Abstract
17p (TP53) deletion identifies patients with chronic lymphocytic leukemia (CLL) who are resistant to chemotherapy. The members of the miR-34 family have been discovered to be direct p53 targets and mediate some of the p53-dependent effects. We studied miR-34a and miR-34b/c expression in a large cohort to define their potential role in refractory CLL. While no expression of miR-34b/c could be detected, we found variable expression levels of miR-34a. miR-34a levels were up-regulated after DNA damage in the presence of functional p53, but not in cases with 17p deletion (P < .001). We found a strong correlation of low miR-34a levels with impaired DNA damage response, TP53 mutations (without 17p deletion), and fludarabine-refractory disease (also in the absence of 17p deletion). Up-regulation of miR-34a after irradiation was associated with induction of Bax and p21, but not Puma. CLL cells with reduced miR-34a expression showed increased viability after DNA damage independently of 17p status. Therefore, low expression of miR-34a in CLL is associated with p53 inactivation but also chemotherapy-refractory disease, impaired DNA damage response, and apoptosis resistance irrespective of 17p deletion/TP53 mutation. The elucidation of mechanisms underlying miR-34a regulation and overcoming its role in chemotherapy resistance warrant further study.
Introduction
Chronic lymphocytic leukemia (CLL) is the most frequent type of leukemia in the Western world and is characterized by a highly variable clinical course.1-3 Traditionally, therapy has been used for advanced stage or symptomatic disease and consists of chemotherapy with alkylating agents, purine analogs, and more recently antibody-chemotherapy combinations. Different molecular prognostic factors have been used to predict time to treatment, likelihood of responding to chemotherapy and survival time.4-6 A central role of the DNA damage-response pathway and particularly p53 has been suggested by the prognostic role of 17p and 11q deletions in CLL. In the critical regions, 2 prominent genes are located that are involved in the cells' response to DNA damage (eg, induced by chemotherapy). ATM and TP53 have been shown to be deleted in virtually all cases with deletion 11q and 17p, respectively. In contrast, TP53 and ATM are mutated in different proportions.7-11 There is growing evidence that mutations of TP53 or ATM also in the absence of deletion of 17p or 11q are associated with poor prognosis as a result of impaired response to chemotherapy.12 Particularly loss of 17p has been associated with failure to respond to chemotherapy and short event-free and overall survival.8,13,14
However, in chemotherapy-refractory CLL, only 30% to 40% percent of cases will have a deletion or mutation of TP53, whereas approximately one-third of the remaining cases have a deletion of 11q.15 This suggests that almost half of the refractory cases cannot be explained by a direct defect of p53 or loss of 11q. A hypothesis explaining resistance in these cases might be defects of the DNA damage/p53 pathway without direct involvement of p53 or ATM by loss or mutation. These defects might have similar clinical consequences, but experimental data for this hypothesis is currently lacking. Functional assessment of p53 may be of help to more precisely define a subgroup of patients with defects in the DNA-damage pathway. The most commonly used assays include fluorescence-activated cell sorting (FACS) measurement of induction of p53/p21 after DNA damage, induction of p53 by nutlins or the measurement of transcriptional targets of p53 by reverse transcription–polymerase chain reaction (RT-PCR) or multiplex ligation-dependent probe amplification (MLPA).16-18 All these techniques have proven to be helpful in identifying abnormal p53 responses, but their precise clinical value is currently unclear.
Recently, microRNAs (miRs) have been implicated in the pathogenesis of CLL. miR-15a/16-1 were shown to be located in the critical region of 13q14, which is lost in more than 50% of patients with CLL.19-21 Interestingly, miR-15a/16-1 have also been shown to be induced in cell lines after induction of p53.22 Tumor cells in general and also CLL cells show an aberrant pattern of miR expression.19,23-25 p53 has recently been shown to target miR-34a and miR-34b/c located in the chromosomal regions 1p36 and 11q23 (the critical region of deletion 11q in CLL), respectively.22,26-29 After treatment with DNA-damaging agents, the expression of miR-34a and miR-34b/c were induced in mouse tissues in a p53-dependent manner, and p53 was shown to directly regulate miR-34s expression through a p53 binding site.26,28,29 Importantly, the miR-34 family and particularly miR-34a have been shown to mediate some of the functional consequences of p53 activation including apoptosis, cell cycle arrest, and senescence in different cell lines and primary cells.28,30,31 Because miRs are expected to regulate the levels of hundreds of different proteins in different tissues, these findings add further complexity to the p53 network.31 Currently, CDK 4/6, CCND1, cMET, cyclin E2, MycN, and BCL2 are candidate targets of the miR-34 family.26,28,30-33
Because very little is known about the function of the miR-34 family in primary human cancer cells and because of the central role of the p53 pathway in affecting prognosis and response to chemotherapy, we set out to define the role of miR-34s in CLL.
