulsed APCs at different peptide doses, alone or in the presence of IL-2 (10 U/mL). For secondary immune response, mice received a peptide injection and were killed after 10 days, when T cells were purified from spleen and lymph nodes. T cells were then activated in vitro. [3H]thymidine incorporation was measured at 48 hours (mean ± SD, n = 3).
indicates peptide injection. (C) For primary in vitro proliferation assays, peripheral T cells from p85β−/− and p85β+/− F5TCRTg mice were activated with peptide-pulsed APCs at different peptide doses, alone or in the presence of IL-2 (10 U/mL). For secondary immune response, mice received a peptide injection and were killed after 10 days, when T cells were purified from spleen and lymph nodes. T cells were then activated in vitro. [3H]thymidine incorporation was measured at 48 hours (mean ± SD, n = 3).  indicates the compared data for P value calculation (2 ng/mL peptide). (D) p85β−/− and p85β+/− F5TCRTg mice received NP366-374 peptide at day 0 (
indicates the compared data for P value calculation (2 ng/mL peptide). (D) p85β−/− and p85β+/− F5TCRTg mice received NP366-374 peptide at day 0 ( ) to induce a primary IR; injection was repeated at day 11 for the secondary IR. Mice were killed at different times after peptide injection. T-cell activation/memory markers were analyzed in the CD8+/Vβ11+ population by flow cytometry. P values were calculated at 6 hours after secondary IR *P < .05).
) to induce a primary IR; injection was repeated at day 11 for the secondary IR. Mice were killed at different times after peptide injection. T-cell activation/memory markers were analyzed in the CD8+/Vβ11+ population by flow cytometry. P values were calculated at 6 hours after secondary IR *P < .05).
Reduced secondary responses in p85β-deficient mice. (A) p85β−/− and p85β+/− mice received an intraperitoneal injection of heat-inactivated C albicans to induce a primary response; 21 days later, some mice received an identical injection to induce the secondary response. Splenocytes or lymph nodes (LNs) were isolated and the cells analyzed by flow cytometry using appropriate antibodies. The figure shows CD4+ cell numbers at different times after injection (mean ± SD; n = 3). P values were calculated at day 5. (B) The NP366-374 influenza peptide was injected into p85β−/− and p85β+/− F5TCRTg mice for a primary response; injection was repeated after 11 days for the secondary response. The figure shows CD8+/Vβ11+ spleen cell numbers at different times after injection (mean ± SD, n = 3).  indicates peptide injection. (C) For primary in vitro proliferation assays, peripheral T cells from p85β−/− and p85β+/− F5TCRTg mice were activated with peptide-pulsed APCs at different peptide doses, alone or in the presence of IL-2 (10 U/mL). For secondary immune response, mice received a peptide injection and were killed after 10 days, when T cells were purified from spleen and lymph nodes. T cells were then activated in vitro. [3H]thymidine incorporation was measured at 48 hours (mean ± SD, n = 3).
indicates peptide injection. (C) For primary in vitro proliferation assays, peripheral T cells from p85β−/− and p85β+/− F5TCRTg mice were activated with peptide-pulsed APCs at different peptide doses, alone or in the presence of IL-2 (10 U/mL). For secondary immune response, mice received a peptide injection and were killed after 10 days, when T cells were purified from spleen and lymph nodes. T cells were then activated in vitro. [3H]thymidine incorporation was measured at 48 hours (mean ± SD, n = 3).  indicates the compared data for P value calculation (2 ng/mL peptide). (D) p85β−/− and p85β+/− F5TCRTg mice received NP366-374 peptide at day 0 (
indicates the compared data for P value calculation (2 ng/mL peptide). (D) p85β−/− and p85β+/− F5TCRTg mice received NP366-374 peptide at day 0 ( ) to induce a primary IR; injection was repeated at day 11 for the secondary IR. Mice were killed at different times after peptide injection. T-cell activation/memory markers were analyzed in the CD8+/Vβ11+ population by flow cytometry. P values were calculated at 6 hours after secondary IR *P < .05).
