Abstract
Differentiating erythroid cells execute a unique gene expression program that insures synthesis of the appropriate proteome at each stage of maturation. Standard expression microarrays provide important insight into erythroid gene expression but cannot detect qualitative changes in transcript structure, mediated by RNA processing, that alter structure and function of encoded proteins. We analyzed stage-specific changes in the late erythroid transcriptome via use of high-resolution microarrays that detect altered expression of individual exons. Ten differentiation-associated changes in erythroblast splicing patterns were identified, including the previously known activation of protein 4.1R exon 16 splicing. Six new alternative splicing switches involving enhanced inclusion of internal cassette exons were discovered, as well as 3 changes in use of alternative first exons. All of these erythroid stage-specific splicing events represent activated inclusion of authentic annotated exons, suggesting they represent an active regulatory process rather than a general loss of splicing fidelity. The observation that 3 of the regulated transcripts encode RNA binding proteins (SNRP70, HNRPLL, MBNL2) may indicate significant changes in the RNA processing machinery of late erythroblasts. Together, these results support the existence of a regulated alternative pre-mRNA splicing program that is critical for late erythroid differentiation.
Introduction
Differentiation of erythroid progenitors into mature red cells requires a carefully orchestrated gene expression program to insure synthesis of the appropriate stage-specific proteome as the cells become progressively more specialized. Previous studies of erythroid gene expression have focused predominantly on quantitative or semiquantitative assays, including Western and Northern blotting, proteomic analysis, cDNA cloning, and microarray analysis. Together, these studies have provided considerable insights into the late erythroid gene expression program.
However, gene-level expression analysis cannot identify important qualitative changes in erythroid gene expression predicted to result from alternative pre-mRNA processing pathways. Exclusion or inclusion of individual exons can substantially alter the structure and function of the encoded protein isoforms independent of changes in transcript expression levels. Given that the majority of genes in the human genome exhibit alternative splicing1,2 and that alternative splicing pathways may be regulated during differentiation and development, it is probable that stage-specific alternative splicing “switches” play an important role in modulating protein function during erythroid differentiation. Such changes in erythroid transcript structure may be regulated at the transcriptional level in the case of alternative first exons3 or at the pre-mRNA splicing level in the case of internal exons.4 To further understand the erythroid gene expression program, the erythroid transcriptome will need to be explored at different stages of erythropoiesis and at the resolution of individual exons.
The best-studied example of regulated pre-mRNA splicing in erythroid cells is the stage-specific splicing switch of protein 4.1R exon 16. Alternative exon 16 is tightly regulated such that it is excluded in early erythroid progenitor cells but efficiently included in late erythroblasts.5,6 This splicing switch is functionally important: exon 16 inclusion leads to synthesis of 4.1R protein isoforms with high affinity for spectrin and actin, and increased ability to mechanically stabilize the erythroid membrane.7-9 Mechanistically, the splicing switch is regulated at least in part by changes in expression of antagonistic splicing factors, in particular, a decrease in expression of the splicing inhibitory factor hnRNP A1 relative to that of stimulatory factors Fox-2 and SF2/ASF.10-13 It seems reasonable to propose that these changes in splicing factor activity would regulate not only protein 4.1R pre-mRNA splicing but also a subset of other alternative splicing events that together may constitute an erythroid alternative splicing program.
To explore the hypothesis that alternative splicing switches in other erythroid transcripts are executed during late erythropoiesis, we have undertaken a genome-wide expression analysis of erythroblast transcripts at the level of individual exons. We used Affymetrix exon microarrays (Santa Clara, CA)1,14,15 to identify erythroid stage-specific changes in exon expression, using RNA isolated from basophilic versus orthochromatic erythroblasts differentiated in vitro from human CD34+ erythroid progenitors.16-18 Candidate alternative splicing events identified by microarray analysis were further validated by reverse-transcription polymerase chain reaction (RT-PCR). Together, these experiments revealed several internal alternative exons that exhibit significant changes in splicing efficiency, as well as differential expression of several alternative first exons that suggest changes in transcriptional promoter usage. These results suggest that an erythroid splicing program mediates stage-specific changes in transcript (and ultimately protein) structure and function that are probably critical for proper erythropoiesis.
Methods
Cell isolation and culture
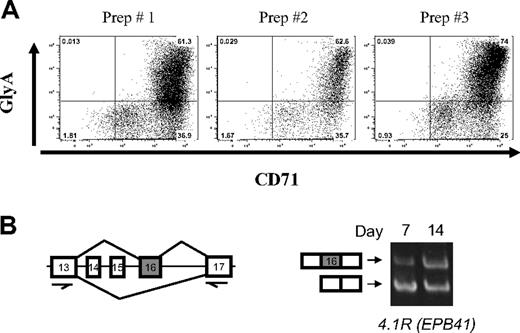
An in vitro primary culture system was used to generate cells at different stages of human erythroid differentiation.16 The culture system used was CD34+ early hematopoietic progenitors isolated from growth factor mobilized peripheral blood (purchased from ALL Cells, Berkeley, CA) to promote the erythroid differentiation program. The culture media contained 15% fetal calf serum, 15% human AB serum, Iscove modified Dulbecco medium, 10 ng/mL interleukin-3, 2 units mL erythropoietin, and 50 ng/mL stem cell factor. Stem cell factor was gradually decreased, and interleukin-3 was not added after day 3. Cells were collected at days 7, 10, and 14, which correspond, respectively, to basophilic (early), polychromatic (intermediate), and orthochromatic (late) erythroblasts. A total of 97% to 99% of these cells were erythroid as determined by flow cytometry for glycophorin A and CD71 (Figure 1A).