Methods
Patients
A cohort of 60 CLL patients from a single institution (University of Ulm, Ulm, Germany) was enrolled in this study after obtaining patients' informed consent and approval of the University of Ulm in accordance with the Declaration of Helsinki. Criteria for patient selection were a high lymphocyte count and a percentage of lymphocytes in excess of 80%. Age ranged from 33 to 81 years (mean, 63 years), and detailed clinical as well as genetic characterization was performed (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The population was selected to represent meaningful numbers of patients from well-defined genetic risk groups. Lymphocytes were isolated from blood samples with Biocoll solution, and aliquots were frozen viably at −196°C.
Genetic analysis
Fluorescence in situ hybridization (FISH) analysis and VH sequencing were performed in all cases as previously described.8,34,35 A germ line homology of 98% was used as cut-off between VH mutated and unmutated cases (Table S1).
Denaturing high-pressure liquid chromatography (DHPLC) was used to identify mutations in exons 2-11 (coding region of TP53). DHPLC analysis is based on the temperature-dependent differences in column-retention time of PCR products generated from homoduplex (wild-type) and heteroduplex (mutated) DNA, resulting in the presence of distorted or additional peaks when mutations are present. To detect homozygous mutations or mutations in samples that had undergone allelic loss, each test sample was mixed with wild-type DNA (20%). Assay conditions and sequencing details were as described previously.36
In vitro conditions and treatment
CLL cells were thawed and seeded in RPMI 1640 medium containing 10% fetal bovine serum (FBS), penicillin and streptamycin at a density of 5 × 106 cells/mL. Cells were treated with γ-irradiation (5Gy IR) and cultured at 37°C for 48 hours.
RNA isolation and real-time RT-PCR
RNA was isolated using the Ambion mirVana PARIS kit (Applied Biosystems, Foster City, CA). Expression of hsa-miR-34a and hsa-miR-34c was analyzed using the quantitative RT-PCR detection kit and specific primer sets (Ambion, Applied Biosystems). Reverse transcription reactions were performed according to the manufacturer's protocol. For real-time PCR, specific PCR-primers were used with the SYBR Green Rox mix (ABgene, Epsom, United Kingdom). Analysis of hsa-miR-34b was performed using the miScript system (QIAGEN, Valencia, CA), consisting of miScript Reverse Transcription kit, miScript Primer assays and miScript SYBR Green PCR kit, according to the protocol provided by the company. Small nuclear RNA U6 was used for normalization. For the analysis of p21 expression, cDNA was synthesized using QuantiTect Reverse Transcription kit (QIAGEN) as described by the manufacturer. The housekeeping gene lamin B1 was used for normalization. Primers were designed as follows: p21 for, 5′-CGCTAATGGCGGGCTG-3′; p21 rev, 5′-CGGTGACAAAGTCGAAGTTCC-3′. Real-time PCR was run on the ABI Prism 7700 Sequence Detector (Applied Biosystems). Relative expression was calculated using the 2−ΔΔCT method, taking one patient sample with a very low expression as a reference unless otherwise stated.37
Detection of apoptosis
Cells were stained with Annexin V–PE (BD Biosciences, San Jose, CA) and 7-aminoactinomycin D (7AAD; Sigma-Aldrich, St Louis, MO) according to the manufacturer's protocol. Stained cells were analyzed on the FACS-Calibur (BD Biosciences). Data were analyzed using the Cellquest Pro software (BD Biosciences).
p53-p21 flow cytometry
Cells were harvested 24 hours after IR, washed with phosphate-buffered saline (PBS) and stained with CD19-PC7 antibody (Beckman Coulter, Fullerton, CA). Cells were then fixed in 2% cold paraformaldehyde (PFA) and kept overnight in 80% ethanol at −20°C. Fixed cells were washed with bovine serum albumin (BSA) in PBS to prevent nonspecific binding and then stained with p53-PE antibody (clone DO-7; BD, Franklin Lakes, NJ) and p21-fluorescein isothiocyanate (FITC; Calbiochem, San Diego, CA) or the corresponding isotype controls. After incubation at 4°C for 1 hour, cells were analyzed on the FACSCalibur, and data were analyzed using the Cellquest Pro software.