) to induce a primary IR; injection was repeated at day 11 for the secondary IR. Mice were killed at different times after peptide injection. T-cell activation/memory markers were analyzed in the CD8+/Vβ11+ population by flow cytometry. P values were calculated at 6 hours after secondary IR *P < .05).
We also examined primary and secondary antigen-specific immune responses. p85β+/− F5TCRTg and p85β−/− F5TCRTg mice were immunized once with the NP366-374 influenza peptide (primary) or challenged at day 0, and treated again 10 days later (secondary). The primary response was moderately enhanced in p85β−/− F5TCRTg mice but the secondary response was significantly lower than in controls (Figure 6B). IL-2 partially restores anergic cell proliferation.7-9 IL-2 addition enhanced T-cell proliferation in p85β−/− mice secondary response (Figure 6C), suggesting that primary activated p85β−/− T cells behave as anergic cells. Confirming a defective secondary response in p85β−/− mice, the number of cells expressing the activation (CD25, CD69) and memory (CD44high, CD62Llow) markers was significantly lower upon secondary challenge in p85β−/− mice (Figure 6D). These data show that p85β deficiency results in reduced CD4+ and CD8+ T-cell secondary immune responses.
CD28-mediated defects upon secondary p85β−/− T-cell stimulation
We examined CBL down-regulation and intracellular pathways in secondary responses. We immunized p85β−/− and p85β+/− F5TCRTg mice with the NP366-374 peptide, and 11 days later mice were reinjected with the peptide; we examined CBL levels in vivo, in extracts of cells collected at different times after injection. Basal levels of CBL-b and c-CBL were moderately higher in p85β−/− T cells compared with controls, as in anergic cells46 ; in addition, restimulation triggered a large CBL down-regulation in controls, and a minor reduction in p85β−/− T cells (Figure 7A). For intracellular signal analysis, mice were immunized with antigenic peptide, and 11 days later purified T cells were restimulated in vitro. PKB activation was reduced in p85β−/− T cells compared with controls (Figure 7B); because CBL-b ubiquitinates p85α and reduces PI3K activation,18,19 this concurs with the higher CBL-b levels in p85β−/− cells. TyrK activation and p44/42MAPK activation were lower and delayed in p85β−/− T cells, as in anergic cells.52 The basal activity of PKC was higher in p85β−/− T cells (Figure 7B), in agreement with the larger c-CBL levels in p85β−/− T cells, because c-CBL enhances PLCγ/PKC activity.21-24,27
CBL down-regulation and pPKB defects upon secondary stimulation of p85β−/− T cells. (A,B) We measured CBL expression levels in antigen-experienced T cells; p85β−/− and p85β+/− F5TCRTg mice were injected with the NP366-374 peptide; 11 days later both mice groups were reinjected with the NP366-374 peptide, and CBL-b or c-CBL was analyzed in WB at different times after injection (A). Alternatively, 11 days after first priming, mice were killed and purified T cells were stimulated in vitro with peptide-pulsed APCs for the times indicated. Cell extracts were examined in WB using antibodies as indicated (B). Figures are representative of at least 3 experiments with similar results. Graphs in panel A show the mean plus or minus SD (n = 3) of CBL-b or c-CBL signal. Graphs in panel B show the mean percentage plus or minus SD (n = 3) of the signal for each of the proteins, compared with the maximal signal in control cells (100%) and normalized in comparison with loading controls (n = 3). P values are shown for the times of maximal signal (*P < .05).