Stage-specific switch of protein 4.1R exon 16 splicing in highly purified erythroblast cultures. (A) Fluorescence-activated cell sorter analysis of day 7 erythroblasts from 3 different preparations indicates that purity is more than or equal to 97%, based on expression of erythroid markers for glycophorin A and CD71. Quantitative analysis demonstrated erythroblast purities as follows: prep 1, 97%; prep 2, 97%; prep 3, 99%. (B) RT-PCR scheme used to analyze 4.1R pre-mRNA splicing in early (day 7) and late erythroblasts (day 14), using primers in the nearest constitutive exons 13 and 17. Gel image shows primarily exclusion of exon 16 in early erythroblasts (bottom band), whereas substantial inclusion of exon 16 was observed in late erythroblasts (top band). Alternative exons 14 and 15 are not expressed in erythroid cells.
Stage-specific switch of protein 4.1R exon 16 splicing in highly purified erythroblast cultures. (A) Fluorescence-activated cell sorter analysis of day 7 erythroblasts from 3 different preparations indicates that purity is more than or equal to 97%, based on expression of erythroid markers for glycophorin A and CD71. Quantitative analysis demonstrated erythroblast purities as follows: prep 1, 97%; prep 2, 97%; prep 3, 99%. (B) RT-PCR scheme used to analyze 4.1R pre-mRNA splicing in early (day 7) and late erythroblasts (day 14), using primers in the nearest constitutive exons 13 and 17. Gel image shows primarily exclusion of exon 16 in early erythroblasts (bottom band), whereas substantial inclusion of exon 16 was observed in late erythroblasts (top band). Alternative exons 14 and 15 are not expressed in erythroid cells.
Total RNA extraction and sample preparation
Total cellular RNA was extracted from each culture at early (day 7), intermediate (day 10), and late (day 14) stages of differentiation. α- and β-globin mRNAs were depleted from the RNA samples using GLOBINclear Kit (Ambion, Austin, TX); 1 μg of total globin-depleted RNA was used to subtract ribosomal RNA using the Ribominus Human/Mouse Transcriptome Isolation kit (Invitrogen, Carlsbad, CA). Globin- and ribosomal RNA–depleted RNA was amplified and labeled for the exon array hybridization according to the Affymetrix WT Sense Target Labeling Assay Manual (http://affymetrix.com/products/arrays/specific/hugene_1_0_st/hugene_reagent_instr_solution.affx). The GeneChip Hybridization Oven 320 and the GeneChip Fluidics Station 450 were used for hybridization and washing, respectively (Affymetrix, Santa Clara, CA). The GeneChip Scanner 3000 was used for scanning.
Exon array analysis
To identify candidate alternative splicing switches in differentiating erythroid cells, we assessed genome-wide changes in exon expression using 2 platforms: the Affymetrix Human Exon 1.0 ST Array1 and a noncommercial exon junction array (described later in this section). The commercial Human Exon 1.0 ST array was designed to be as inclusive as possible; it contains probes for virtually all known and predicted exons in the human genome and has the potential to discover novel splicing events.1 In contrast, the junction array focuses on a smaller set of well-annotated exons and includes probes not only to each exon, but also to exon-exon junctions. This latter feature provides additional information that aids in predicting splicing differences between RNA samples because a change in alternative splicing should yield reciprocal changes in intensity of junction probes for the exon-skipping and exon inclusion event, respectively.
For the Human Exon 1.0 ST array, comparison of exon expression patterns in early versus late erythroblasts was performed using XRAY software, version 2.5 Excel Add-In (Biotique Systems, Reno, NV). The 8 input CEL files (3 replicates of day 7 samples, 2 replicates of day 10 samples, and 3 replicates of day 14 samples) were analyzed to identify genes that were significantly differentially expressed or displayed significant differential alternative splicing between the groups of interest. Replicate 1 of day 10 was removed because of poor data quality.
For the tissue-specific exon expression analysis of the beta spectrin gene, we compared 3 replicates of erythroid day 14 to 3 replicates of the muscle tissue Exon Array data.1 The splicing index1,15 was calculated to create figures for the candidate genes showing alternative exon usage in the array data.
The exon junction array contains probe sets that were designed to interrogate both exons and the observed exon-exon junctions for approximately 35 000 genes in the human genome. The design uses transcript annotations from RefSeq, Ensembl, and ExonWalk (available at http://genome.ucsc.edu). Approximately 250 000 exon clusters are represented. Additional details on the junction array design are included in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Labeled target generated from 5 biologic replicates of total RNA extracted from day 7 and day 14 was hybridized to the junction array. Target was prepared using the same method as the Human Exon 1.0 ST arrays. One replicate of the day 14 samples was deemed an outlier and removed from subsequent analyses. Data from the junction arrays were processed using the Affymetrix Power Tools. Candidate alternative splicing events were established using the splicing index approach.1 Each exon or junction probe set was analyzed independently. We then selected splicing events for which one informative probe set showed a significant change in use between the day 7 and day 14 samples. In particular, we selected cassette exons that exhibited reciprocal behavior of the probe set specific to the skip junction and one or more of the probe sets monitoring inclusion of the exon. Additional details regarding analysis of the junction array data are included in Document S1.
RT-PCR
Validation of splicing changes predicted by microarray experiments was performed by RT-PCR analysis of the same RNA samples used for the array hybridizations. Total RNA was DNase-treated, and 1 μg was used to synthesize first-strand cDNA with SuperScript III (Invitrogen) and random hexamer primers in a total volume of 20 μL; 1 μL of the cDNA was used for standard 20 μL total volume PCR. A total of 25 to 35 cycles of amplification were performed under the following conditions: denaturation for 30 seconds at 94°C, annealing for 30 seconds at 55°C, and extension for 30 seconds at 72°C. Primer sequences are shown in Table S1. PCR products were analyzed on 5% polyacrylamide gels. Densitometry was carried out to compare bands using Alpha Imager 2200, version 5.5 (Alpha Innotech, San Leandro, CA). At least 2 replicates were used to validate the array predictions by RT-PCR, and the identity of all PCR fragments was confirmed by DNA sequence analysis. It is worth noting that alternative splicing events supported by reciprocal changes in probe sets for both exon inclusion versus exclusion events had a higher validation rate than thosepredicted by only one or the other. The microarray data have been deposited with a GEO accession number of GSE14588 (http://www.ncbi.nlm.nih.gov/geo).