RT-MLPA assay
RT-MLPA assay procedure was performed as described previously.17 Briefly, 100 ng total RNA was reverse-transcribed using a gene-specific probe mix. The resulting cDNA was annealed overnight at 60°C to the MLPA probes. Annealed oligonucleotides were covalently linked by Ligase-65 (MRC, Amsterdam, The Netherlands) at 54°C. Ligation products were amplified by PCR (33 cycles of 30 seconds at 95°C, 30 seconds at 60°C, and 1 minute at 72°C) using one unlabeled and one 6-carboxy–fluorescein-labeled primer (10 pM). PCR products were run on an ABI 3100 capillary sequencer in the presence of 1 pM ROX 500 size standard (Applied Biosystems). Results were analyzed using the Genescan analysis and Genotyper software (Applied Biosystems).
Statistical analysis
Statistical analyses were performed with GraphPad Prism version 3.00 (GraphPad Software, San Diego, CA). Group-wise comparisons of distributions of clinical, laboratory, and genetic data were performed with the Fisher exact test (categorical variables). All tests were 2-sided. An effect was considered statistically significant at P values less than or equal to .05.
Results
Expression of miR-34a and miR-34b/c in CLL
To activate the DNA damage pathway and p53, we exposed the cells to 5Gy IR and harvested RNA from CLL cells after in vitro culture to assess expression levels over time. While we could not detect miR-34b/c expression in any of the CLL samples and time points investigated (with and without IR; n = 3-6), we found detectable basal levels of miR-34a. miR-34a levels were highly induced at 16 hours after IR and peaked at 32 hours (Figure 1A). Interestingly, miR-34a peaked later than p21 mRNA levels (data not shown). For our further studies, we focused on the 16-hour time point to avoid secondary effects.
miR-34a is induced in CLL after IR, and 17p deletion is associated with defective induction. (A) CLL with 17p deletion (n = 3) show impaired miR-34 up-regulation over time. In comparison, CLL with favorable genetics (13q deletion as sole abnormality; n = 3) show miR-34a up-regulation peaking around 32 hours after IR. Values are expression levels relative to the nonirradiated sample (normalized for RNA by U6 control). (B) Relative expression levels of miR-34a were low in cases with 17p deletion with and without IR. The comparison to all other cases showed significantly lower expression of miR-34a in cases with 17p deletion (n = 12) versus all other cases (n = 48; median, 2.5 vs 32.8; P < .001). Similarly, up-regulation of miR-34a was negligible compared with the majority of cases without 17p deletion (median, 5.8 [1.38-12.9] vs 121.8 [1.79-608.9]; P < .001).
miR-34a is induced in CLL after IR, and 17p deletion is associated with defective induction. (A) CLL with 17p deletion (n = 3) show impaired miR-34 up-regulation over time. In comparison, CLL with favorable genetics (13q deletion as sole abnormality; n = 3) show miR-34a up-regulation peaking around 32 hours after IR. Values are expression levels relative to the nonirradiated sample (normalized for RNA by U6 control). (B) Relative expression levels of miR-34a were low in cases with 17p deletion with and without IR. The comparison to all other cases showed significantly lower expression of miR-34a in cases with 17p deletion (n = 12) versus all other cases (n = 48; median, 2.5 vs 32.8; P < .001). Similarly, up-regulation of miR-34a was negligible compared with the majority of cases without 17p deletion (median, 5.8 [1.38-12.9] vs 121.8 [1.79-608.9]; P < .001).
When assessing basal miR-34a levels in a cohort of 60 CLL samples, we found highly variable expression with a median level of 23.5 (0.89-337.8). After IR, expression of miR-34a was increased to a median level of 83 (1.4-608.9; P < .001; Table S1). We found similar levels of basal miR-34a expression and induction when limiting the analysis to CLL cases that had no prior therapy (n = 35; data not shown).
Deletion of 17p is associated with low basal expression of miR-34a and failure to induce expression of miR-34a after DNA damage
To assess whether miR-34a is a bona fide p53 target in primary CLL cells, we compared the basal level and induction of miR-34a between samples with or without 17p deletion. We found significant differences in basal and induced levels, suggesting that miR-34a is a target of p53 in CLL (Figure 1A,B). All cases with deletion of 17p (n = 12) exhibited low or very low basal expression (median basal expression 2.5 [1.0-15.78] vs median of 32.8 [0.89-337.8] in cases without 17p deletion, P < .001; n = 48; Figure 1B). Similarly, cases with deletion of 17p showed only a subtle up-regulation of miR-34a after IR (median 5.8 [1.38-12.9] vs 121.8 [1.79-608.9], P < .001; Figure 1B). When separating groups according to expression levels of miR-34a (below and above median expression), all patients with deletion of 17p were in the group with a low expression of miR-34a (P = .001).