CBL down-regulation and pPKB defects upon secondary stimulation of p85β−/− T cells. (A,B) We measured CBL expression levels in antigen-experienced T cells; p85β−/− and p85β+/− F5TCRTg mice were injected with the NP366-374 peptide; 11 days later both mice groups were reinjected with the NP366-374 peptide, and CBL-b or c-CBL was analyzed in WB at different times after injection (A). Alternatively, 11 days after first priming, mice were killed and purified T cells were stimulated in vitro with peptide-pulsed APCs for the times indicated. Cell extracts were examined in WB using antibodies as indicated (B). Figures are representative of at least 3 experiments with similar results. Graphs in panel A show the mean plus or minus SD (n = 3) of CBL-b or c-CBL signal. Graphs in panel B show the mean percentage plus or minus SD (n = 3) of the signal for each of the proteins, compared with the maximal signal in control cells (100%) and normalized in comparison with loading controls (n = 3). P values are shown for the times of maximal signal (*P < .05).
Discussion
The presented observations show that p85β associates with CD28 and regulates CD28 signaling delivery. p85β binds to CD28, CBL-b, and c-CBL more efficiently than p85α. This function of p85β explains the reduced transmission of signals by CD28 in p85β−/− T cells, including a lower PKB activation, as well as a defective down-regulation of CBL-b and c-CBL. Because PI3K/PKB activity is required for long-term survival,30 and CBL down-regulation by CD28 is required for activated T cells to escape the anergy program,15 CBL and PKB defects likely contribute to the impaired secondary immune response in p85β−/− mice.
Effective activation of naive T cells requires a signal dependent on TCR engagement by the peptide MHC and a secondary signal provided by interaction between the antigen-presenting cell (APC) and additional receptors in the T cell. CD28 provides a potent costimulatory signal after engagement of its B7 ligands on the APC. CD28 contributes to enhancing and prolonging the signals induced by the TCR, synapse stability and long-time survival5-15 ; these events regulate differentiation of primary stimulated T cells, as activation of CD28−/− T cells induces anergy.7-9 p85, the regulatory subunit of PI3K, is one of the signaling molecules that binds to CD28.36-40 We show that the p85β isoform exhibits preferential binding to CD28 receptor compared with p85α, as examined in Jurkat cells expressing similar levels of rp85α or β, or in transformed and normal T cells analyzing endogenous p85α and p85β subunits. Greater p85β binding to CD28 was confirmed in vitro using purified p85 and CD28 peptides.
p85β binding to CD28 contributed to CD28-mediated early signals as determined examining antigenic stimulation of normal murine T cells, occurring in the context of CD28 stimulation,5-15 which resulted in lower PKB activation and less sustained MAPK activation in p85β−/− T cells. The CD28 signaling defects in p85β−/− T cells were confirmed examining activation of the PI3K effectors PKB and p70S6K after CD3 or CD28 cross-linking, which showed greater signaling defects upon CD28 activation than after CD3 activation.
T-cell activation induces phosphorylation of CD28 on Tyr170 (on the M1 peptide, Figure 1). This phosphorylated site was initially considered the main residue responsible for p85 binding.37 However, reconstitution of CD28-deficient mice with the transgenic Y170F-CD28 mutant rescued the T-cell anergy phenotype of CD28−/−mice,53 arguing against the relevance of this residue or of PI3K for anergy prevention. Nevertheless, whereas CD28 deletion reduces PI3K activity after T-cell antigenic stimulation, the Y170F-CD28 mutant did not reduce CD28-mediated PI3K activation,54 showing that PI3K binds to CD28 on a different site(s). There is a second, noncanonical CD28 binding site for p85 at the C-terminus of CD28.40 This motif is in the last 10 residues on CD28 (included in peptide M2, Figure 1). Although in vitro the C-terminal CD28 region bound a lower amount of p85 regulatory subunit than the Tyr170-containing peptide, in vivo the scenario might differ, as deletion of the C-terminal motif induced a 90% reduction on PI3K activation and a defective CD28 costimulatory function,40 suggesting that binding of PI3K to this motif is critical for CD28 function. We showed that this region exhibits a high selectivity for p85β binding, pointing at binding of p85β to CD28 as a critical event for CD28 costimulation.