Results
Analysis of differentiation stage-specific alternative splicing switches in human erythroblasts
Human CD34+ primary erythroid progenitors from growth factor-mobilized peripheral blood can differentiate in culture and undergo many of the programmed changes in gene expression that are characteristic of late erythropoiesis.16-18 To investigate changes in alternative splicing that occur specifically in differentiating erythroblasts, we analyzed cultures highly enriched (≥ 97%) for erythroid cells as determined by fluorescence-activated cell sorter analysis using CD71 and glycophorin A antibodies (Figure 1A). RNA from cells cultured for 7 days (basophilic erythroblasts) and 14 days (orthochromatic erythroblasts) was then examined by exon microarray analysis and by RT-PCR to identify changes in gene expression at the level of alternative pre-mRNA splicing.
A key alternative splicing event during late erythropoiesis involves protein 4.1R exon 16, which is excluded in early erythroblasts but included efficiently in late erythroblasts5,8,10 (Figure 1A). Consistent with RNA studies, isoforms of 4.1R protein that include the exon 16-encoded peptide are increased in these cells.5,6 To confirm that the exon 16 alternative splicing switch occurs during differentiation of these primary erythroid cultures, we used RT-PCR to assay the abundance of transcripts that either include or exclude exon 16. Figure 1B shows that exon 16 was included much more efficiently in late erythroblasts (day 14) than in early erythroblasts (day 7). This result validates the use of these cultures for analysis of stage-specific splicing changes in late erythropoiesis.
Exon microarray analysis of changes in exon expression during late erythroid differentiation
To identify new cases of alternative splicing in differentiating erythroid cells, we assessed genome-wide changes in exon expression using the Affymetrix Human Exon 1.0 ST Array. This microarray was designed to be as inclusive as possible, containing probes designed to detect expression of virtually all known and predicted exons in the human genome.1 We hybridized these arrays with probes prepared from erythroblast RNA at 3 stages of differentiation (basophilic, polychromatic, and orthochromatic erythroblasts) from 3 independent cultures. Array data from these 3 groups were then analyzed using XRAY software to look for differentiation-associated changes in expression, both at the level of whole transcripts and at the level of individual exons. For most comparisons, we focused on basophilic versus orthochromatic samples to maximize potential differences in gene expression.
First, gene-level changes in expression were deduced by combining the hybridization data for all informative probes from each annotated transcript. Previous studies have shown that differentiation in this erythroid culture system is characterized by increased expression of known red cell proteins, including band 3, ankyrin, and protein 4.1, whose mRNAs begins to accumulate around day 8.17 As expected, exon expression analysis confirmed the up-regulation of these transcripts, as well as others encoding enzymes involved in heme biosynthesis (eg, ALAS2) and proteins associated with the membrane skeletal network (eg, EPB49, EPB42, AQP1, GYPB). Conversely, many other transcripts exhibit a significant decrease in expression in late erythroblasts. For example, the transcript encoding HNRPA1, a negative regulator of 4.1 exon 16 splicing known to be down-regulated in late erythroblasts,10 was lower at late time points (eg, day 14) compared with the early time point (day 7). Tables S2 and S3 list the transcripts that show greater than 2-fold increase or decrease in expression in early versus late erythroid progenitor cells.
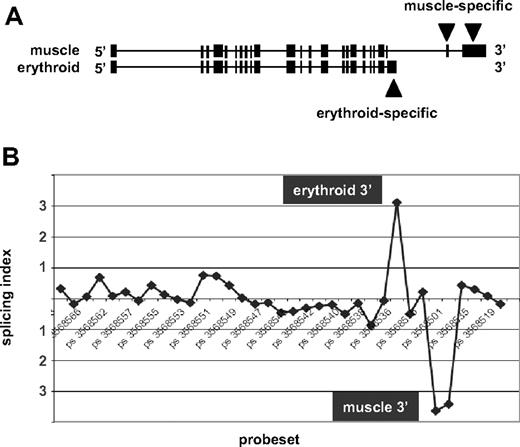
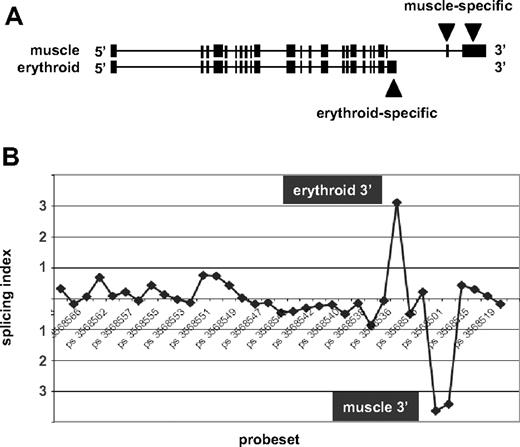
Next, to determine whether the exon array data could detect known alternative splicing events, we considered the case of beta spectrin (SPTB). Beta spectrin mRNAs are known to possess distinct 3′ terminal exons in eythroid cells and in muscle cells resulting from alternative splicing and alternative polyadenylation19 (Figure 2A). Comparison of the new erythroid exon expression data to publicly available Exon Array data for human skeletal muscle1 was performed using XRAY analysis software to identify predicted splicing differences. As expected, β-spectrin ranked near the top of the list of genes predicted to splice differently in muscle versus erythroid cells. To visualize these differences, the array data were used to calculate the splicing index, a measure for detection of alternative splicing differences by identifying probe sets that exhibit differential expression between RNA samples after normalizing for transcript levels1 (Document S1). The splicing index is shown in Figure 2B. Probe sets representing the 5′ and central regions of the gene exhibited no significant differences in relative expression in erythroblasts versus muscle, as indicated by consistent values for the splicing index close to zero. In contrast, substantial differences were observed at the 3′ end. The upward peak labeled “erythroid” indicates enriched expression of probe sets corresponding to the known erythroid-specific 3′ end, whereas the downward peak “muscle” represents probe sets that map to the muscle-specific 3′ exons and are underexpressed in erythroid RNA relative to their higher expression in muscle. This finding confirms exon array detection of a well-known erythroid-specific gene expression event.