Low expression of miR-34a is associated with impaired DNA damage response and chemotherapy resistance in cases without 17p deletion/TP53 mutation
Because basal and induced expression levels of miR-34a varied widely in cases without deletion of 17p (Figure 1B), we hypothesized that low levels of miR-34a might be associated with an impaired DNA damage response and thus clinical resistance to fludarabine also in the absence of 17p deletion. We therefore analyzed the group of CLL cases showing low expression of miR-34a, defined as levels below the median (32.83 ± 10.45), and failure to up-regulate miR-34a upon IR (lower than the median [121.8 ± 19.69] up-regulation of the cohort). Using this differentiation, 15 of 48 patients (31.5%) without 17p deletion were identified (“low miR-34a” group). The clinical and genetic characteristics of this group, compared with cases with high expression, showed a similar distribution of VH mutated and unmutated cases (P = .2) and no significant differences in the distribution of genomic aberrations. Cases with 11q deletion were distributed equally in the 2 groups. Specifically, the basal and induced levels of miR-34a in cases with 11q- were not significantly different from patients without 17p deletion or without TP53 mutation (median 31.7/116.2 [no IR/IR with 5Gy] vs median 38.4/152.1; Figure 2). This suggests that miR-34a is directly involved in the p53-dependent response to DNA damage in primary CLL cells. However, reduced expression may be related to impaired DNA damage response and chemotherapy resistance in cases without 17p deletion/TP53 mutation.
CLL exhibits differential miR-34a expression based on genetic background. Relative expression levels of miR-34a are shown in cases stratified by genomic aberrations and TP53 mutation without (■) and with IR (▲). CLL cases with a deletion of 17p show a severely reduced level of miR-34a expression with and without IR. Similarly, cases with TP53 mutation but no 17p deletion showed failure to up-regulate miR-34a after IR. The 2 cases with “normal” up-regulation had a wild-type TP53 allele in 50% to 70% of cells (* case with 70% wild-type allele [c.455C > T], ** case with 50% wild-type allele [c.847C > T]), likely explaining the up-regulation of miR-34a after IR. Cases with 11q deletion did not show different basal expression or induction compared with cells without 17p-, 11q-, or TP53 mutation.
CLL exhibits differential miR-34a expression based on genetic background. Relative expression levels of miR-34a are shown in cases stratified by genomic aberrations and TP53 mutation without (■) and with IR (▲). CLL cases with a deletion of 17p show a severely reduced level of miR-34a expression with and without IR. Similarly, cases with TP53 mutation but no 17p deletion showed failure to up-regulate miR-34a after IR. The 2 cases with “normal” up-regulation had a wild-type TP53 allele in 50% to 70% of cells (* case with 70% wild-type allele [c.455C > T], ** case with 50% wild-type allele [c.847C > T]), likely explaining the up-regulation of miR-34a after IR. Cases with 11q deletion did not show different basal expression or induction compared with cells without 17p-, 11q-, or TP53 mutation.
When we correlated the likelihood of having received prior therapy with expression of miR-34a, we found an overrepresentation of chemotherapy-naive patients in the subgroup with high expression levels of miR-34a. More importantly, 53% of patients with low miR-34a expression in the absence of 17p deletion had fludarabine-refractory CLL at the time the sample was taken (8/15). In contrast, fludarabine-refractory CLL was observed in only 3 of 33 patients (9%) with high miR-34a expression (P < .002; Table 1). Also when excluding patients with 17p deletion, we found significantly lower levels of miR-34a expression (basal and induced) in the fludarabine-refractory group compared with cases without refractory disease (median basal/induced level 18.96/39.49 vs 39.53/143.0; P = .04/.004).