p85β binding to CD28 also modulated CBL recruitment and down-regulation. We show that rHAp85β bound better than p85α to CBL-b and c-CBL. p85β contribution to CD28-induced CBL down-regulation was revealed examining antigen- or CD3+CD28-induced CBL down-regulation, which was markedly impaired in p85β−/− T cells. We show that p85β regulated not only CBL-b down-regulation, but also that of c-CBL, which was not previously reported. A less marked defective down-regulation was also observed upon CD3 stimulation, however, as CD3-induced CBL-b down-regulation also involves CD28,16 this defect might be secondary to the CD28 signaling defect. Alternatively, p85β might also affect the signaling properties of TCRζ, Lck, LAT, and TRIM, which also associate with p85 subunits.2,55,56 Nonetheless, comparison of the signaling defects of CD3- or CD28-stimulated p85β−/− T cells (Figure 2C) supports a greater contribution of p85β to CD28-induced signal transduction. The requirement of p85β for CD28-induced CBL down-regulation was not the consequence of the reduced PI3K/PKB activity in p85β−/− T cells, because PI3K activity was not required for CBL down-regulation.
We investigated the p85β regions responsible for association to CD28, c-CBL, and CBL-b. The principal regions involved in binding to CD28 were the SH2 domains, with a lower contribution of the SH3 domain. Because p65β, a mutant including the BcR, SH3, and N-SH2 domains, did not bind CD28 efficiently, either the 2 SH2 domains are required for p85β binding to CD28 or the p85β C-SH2 domain (which is not present in p65β) is critical for this interaction. Studies in vitro using p85α domains support the contribution of C-SH2 and SH3 regions for the binding of CD28 to p85.40 As the SH3 regions are less conserved between p85α and p85β (∼ 50% homology) than the SH2-SH2 domains (∼ 82%), the greater p85β association might result from the contribution of the SH3 domain.
In the case of the CBL, both c-CBL and CBL-b bound to p85β N-SH2 and SH3 regions; nonetheless, the relative contribution of these domains was distinct. Whereas c-CBL exhibited a greater binding to N-SH2 region, CBL-b bound preferentially to the SH3. Therefore, one might envision a quadruple complex involving p85β C-SH2 domain associating with CD28, the N-SH2 domain binding c-CBL, and the SH3 region bringing CBL-b into complex. More likely, as association of CD28 and CBL to p85β was not exclusively dependent on single domains, it is possible that several p85β molecules associated with the T-cell synapse (through Lck, TCRζ, LAT, TRIM, etc) establish a more complex network of interactions in which different p85β associate with CD28, c-CBL, or CBL-b. We found that isolated domains bound better to CD28, c-CBL, and CBL-b than the full-length p85, supporting the idea that the binding of p85/p110 to receptor(s) or its activation induced conformational changes facilitating subsequent interactions with p85.
p85α−/−/p85β−/− double-deficient mice developed a rare autoimmune disorder similar to Sjögren syndrome.57 T cells from these mice exhibit a dramatic reduction in PI3K activation58 because p85 molecules protect p110 from degradation.59 Considering that PI3K activity is required for regulatory T-cell differentiation,60 it is possible that defective development of this population causes the Sjögren syndrome in p85α−/−/p85β−/− mice.