Exon array detection of erythroid-specific beta spectrin mRNA 3′ end. (A) The exon structure of human beta spectrin transcripts in muscle versus erythroid cells, which express distinct 3′ terminal exons. (B) The splicing index of probe sets across the full length of the beta spectrin gene. Numbers along the horizontal axis represent probe set IDs. Positive values for the splicing index represent higher relative probe set expression in erythroblasts, whereas negative values indicate higher relative expression in muscle. The significant upward peak maps to the known erythroid-specific 3′ end, whereas the downward peak represents the muscle-specific 3′ end.
Exon array detection of erythroid-specific beta spectrin mRNA 3′ end. (A) The exon structure of human beta spectrin transcripts in muscle versus erythroid cells, which express distinct 3′ terminal exons. (B) The splicing index of probe sets across the full length of the beta spectrin gene. Numbers along the horizontal axis represent probe set IDs. Positive values for the splicing index represent higher relative probe set expression in erythroblasts, whereas negative values indicate higher relative expression in muscle. The significant upward peak maps to the known erythroid-specific 3′ end, whereas the downward peak represents the muscle-specific 3′ end.
Identification of stage-specific changes in use of alternative first exons
Array data were next examined for evidence of differential exon expression in late erythropoiesis, as manifested by significant changes in probe set expression between basophilic (day 7) and orthochromatic (day 14) erythroblasts. After filtering out probe sets with low expression and low variance, transcripts with highly significant differences in alternative splicing were manually inspected by mapping to the human genome to eliminate those with ambiguous transcript cluster assignments.1 Transcripts that displayed consistent day 7/day 14 expression ratios across most of their probe sets, but differed markedly in one discrete region, were chosen for further characterization. This process eliminated many transcripts with poor annotation, inconsistent probe behavior, and/or overly complex splicing patterns.
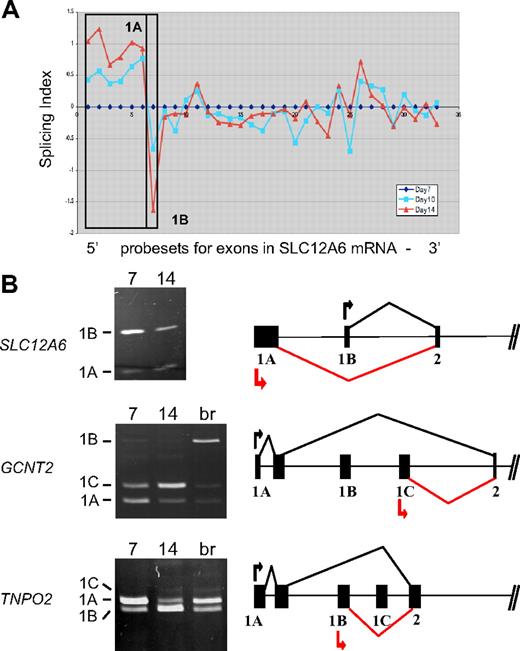
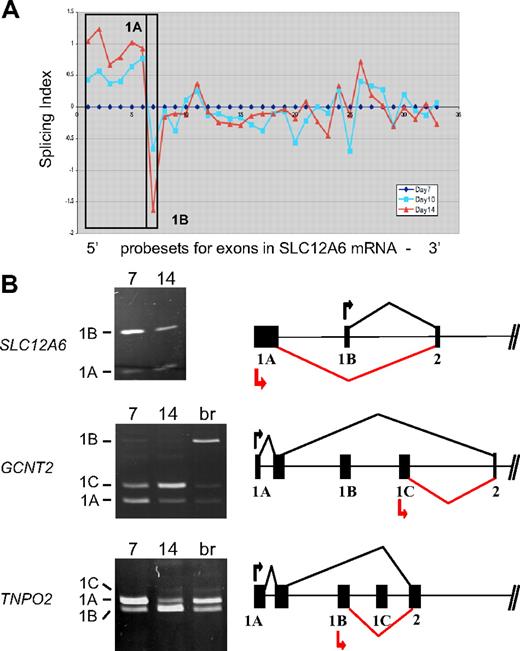
Among the probe sets exhibiting stage-specific changes in expression, several mapped to the 5′ ends of transcripts and were considered to be candidate alternative first exons. Many erythroid genes are known to possess alternative first exons,3 but few have been analyzed for changes in expression during late erythropoiesis. Figure 3A depicts exon microarray data for probe sets in the SLC12A6 transcript that encodes KCC3, a KCl cotransporter. The microarray data suggest a stage-specific decrease in expression of exon 1B relative to exon 1A, as cells differentiate from day 7 to day 14. To validate this prediction, RT-PCR analysis was performed with 2 forward primers (in exons 1A and 1B) and a single reverse primer (in exon 2). Competitive PCR reactions of this nature have been used previously to detect variations in expression of terminal exons.20 As shown in Figure 3B, the relative amounts of amplified exon 1B product declined significantly relative to that of exon 1A at day 14 (top left), confirming the microarray prediction. The gene model to the right depicts the 5′ exon/intron structure of SLC12A, based on data from the Refseq track in the UCSC genome browser. It illustrates the predominant expression pattern of SLC12A transcripts from exon 1A at day 7 (black lines) and the relative shift toward use of exon 1B at day 14 (red lines).