Low expression of miR-34a is associated with TP53 mutations in the absence of 17p deletion
Because of growing evidence that mutations of TP53 affect clinical outcome in CLL even in the absence of 17p deletions, we looked for mutations of the TP53 gene in 34 patients of the cohort. We detected mutations of TP53 in the group with a low expression of miR-34a in 5 of 15 patients (33%) compared with only 3 of 33 patients (9%) in the cases with high miR-34a expression. Restricting the analysis only to cases with a mutation analysis, the observed difference was not significant, as we did not analyze all the cases with normal p53/p21 induction after IR (see below). However, these CLL cases are unlikely to harbor mutations of TP53, which would have been detected by p21/p53 FACS. Interestingly, the 2 cases with a TP53 mutation that showed highest basal expression and intact induction of miR-34a had remaining wild-type signal of 50% to 70% (indicative of only a small p53 deficient subclone), which might explain the findings (Figure 2).
miR-34a expression is associated with impaired DNA damage response
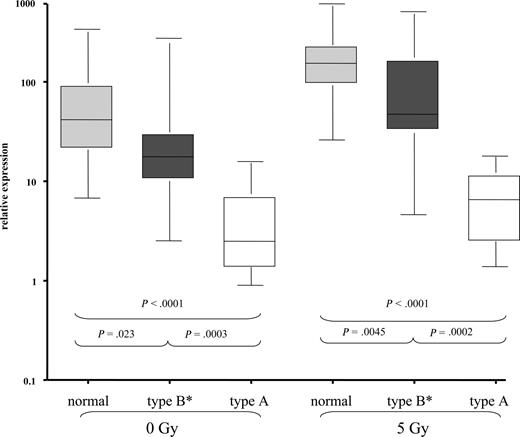
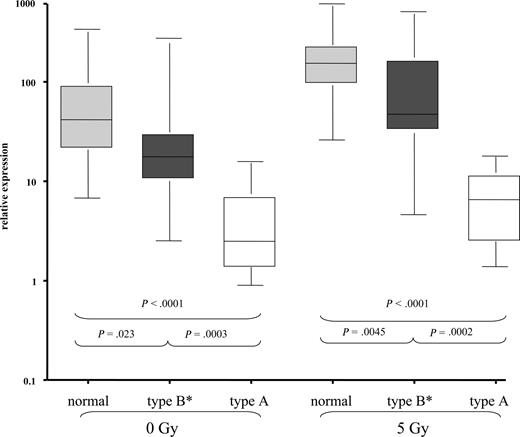
To get a more precise picture of the relation of miR-34a to the DNA damage pathway, we analyzed induction of p53 and p21 by FACS after IR. Similar to what has been described before, we detected “type A” dysfunction (high basal p53, failure to up-regulate p21 after IR) in 11 of 12 cases with 17p deletion as well as 2 cases without 17p deletion, but with TP53 mutation. Thirty-two cases showed a normal induction of p53 and p21 after IR, and this group included 7 patients with 11q deletion, suggesting again that cases with 11q deletion are heterogeneous with respect to their DNA-damage response. The remaining 15 cases displayed an abnormal pattern with failure to induce both p21 and p53 (n = 4), failure to induce p53 [n = 2 (both with TP53 mutation)], or failure to induce p21 (n = 9). We grouped these later cases together (“type B* defect”).18 When we compared the miR-34a expression levels, we found a hierarchy between normal cases (median expression with and without IR: 41.7/153.4), cases with type B defect (median expression: 17.6/47.2), and cases with p53 defect (“type A”), which showed the lowest levels of expression and induction of miR-34a (median expression: 2.5/6.5; Figure 3). The comparison of basal and induced levels was significantly different between the normal group, type B, and type A group. Similarly, when restricting the analysis to cases without 17p deletion as defined above, the group with low miR-34a expression was highly enriched for cases with p53/p21 dysfunction as detected by FACS (Table 1; P < .002).
CLL exhibits differential miR-34a expression based on the response to DNA damage as measured by p53 and p21 induction after IR. Based on the induction of p53 and p21 protein by FACS, CLL cases were stratified into 3 groups as previously described.18,46 Type A dysfunction (p53 defect) shows high basal p53 expression and failure to up-regulate p53 and p21 after IR. Type B defect shows normal (no) basal expression of p53 and failure to induce p53 and p21 protein expression. Since we saw a heterogeneous pattern with cases that up-regulated p53 or p21 only, but exhibited an abnormal pattern, we grouped these patients as type B* defects. In the “normal” group, no baseline expression of p21 and p53 could be detected, but both p21 and p53 were induced by IR. When we assessed miR-34a levels in these subgroups, we saw significant differences in miR-34a expression not only after IR but also at basal levels.