Together, we conclude that the p85β regulatory subunit of class IA PI3K binds to CD28 more efficiently than to CD3, and with greater affinity than p85α, thereby regulating CD28-mediated downstream signaling. In the absence of the p85β, primary T-cell activation induces a flawed PI3K pathway activation, and an impaired c-CBL and CBL-b down-regulation, an effect that is independent of p85β-associated PI3K activity. These defects result in an impaired differentiation of activated T cells, which are then unable to develop an efficient secondary immune response. Considering the contribution of PI3K/PKB for long-term T-cell survival30 as well as the need of CBL down-regulation for CD28-mediated differentiation of activated T cells to functionally competent memory T cells,15 we propose that impaired PKB activation and CBL down-regulation in p85β−/− cells contributes to cause the defective secondary responses in these mice, implying that p85β plays an important role in CD28-mediated T-cell activation and differentiation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank A. Beloso, M. Fernandez, and L. Sanz for excellent technical assistance; Dr D. Kioussis for F5TCRTg mice; L. Williams and J. Janssen for constructs; the CNB animal facility for continuous help; L. Kremer for Ab production; B. Vanhaesebroeck for p110δ plasmid; and C. Mark for editorial assistance.
This work was supported by grants from the Ramon Areces Foundation (D.F.B., A.C.C.), the Spanish Dirección General de Ciencia y Desarrollo Tecnológico (Madrid, Spain; SAF2004-00815; SAF2008-00471; D.F.B.); (SAF2004-05955-C02-01; SAF2007-63624; A.C.C.), and NIH (Bethesda, MD; AI50831; D.A.F.). The Department of Immunology and Oncology was founded and is supported by the Spanish National Research Council (CSIC) and by Pfizer (New York, NY). I.A. held a predoctoral fellowship from the Association for International Cancer Research (St. Andrews, United Kingdom), and holds a research support contract from the Madrid regional government (CAM, Madrid, Spain). I.C. holds a predoctoral fellowship from the Spanish Ministry of Education and Science FPU program. D.F.B. held a Ramón y Cajal contract from the Spanish Ministry of Education and Science.
National Institutes of Health
Authorship
Contribution: I.A., D.F.B., A.Z., and I.C. performed experiments; I.A., D.F.B., and A.C.C. analyzed the results and made the figures; D.A.F. contributed vital reagents, participated in discussion of the results, and reviewed the paper; C.H. analyzed animal phenotypes; D.F.B. and A.C.C. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ana C. Carrera, Department of Immunology and Oncology, Centro Nacional de Biotecnología/CSIC, Darwin 3, Campus de Cantoblanco, Madrid E-28049, Spain; e-mail: acarrera@cnb.csic.es.
References
Author notes
*I.A. and D.F.B. contributed equally to the experiments done in this study.

![Figure 1. CD28 preferentially associates with p85β. (A) Ratio of p85α/p85β molecules per nanogram of total mRNA determined by QRT-PCR. (B) Jurkat cells transfected with control, rHAp85α, or rHAp85β vectors (36 hours), were activated via CD3+CD28 (15 minutes). WB examined protein expression in total extracts, extracts were also immunoprecipitated with anti-CD28 Ab, and associated p85 was analyzed in Western blot (WB) using anti-HA Ab (long and short exposure). (C) Number of p85α or p85β molecules per nanogram of total mRNA (from p85β−/− and p85β+/+ T cells) by QRT-PCR; each dot represents a mouse. Western blot analysis of p85 in extracts of a representative mouse (vertical line indicate a repositioned gel lane). (D) T