Stage-specific change in expression of alternative first exons. (A) Splicing index shows normalized changes in probe set expression along the entire SLC12A6 transcript for day 10 (blue curve) and day 14 erythroblasts (red curve), relative to expression at day 7 (black line). Results show a stage-dependent increase in expression of probe sets representing the 1A region (boxed) and a decrease in expression of probe sets for exon 1B. (B) RT-PCR validation of exon array predictions for SLC12A6 and 2 additional genes. Shown are gels of PCR products validating alternative splicing switches in first exon expression (left) and diagrams of the relevant pre-mRNA regions (right). Black arrow/black lines represent predominant pattern in day 7 erythroblasts; red arrow/red lines, predominant pattern in day 14 erythroblasts. Alternative first exons are indicated by 1A, 1B, 1C, and a shared constitutive exon indicated as exon 2. Common names of the alternatively spliced transcripts are as follows: SLC12A6 indicates KCl cotransporter 3 (KCC3); TNPO2, transportin 2 (a nuclear import protein); GCNT2, glucosaminyl (N-acetyl) transferase 2 (generates the branched chain carbohydrate structure that constitutes the I antigen).
Stage-specific change in expression of alternative first exons. (A) Splicing index shows normalized changes in probe set expression along the entire SLC12A6 transcript for day 10 (blue curve) and day 14 erythroblasts (red curve), relative to expression at day 7 (black line). Results show a stage-dependent increase in expression of probe sets representing the 1A region (boxed) and a decrease in expression of probe sets for exon 1B. (B) RT-PCR validation of exon array predictions for SLC12A6 and 2 additional genes. Shown are gels of PCR products validating alternative splicing switches in first exon expression (left) and diagrams of the relevant pre-mRNA regions (right). Black arrow/black lines represent predominant pattern in day 7 erythroblasts; red arrow/red lines, predominant pattern in day 14 erythroblasts. Alternative first exons are indicated by 1A, 1B, 1C, and a shared constitutive exon indicated as exon 2. Common names of the alternatively spliced transcripts are as follows: SLC12A6 indicates KCl cotransporter 3 (KCC3); TNPO2, transportin 2 (a nuclear import protein); GCNT2, glucosaminyl (N-acetyl) transferase 2 (generates the branched chain carbohydrate structure that constitutes the I antigen).
Two additional examples of alternative first exon usage were similarly predicted by the exon array data and verified by RT-PCR, in the GCNT2 (an acetylglucosaminyltransferase important in the formation of human blood group I antigen) and TNPO2 (transportin-2) genes (Figure 3B middle and bottom panels). PCR clearly showed a substantial difference in relative expression of alternative first exons between basophilic and orthochromatic erythroblasts for GCNT2, with exon 1A predominating at day 7 and exon 1C relatively much more abundant at day 14. Exon 1B was barely expressed in these erythroblast cultures; however, it could be amplified efficiently from brain cDNA (lane br), providing further evidence for tissue-specific regulation of first exon usage in the GCNT2 gene. For TNPO2, there was a substantial switch in relative use of exons 1A and 1B between day 7 and day 14 erythroblasts (bottom panel). Relatively little expression of exon 1C was observed, with only small amounts detected in day 14 erythroblasts and in brain. For both of these genes, the RT-PCR data were consistent with the exon microarray predictions; thus, 2 independent assays confirm the switch in expression of alternative first exons.
Stage-specific switches in alternative pre-mRNA splicing of internal cassette exons
The Human Exon 1.0 ST Array can also detect alternative splicing changes within internal regions of transcripts.1,14,15 We used this array in conjunction with a prototype exon junction array, which focuses on a smaller set of well-annotated exons, to identify additional splicing changes in differentiating erythroid cells. Junction probes provide additional information that can aid in predicting splicing differences between RNA samples because a change in alternative splicing should yield reciprocal changes in intensity of junction probes for the exon-skipping and exon inclusion event, respectively.
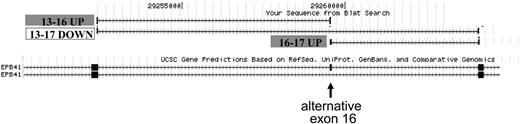
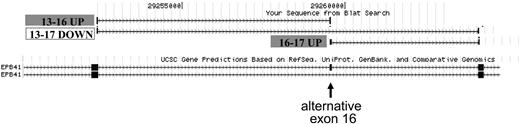
Figure 4 illustrates detection of the protein 4.1R exon 16 splicing switch by the junction array. The structures of 2 Refseq transcripts that include or exclude exon 16 are shown in the lower part of the figure, with the position of alternative exon 16 indicated by the arrow. The position of junction probe sets exhibiting a significant change in expression, normalized to transcript levels, is shown above. The 2 probe sets representing exon 16 inclusion products exhibited strong up-regulation in day 14 orthochromatic erythroblasts (probe set ID 504900 for the exon 13/16 junction, P = 7.5E-07, and probe set 224956 for the exon 16/17 junction, P = 1.6E-09). Importantly, the probe set interrogating the exon 16 skipping event demonstrated a significant decrease in expression in these more mature cells (probe set ID 366937, spanning the exon 13/17 junction, P = 1.2E-04). Thus, the reciprocal increase in inclusion probes and decrease in the exclusion probe validates the ability of the junction array to detect a known splicing switch, the activation of exon 16 splicing in late erythroblasts.
Exon junction array detection of the 4.1R exon 16 splicing switch. Probe sets in the exon 16 region that exhibit significant changes in relative expression between day 7 and day 14 were mapped to the human genome using the UCSC BLAT alignment tool. (Bottom) The exon structure of the 2 mRNA isoforms expressed from the 4.1R gene; arrow represents alternative exon 16. (Top) Probe sets interrogating the exon 16 inclusion event were up-regulated at day 14, whereas a reciprocal decrease in expression of the exons 13 to 17 skipping event was observed. RT-PCR validation of this splicing switch is shown in Figure 1. Alternative exons 14 and 15 are not expressed in these cultures.