CLL exhibits differential miR-34a expression based on the response to DNA damage as measured by p53 and p21 induction after IR. Based on the induction of p53 and p21 protein by FACS, CLL cases were stratified into 3 groups as previously described.18,46 Type A dysfunction (p53 defect) shows high basal p53 expression and failure to up-regulate p53 and p21 after IR. Type B defect shows normal (no) basal expression of p53 and failure to induce p53 and p21 protein expression. Since we saw a heterogeneous pattern with cases that up-regulated p53 or p21 only, but exhibited an abnormal pattern, we grouped these patients as type B* defects. In the “normal” group, no baseline expression of p21 and p53 could be detected, but both p21 and p53 were induced by IR. When we assessed miR-34a levels in these subgroups, we saw significant differences in miR-34a expression not only after IR but also at basal levels.
miR-34a expression levels correlate with induction of apoptosis upon IR
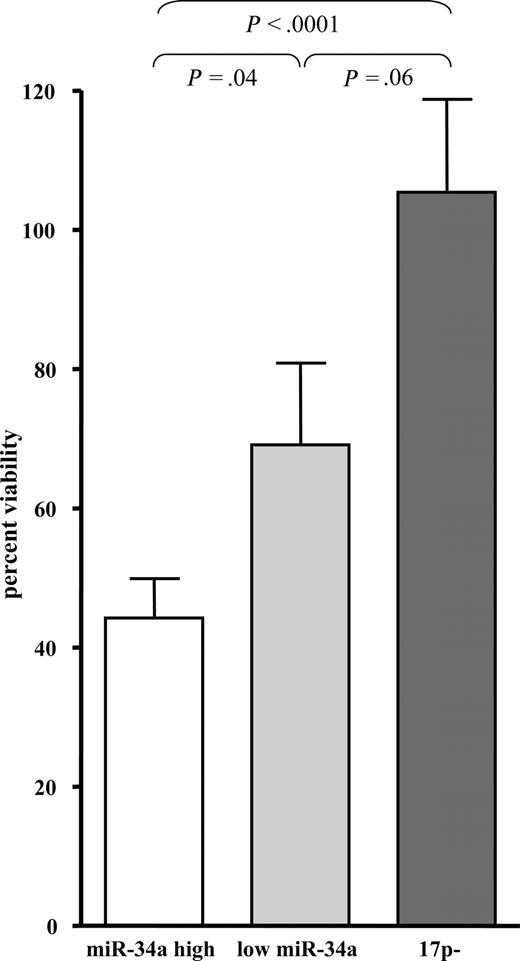
We assessed viability and apoptosis of primary CLL cells by annexin and 7-AAD staining 16 and 48 hours after IR. As expected, we found significantly less induction of apoptosis after 48 hours in cases with 17p deletion compared with CLL cells without deletion (median viability 91% of control vs 51%; P = .001; Figure 4). To assess a possible association between miR-34a expression and rate of cell death in cases without 17p deletion, we compared CLL cases with low miR-34a expression and cases with high expression of miR-34a as specified. CLL cases with low expression of miR-34a showed decreased sensitivity to DNA damage induced by IR compared with cases with high expression of miR-34a (Figure 4 and Table 1; median viability 68% vs 45%; P = .04). miR-34a levels were inversely correlated to viability after IR, suggesting that miR-34a may mediate cell death in response to IR (R2 = 0.1901, P = .009). The correlation was more pronounced when we correlated induced miR-34a levels to viability (R2 = 0.3486; P < .001). The separation into groups based on miR-34a expression predicted viability better than stratification according to p53 and p21 expression as assessed by FACS assay or cytogenetic subgroups (data not shown).
CLL exhibits differential apoptosis 48 hours after IR, which correlates with the presence of deletion of 17p, but also with the level of expression of miR-34a. Apoptosis was assessed by annexin/7-AAD staining 48 hours after IR in primary CLL cells (n = 35; graph shows mean and standard error). We compared the rate of cell death in the patient groups harboring a 17p deletion, CLL patients with low miR-34a expression but no deletion of 17p, and high miR-34a expression but no 17p deletion. We found significantly decreased apoptosis in the patients with 17p deletion (P < .001) or with low expression of miR-34a (P = .04), whereas the difference between 17p deletion and the group with low miR-34a expression was not significant.