cells from p85β−/− and p85β+/+ mice and Jurkat cells were activated as in panel A. Total extracts or CD28 immunoprecipitates were resolved in 2-dimensional electrophoresis and analyzed in WB using anti–pan-p85 antibody. β indicates the p85β spot (absent in p85β−/− mice). The isoelectric point (pI) of mp85β was 5.74, and the pIs were 5.96, 5.9, and 5.8 for mp85α and its phosphorylated forms; the pI for hp85β was 6.03, and the pIs were 5.84, 5.77, and 5.71 for hp85α and its phosphorylated forms. Graphs show the mean plus or minus SD (n = 3) of the β spot signal relative to the combined signal for all spots (100%). (E) Jurkat cells were activated as in panel B; WB shows the amount of p85 present in CD3 or CD28 immunoprecipitates. Graphs show the mean signal plus or minus SD (AU indicates arbitrary unit) of p85 bound to CD3 or CD28. (F) cDNA encoding human (h) or mouse rp85α or rp85β was transcribed/translated in vitro (rhp85α was fused to GFP) in the presence of 35S-Met. Purified proteins were examined by SDS-PAGE and autoradiography (top left). Sequence of the hCD28 peptides; the dots represent the nonconserved amino acids in mCD28 (H > P and P > A). In the bottom, p85 bound to the different CD28 peptide columns analyzed by SDS-PAGE and autoradiography. PDGFR (P) peptide was used as a positive control; column with no peptide and mp85α was used as the negative control. The histograms show the absolute binding of each rp85 form to CD28 motifs (mean signal ± SD in arbitrary units [AU]) on the right, and rp85 relative binding to M peptides and to P column (n = 3). *t test, P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/14/10.1182_blood-2008-04-152942/4/m_zh80170934080001.jpeg?Expires=1769314099&Signature=lsjLS5G9HMU1ePexuv~z0gwI8twGcPojPxJE-1zi0UQ2bBJw4dWeJ1lPwkIUu9VOCBks2xB5kgr1ykJxf~yQkLzKCFkQbFBG0KDvSfV6VeOscZOWhHI3QUFg2RBUZ8MkB9nbDxNjwWRYbHAHNwfucYxmKa3jfWSxnbvC9wpV~HRoRmSqbGqa~TTSry~65PwAes~ac90joH-QE8pF0pwGJNMCB-9Y3xshl~PDTwKiJ3pb4LGN3OAnsWk2M9pME6qg3NZbRoqhuOYiGOBnhItgz3EeR3S45RHST0DhfinFSBWMax3Qp8g2RJcXC54psHecGqZN3~t9kH7Eh6o5Fm0IcQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Reduced secondary responses in p85β-deficient mice. (A) p85β−/− and p85β+/− mice received an intraperitoneal injection of heat-inactivated C albicans to induce a primary response; 21 days later, some mice received an identical injection to induce the secondary response. Splenocytes or lymph nodes (LNs) were isolated and the cells analyzed by flow cytometry using appropriate antibodies. The figure shows CD4+ cell numbers at different times after injection (mean ± SD; n = 3). P values were calculated at day 5. (B) The NP366-374 influenza peptide was injected into p85β−/− and p85β+/− F5TCRTg mice for a primary response; injection was repeated after 11 days for the secondary response. The figure shows CD8+/Vβ11+ spleen cell numbers at different times after injection (mean ± SD, n = 3). indicates peptide injection. (C) For primary in vitro proliferation assays, peripheral T cells from p85β−/− and p85β+/− F5TCRTg mice were activated with peptide-pulsed APCs at different peptide doses, alone or in the presence of IL-2 (10 U/mL). For secondary immune response, mice received a peptide injection and were killed after 10 days, when T cells were purified from spleen and lymph nodes. T cells were then activated in vitro. [3H]thymidine incorporation was measured at 48 hours (mean ± SD, n = 3). indicates the compared data for P value calculation (2 ng/mL peptide). (D) p85β−/− and p85β+/− F5TCRTg mice received NP366-374 peptide at day 0 () to induce a primary IR; injection was repeated at day 11 for the secondary IR. Mice were killed at different times after peptide injection. T-cell activation/memory markers were analyzed in the CD8+/Vβ11+ population by flow cytometry. P values were calculated at 6 hours after secondary IR *P < .05).