Exon junction array detection of the 4.1R exon 16 splicing switch. Probe sets in the exon 16 region that exhibit significant changes in relative expression between day 7 and day 14 were mapped to the human genome using the UCSC BLAT alignment tool. (Bottom) The exon structure of the 2 mRNA isoforms expressed from the 4.1R gene; arrow represents alternative exon 16. (Top) Probe sets interrogating the exon 16 inclusion event were up-regulated at day 14, whereas a reciprocal decrease in expression of the exons 13 to 17 skipping event was observed. RT-PCR validation of this splicing switch is shown in Figure 1. Alternative exons 14 and 15 are not expressed in these cultures.
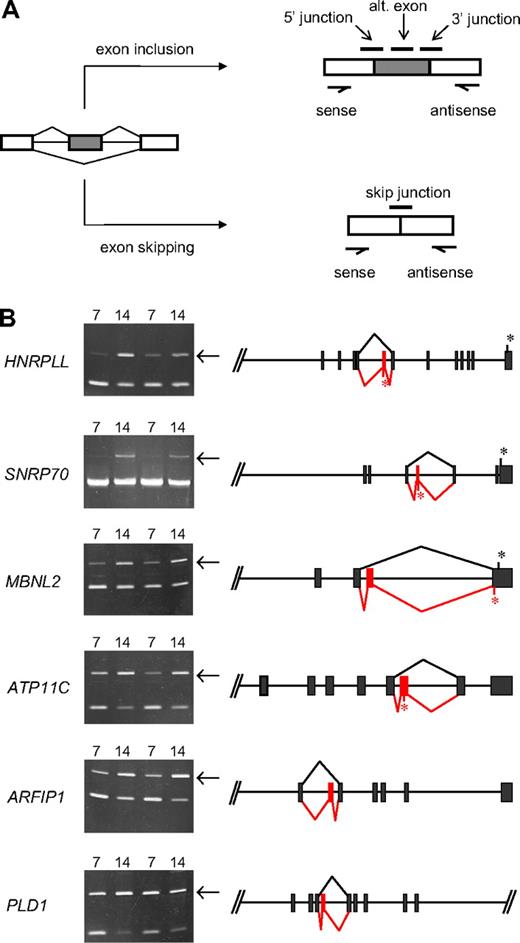
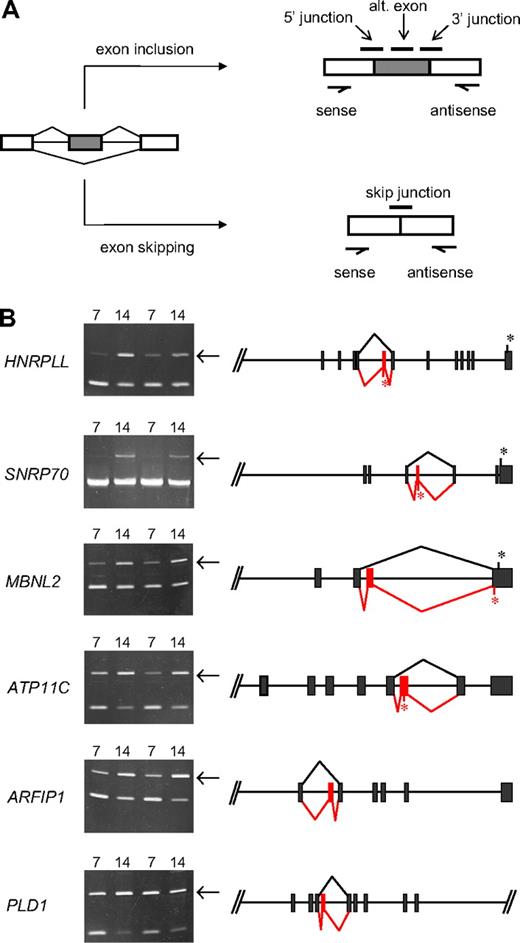
Further analysis of junction array data predicted several novel erythroid stage-specific alternative splicing switches. All of these cases exhibited a significant decrease at day 14 in the junction probe set representing the exon-skipping event and a reciprocal increase for at least one probe set representing the alternative exon inclusion event. The diagram in Figure 5A illustrates schematically the locations of these diagnostic probe sets for a model alternative splicing event. Table 1 summarizes hybridization data of specific probe sets for 6 candidate stage-specific exons and also provides some information regarding the identity of the genes/exons undergoing the putative splicing switches. To evaluate the performance of the microarray, we sought independent verification of these splicing predictions using PCR methods to amplify across the relevant region of each mRNA using primers in the flanking exons. Alternative splicing switches should be manifested by changes in the relative amounts of exon inclusion/exon exclusion products comparing PCR products derived from basophilic and orthochromatic erythroblast samples. As shown in Figure 5B, this was indeed the case, as 6 new exons were validated by RT-PCR analysis to have substantial erythroid stage-specific changes in alternative splicing in 2 independent biologic replicates. In each case, consistent with the microarray predictions, the relative efficiency of alternative exon inclusion was higher in the more mature day 14 (orthochromatic) erythroblasts. Control experiments confirmed the expected sequence of these PCR products and demonstrated that exon inclusion/exclusion ratios were independent of PCR cycle number within the range examined (Figure S1). The latter is an important criterion typically used in splicing studies to validate cell-specific differences in alternative splicing.21-24 Together, the combined analysis of erythroblast mRNA by exon microarrays and RT-PCR strongly supports the hypothesis that these splicing switches are bona fide features of the gene expression program in late erythropoiesis.