CLL exhibits differential apoptosis 48 hours after IR, which correlates with the presence of deletion of 17p, but also with the level of expression of miR-34a. Apoptosis was assessed by annexin/7-AAD staining 48 hours after IR in primary CLL cells (n = 35; graph shows mean and standard error). We compared the rate of cell death in the patient groups harboring a 17p deletion, CLL patients with low miR-34a expression but no deletion of 17p, and high miR-34a expression but no 17p deletion. We found significantly decreased apoptosis in the patients with 17p deletion (P < .001) or with low expression of miR-34a (P = .04), whereas the difference between 17p deletion and the group with low miR-34a expression was not significant.
Induction of miR-34a is related to a subgroup of other p53 transcriptional target genes
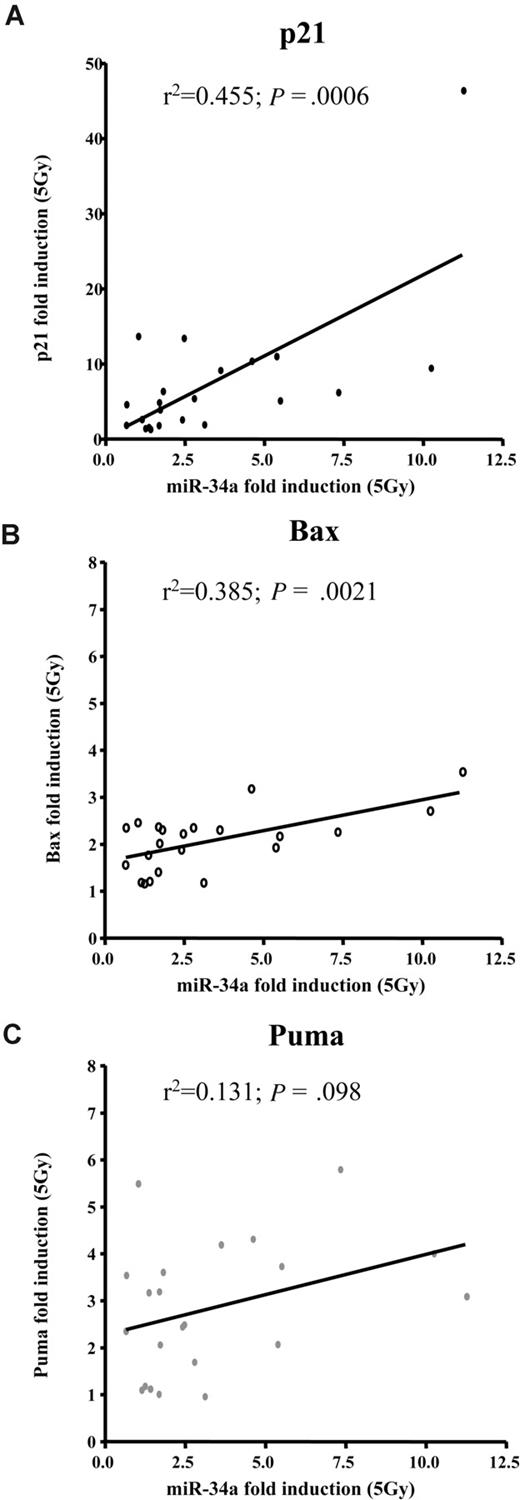
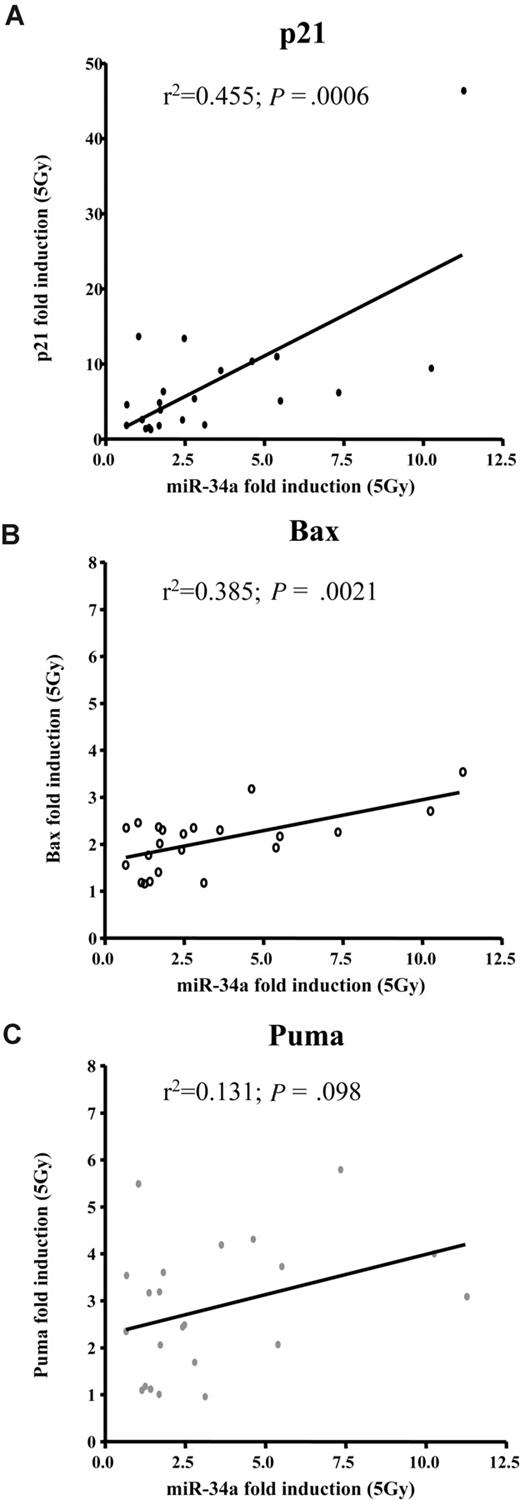
To get a more precise picture of how miR-34a expression and induction relates to other known p53 targets, we assessed transcriptional activation of p21, Bax, and Puma by MLPA (n = 22). Sixteen hours after IR, p21, Bax, and Puma levels increased by a median of 5, 2.2, and 2.8 times, respectively. Cases with deletion of 17p showed impaired induction of these p53 targets. We assessed whether expression of these target genes correlated to the basal expression levels of miR-34a or to induction of miR-34a levels and observed no significant association with basal miR-34a levels. In contrast, the up-regulation of miR-34a was significantly associated with induction of p21 (R2 = 0.595; P < .001) and Bax (R2 = 0.51; P < .002). We found no correlation of miR-34a induction to Puma (Figure 5, Table 2).
miR-34a induction after DNA damage correlates well with induction of p21 and Bax, but not Puma. Regression analyses of miR-34a induction 16 hours after irradiating CLL cells with 5Gy in relation to (A) p21 (P < .001; R2 = 0.455); (B) Bax (P = .002; R2 = 0.385); and (C) Puma (P = .1; R2 = 0.131).
miR-34a induction after DNA damage correlates well with induction of p21 and Bax, but not Puma. Regression analyses of miR-34a induction 16 hours after irradiating CLL cells with 5Gy in relation to (A) p21 (P < .001; R2 = 0.455); (B) Bax (P = .002; R2 = 0.385); and (C) Puma (P = .1; R2 = 0.131).