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/14/10.1182_blood-2008-04-152942/4/m_zh80170934080006.jpeg?Expires=1769314099&Signature=rnbe9~wtEbKOaGc0PvcvmLX9kVWx5wtgLIVwzEk-TTxoecv9oMt~Zh1e7aDzWUAmB-rIM543zfVnqGaMmF27rJuQdcPxu5cQFmbGOS-IsW~kDPuoy3~Msw5LRlgGYolglS54xdFmptQxb0FiXKih~ycglZXLhR8RQFZLsuUaqSQbgo8r0oLZjSc1nFYQ~aCWID7~aRmq45MT-s8Owb3trrmxl37-PCRa~0Ycr2ZD1JqCzjB0-XFBw5HlZgiA7pRVoyUrcFYK5hsSBv6k98jQbq8fHdMYGXttZk9im0WbCc-CRp5fQ6LYNoPCsNF4DbSGcZX5OoWeIAGqhnjR5BcL8Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 1. CD28 preferentially associates with p85β. (A) Ratio of p85α/p85β molecules per nanogram of total mRNA determined by QRT-PCR. (B) Jurkat cells transfected with control, rHAp85α, or rHAp85β vectors (36 hours), were activated via CD3+CD28 (15 minutes). WB examined protein expression in total extracts, extracts were also immunoprecipitated with anti-CD28 Ab, and associated p85 was analyzed in Western blot (WB) using anti-HA Ab (long and short exposure). (C) Number of p85α or p85β molecules per nanogram of total mRNA (from p85β−/− and p85β+/+ T cells) by QRT-PCR; each dot represents a mouse. Western blot analysis of p85 in extracts of a representative mouse (vertical line indicate a repositioned gel lane). (D) T cells from p85β−/− and p85β+/+ mice and Jurkat cells were activated as in panel A. Total extracts or CD28 immunoprecipitates were resolved in 2-dimensional electrophoresis and analyzed in WB using anti–pan-p85 antibody. β indicates the p85β spot (absent in p85β−/− mice). The isoelectric point (pI) of mp85β was 5.74, and the pIs were 5.96, 5.9, and 5.8 for mp85α and its phosphorylated forms; the pI for hp85β was 6.03, and the pIs were 5.84, 5.77, and 5.71 for hp85α and its phosphorylated forms. Graphs show the mean plus or minus SD (n = 3) of the β spot signal relative to the combined signal for all spots (100%). (E) Jurkat cells were activated as in panel B; WB shows the amount of p85 present in CD3 or CD28 immunoprecipitates. Graphs show the mean signal plus or minus SD (AU indicates arbitrary unit) of p85 bound to CD3 or CD28. (F) cDNA encoding human (h) or mouse rp85α or rp85β was transcribed/translated in vitro (rhp85α was fused to GFP) in the presence of 35S-Met. Purified proteins were examined by SDS-PAGE and autoradiography (top left). Sequence of the hCD28 peptides; the dots represent the nonconserved amino acids in mCD28 (H > P and P > A). In the bottom, p85 bound to the different CD28 peptide columns analyzed by SDS-PAGE and autoradiography. PDGFR (P) peptide was used as a positive control; column with no peptide and mp85α was used as the negative control. The histograms show the absolute binding of each rp85 form to CD28 motifs (mean signal ± SD in arbitrary units [AU]) on the right, and rp85 relative binding to M peptides and to P column (n = 3). *t test, P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/14/10.1182_blood-2008-04-152942/4/m_zh80170934080001.jpeg?Expires=1769565436&Signature=J0pcpLSLMYazaZw6SzfIL1iglO0TysIArrVHaA4Ennm47J13WOgvap2Eum0ILvCSmrFWS6lCamHjY~I0UDUd6VFT8lV8eRuTOBiwRb-QoAbO9EVcH3pVPn-UoeAXmsgIWG3wjztnycvOsJnKfqKyrV-rTgycbQqzvZ3EOSc5abpgxiTFC1KkGdjUKoCSsWqNKirFn31UBSw2k8wkfKgh12Rh~DUaDG55dfve1EfanTeiMmsrhywiT6paVkexqMPw-UqaqdbJrNVwI3vRjU92qIJ5mco-qSXJ83BYyhdVzhYpPE8llqW1V~kGOg-LtV9r~-P03YJ2Ww8kveoD0Jg3Nw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Reduced