Novel stage-specific alternative splicing switches in erythroid genes. (A) General scheme for detection of alternative splicing of a pre-mRNA (left) into the mRNA isoform including the alternative exon (top right) or the mRNA skipping this exon (bottom right). Diagnostic isoform-specific microarray probes are indicated above the spliced mRNAs, whereas PCR primers used for validation are shown below the mRNAs (arrows). In addition, there are exon probes for the first and third exons that hybridize equally to both isoforms and are useful for determining overall transcript levels. (B) Shown are gels of PCR products validating alternative splicing switches in late erythropoiesis (left) and diagrams of the relevant pre-mRNA regions (right). Gels demonstrate substantial increases in exon inclusion products (top bands, indicated by arrows), relative to exon-skipping products (bottom bands), at day 14. The deduced splicing patterns are indicated at the right, with black lines indicating major splice patterns at day 7 and red lines indicating predominant splice pattern at day 14. Asterisks indicate positions of stop codons (not shown for ARFIP1 and PLD1 because they are located farther downstream). Common names of the alternatively spliced transcripts are as follows: HNRPLL indicates heterogeneous nuclear ribonucleoprotein L-like (an hnRNP protein); SNRP70, U1 small nuclear ribonucleoprotein 70K (a component of the U1 snRNP); MBNL2, muscleblind 2 (RNA binding proteins with known splicing regulatory activity); ATP11C, ATPase class VI type 11C; ARFIP1, ADP-ribosylation factor interacting protein 1; PLD1, phospholipase D1.
Novel stage-specific alternative splicing switches in erythroid genes. (A) General scheme for detection of alternative splicing of a pre-mRNA (left) into the mRNA isoform including the alternative exon (top right) or the mRNA skipping this exon (bottom right). Diagnostic isoform-specific microarray probes are indicated above the spliced mRNAs, whereas PCR primers used for validation are shown below the mRNAs (arrows). In addition, there are exon probes for the first and third exons that hybridize equally to both isoforms and are useful for determining overall transcript levels. (B) Shown are gels of PCR products validating alternative splicing switches in late erythropoiesis (left) and diagrams of the relevant pre-mRNA regions (right). Gels demonstrate substantial increases in exon inclusion products (top bands, indicated by arrows), relative to exon-skipping products (bottom bands), at day 14. The deduced splicing patterns are indicated at the right, with black lines indicating major splice patterns at day 7 and red lines indicating predominant splice pattern at day 14. Asterisks indicate positions of stop codons (not shown for ARFIP1 and PLD1 because they are located farther downstream). Common names of the alternatively spliced transcripts are as follows: HNRPLL indicates heterogeneous nuclear ribonucleoprotein L-like (an hnRNP protein); SNRP70, U1 small nuclear ribonucleoprotein 70K (a component of the U1 snRNP); MBNL2, muscleblind 2 (RNA binding proteins with known splicing regulatory activity); ATP11C, ATPase class VI type 11C; ARFIP1, ADP-ribosylation factor interacting protein 1; PLD1, phospholipase D1.
Effects of stage-specific splicing switches on protein structure
In cases where alternative first exons contain coding information or influence expression of downstream coding sequences, switches in alternative first exon expression can directly impact N-terminal protein structure. The late erythroid switches in first exon use for SLC12A6 and GCNT2 both fall into this category because these exons contain translation initiation sites. The GCNT2 gene has an unusual structure in which 3 long alternative first exons encode alternative but structurally similar N-terminal domains of approximately 308 amino acids each.25 Our finding that exon 1C represents the major isoform in late erythroblasts is consistent with previous reports that exon 1C is strongly expressed in terminally differentiating K562 cells.26 In contrast, first exons in the TNPO2 gene do not possess start codons; thus, switching does not affect protein structure.
All erythroid stage-specific splicing switches involve internal cassette exons that exhibit enhanced inclusion at day 14, relative to less efficient or undetectable inclusion at day 7. The exons switched on in late erythroblasts range in size from 66 to 114 nt, significantly smaller than the average human exon but consistent with previous observations that alternative exons are generally shorter than average.27,28 In each case, the splicing switch is within the coding portion of the transcript; thus, it will alter the protein product. For PLD1 (phospholipase D1) and ARFIP1 (arfaptin1, or ADP-ribosylation factor interacting protein 1), the splicing switches are predicted to insert in-frame peptides of approximately 3 to 4 kDa, without alteration of C-terminal sequences. There are no obvious functional motifs encoded in these alternative peptides. Interestingly, ARFIP1 has been reported to inhibit PLD1 activity,29 suggesting the possibility that splicing changes in both genes might coordinately affect some aspect of signaling or membrane trafficking. The 4 remaining splicing switches differ in that they should induce C-terminal alterations or truncations in the encoded proteins. In MBNL2 (muscleblind2, an RNA binding protein), the switch introduces a new penultimate coding exon that inserts a new peptide and also changes the reading frame. The resulting mRNA should terminate translation at an alternative site in the final exon. In 3 other genes, the inserted exon contains an in-frame stop codon so that the splicing switch will result in C-terminal truncations of the predicted protein product. This is the predicted outcome of splicing switches in HNRPLL (heterogeneous nuclear ribonucleoprotein L-like), SNRP70 (U1 small nuclear ribonucleoprotein 70K, a component of the U1 snRNP), and ATP11C (ATPase class VI type 11C). Whether the truncated proteins have any physiologic role is unknown. However, because the truncations would delete one of the 2 RNA binding motifs in HNRPLL and half of the single RNA binding domain in SNRP70, any function would be quite different from, and potentially antagonistic to, the full-length proteins. Because MBNL2, HNRPLL, and SNRP70 have known or predicted effects on pre-mRNA splicing, these results suggest that alternative splicing of selected factors may potentially affect a wider range of RNA processing events in late erythropoiesis.
Discussion
High-resolution analysis of the erythroid transcriptome provides new insights into the mechanisms by which gene regulatory mechanisms orchestrate expression of a highly dynamic proteome in a developmentally appropriate, tissue-specific manner. The current study complements and extends previous analyses of the erythroid transcriptome30,31 by exon-level analysis of switches in alternative splicing during late erythropoiesis. Based on the finding of several novel stage-specific splicing switches in erythroid gene expression, we propose the existence of an alternative splicing program that is essential for proper differentiation of late erythroid cells. All of the switches involve bona fide splicing events that occur precisely at exon-intron boundaries of annotated exons, supporting the hypothesis that they represent programmed splicing changes rather than a general deregulation of the splicing machinery in late erythropoiesis. Our results indicate that this program is composed of distinct regulatory mechanisms involving transcription events at alternative promoters, and splicing events at internal alternative exons, as both processes contribute to modulation of gene expression in important ways that are generally not detectable by standard expression analysis.
Alternative first exons are common among erythroid genes,3,32-37 and indeed among the whole spectrum of genes in the human genome.38,39 In this report, we show that switches in expression of alternative first exons can occur even late in erythroblast differentiation. Such changes can affect protein levels quantitatively and/or qualitatively, depending on the relative expression levels of the transcripts and the location of translation start codons. Mechanistically, it is probable that these switches are regulated at the transcriptional level because alternative first exons generally possess their own promoters,32 but changes in mRNA stability of alternative isoforms could also play a role in some transcripts. Our finding that GCNT2 exon 1C represents the major isoform in late erythroblasts is consistent with previous reports that exon 1C is up-regulated in terminally differentiating K562 cells by activity of transcription factor CCAAT/enhancer binding protein α (C/EBPα).26 The switches in SLC12A6 and TPNO2 first exons could potentially be determined by a related mechanism, perhaps operating so as to allow the genes to escape down-regulation of expression in late erythroblasts.
A second class of gene expression changes involves internal cassette exons that are regulated at the level of alternative pre-mRNA splicing. Interestingly, in each of the 6 new cases described here, alternative exon inclusion was increased in late erythroblasts. This observation suggests that a common pathway or shared splicing program may be coordinating these splicing events. Preliminary analysis of potential splicing regulatory motifs has revealed that 5 of the 6 stage-specific exons possess, in the flanking proximal introns, a highly specific UGCAUG binding site for the alternative splicing regulator Fox-2 (data not shown). This frequency is much higher than expected by chance and is consistent with a coordinating role of Fox-2 in mediating some of these late erythroid alternative splicing switches. Indeed, Fox-2 has already been reported to play an important role in activating the protein 4.1R exon 16 splicing switch.12,13 Moreover, Fox-1/Fox-2 binding sites are highly overrepresented in the introns near many muscle-enriched exons14 and also near brain-enriched exons,40,41 but not near non–tissue-specific alternative exons or constitutive exons. However, the erythroid-, muscle-, and brain-enriched exon datasets identified in these studies are largely unique. We speculate that Fox proteins are necessary to activate splicing switches of many tissue-specific exons, but not sufficient to determine the spatial or temporal patterns of these switches. Tissue-specific differences in Fox protein structure (mediated by regulated alternative splicing of the Fox pre-mRNAs themselves42 ) or in the complement of putative Fox-interacting proteins43 are presumably required to execute the appropriate alternative splicing programs through development. Future studies will be required to test this hypothesis experimentally.
It is noteworthy that 3 of the alternative splicing switches in late erythropoiesis involve exons containing premature termination codons (PTC), such that the resulting mRNAs would encode truncated proteins. Although it is possible that the truncated proteins have unique functions, recent studies suggest an alternative purpose for this type of regulated splicing. Premature stop codons often induce rapid mRNA degradation via nonsense-mediated decay, and alternative splicing coupled to nonsense-mediated decay can represent an evolutionarily conserved strategy for regulating gene expression.44,45 This process, termed “regulated unproductive splicing and translation,” provides a control mechanism for modulating gene expression by regulating the balance between functional mRNAs that encode full-length proteins and PTC-containing mRNAs that do not. Regulated unproductive splicing and translation has been demonstrated most convincingly in the case of RNA-binding proteins with roles in pre-mRNA splicing,44,45 and it may therefore be very relevant that 2 of the PTC-containing erythroid splicing events involve RNA binding proteins (HNRPLL and SNRP70).
The modest number of alternative splicing switches identified in this study is comparable with other recent reports,15,24 but probably represents only a fraction of the total changes in splicing that occur in differentiating erythroblasts. Here we examined only splicing switches that occur in late stages of erythropoiesis; only annotated alternative splicing events are represented on the microarray; and we can detect only exons represented by “good” oligonucleotide probes that hybridize well under standard conditions. Moreover, given the observation that some splicing switches introduced premature stop codons, nonsense-mediated decay may have masked splicing changes that result in rapidly degraded mRNAs. We anticipate that further advances in array technology, together with application of high throughput sequencing46 and analyses of earlier stages of erythroid differentiation, will lead to discovery of many additional switches in promoter use and alternative splicing that have important implications for erythroid physiology.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Thomas Gallagher for careful review of the manuscript.
This work was supported by the National Institutes of Health (grants DK75021 and HL45182) and by the Director, Office of Biological and Environmental Research, US Department of Energy (contract DE-AC03-76SF00098).
National Institutes of Health
Authorship
Contribution: M.L.Y., T.A.C., and A.W. designed and performed research and wrote the paper; S.L.G. and J.-A.K. performed research; A.C.S. designed research; and J.G.C. designed research and wrote the paper.
Conflict-of-interest disclosure: T.A.C. and A.C.S. are employees of Affymetrix. The remaining authors declare no competing financial interests.
Correspondence: John G. Conboy, Lawrence Berkeley National Laboratory, Life Sciences Division, Building 84-171, One Cyclotron Rd, Berkeley, CA 94720; e-mail: jgconboy@lbl.gov.