secondary responses in p85β-deficient mice. (A) p85β−/− and p85β+/− mice received an intraperitoneal injection of heat-inactivated C albicans to induce a primary response; 21 days later, some mice received an identical injection to induce the secondary response. Splenocytes or lymph nodes (LNs) were isolated and the cells analyzed by flow cytometry using appropriate antibodies. The figure shows CD4+ cell numbers at different times after injection (mean ± SD; n = 3). P values were calculated at day 5. (B) The NP366-374 influenza peptide was injected into p85β−/− and p85β+/− F5TCRTg mice for a primary response; injection was repeated after 11 days for the secondary response. The figure shows CD8+/Vβ11+ spleen cell numbers at different times after injection (mean ± SD, n = 3). indicates peptide injection. (C) For primary in vitro proliferation assays, peripheral T cells from p85β−/− and p85β+/− F5TCRTg mice were activated with peptide-pulsed APCs at different peptide doses, alone or in the presence of IL-2 (10 U/mL). For secondary immune response, mice received a peptide injection and were killed after 10 days, when T cells were purified from spleen and lymph nodes. T cells were then activated in vitro. [3H]thymidine incorporation was measured at 48 hours (mean ± SD, n = 3). indicates the compared data for P value calculation (2 ng/mL peptide). (D) p85β−/− and p85β+/− F5TCRTg mice received NP366-374 peptide at day 0 () to induce a primary IR; injection was repeated at day 11 for the secondary IR. Mice were killed at different times after peptide injection. T-cell activation/memory markers were analyzed in the CD8+/Vβ11+ population by flow cytometry. P values were calculated at 6 hours after secondary IR *P < .05).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/14/10.1182_blood-2008-04-152942/4/m_zh80170934080006.jpeg?Expires=1769565436&Signature=DvPZEFkCtpDf9ROPFB8odRU~L6BxJhp5EE5vED41XE3AUPAkSK0KQ1yX5~Jus5YEZaMJvfVENuFYOtAtVnz~lTF1cUINluJIV4IcygypfOPcxg66WfjcK07uzZzSVsgGso3js8Z3fdzJiQ8Bg0uhJ0X3nIXBUkK31gTEw3nIRI1NItF64HHjRA8YuIAoy7nU2mM701ojbscQP0EZGZmO0ykmnDS9BCsjdA9UIeiuUlqMt4bx50c~oYO47PajVXd~I3EyLLghV3~MdohoWCEJH7fWqrl7n20KpC9BCydKHm~oPWhXbRhA-usfFdoL3s63Yv3WKqhVS4ktu8ARstHNeg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
 indicates peptide injection. (C) For primary in vitro proliferation assays, peripheral T cells from p85β−/− and p85β+/− F5TCRTg mice were activated with peptide-pulsed APCs at different peptide doses, alone or in the presence of IL-2 (10 U/mL). For secondary immune response, mice received a peptide injection and were killed after 10 days, when T cells were purified from spleen and lymph nodes. T cells were then activated in vitro. [3H]thymidine incorporation was measured at 48 hours (mean ± SD, n = 3).
indicates peptide injection. (C) For primary in vitro proliferation assays, peripheral T cells from p85β−/− and p85β+/− F5TCRTg mice were activated with peptide-pulsed APCs at different peptide doses, alone or in the presence of IL-2 (10 U/mL). For secondary immune response, mice received a peptide injection and were killed after 10 days, when T cells were purified from spleen and lymph nodes. T cells were then activated in vitro. [3H]thymidine incorporation was measured at 48 hours (mean ± SD, n = 3).  indicates the compared data for P value calculation (2 ng/mL peptide). (D) p85β−/− and p85β+/− F5TCRTg mice received NP366-374 peptide at day 0 (
indicates the compared data for P value calculation (2 ng/mL peptide). (D) p85β−/− and p85β+/− F5TCRTg mice received NP366-374 peptide at day 0 (