Discussion
Similar to mRNA-encoding genes, several miRNA-encoding genes have been classified as oncogenes or tumor-suppressors according to their function in cellular transformation and their expression in tumors.24,38 miR-15-a/16-1 have been implicated in the pathogenesis of CLL.19,39 In addition, miR-21 and miR-155 have been shown to be overexpressed in CLL, and a recent study found up-regulation of miR-331, miR-29a, miR-195, miR-34a, and miR-29c in 9 patients with CLL.25,40 A precise picture of how these miRs affect disease characteristics is missing, and their functional role in CLL is currently unclear.
In the current study, we have measured the expression levels of the miR-34 family in CLL. The key findings are (1) lack of expression of miR-34b/c in CLL; (2) variable expression level of miR-34a; (3) p53-dependent up-regulation of miR-34a after DNA damage; (4) a strong association of deletion of 17p/mutations of TP53 with decreased miR-34a levels; but also (5) a strong correlation of low miR-34a levels with impaired DNA damage response, apoptosis, and, most importantly, (6) presence of fludarabine-refractory disease even in the absence of 17p deletion. These findings suggest that miR-34a may play an important role in response to DNA damage and thus response to therapy in CLL.
The prognostic and clinical impact of deletion of 17p in patients with CLL in need of treatment are undisputed.8,13,14 In contrast, patients with deletion of 11q may respond to therapy but may relapse early and have a shorter overall survival compared with patients without 11q and 17p deletion. Nonetheless, these markers of bad prognosis account only for a proportion of cases with poor response to therapy and a more detailed knowledge of factors associated with refractory disease could have immediate consequences on treatment stratification. Several agents currently used in the treatment of CLL have been shown to act independently of the DNA damage machinery. In this respect, the current findings are important, as we can show that low miR-34a expression is not only associated with 17p deletion but also with fludarabine-refractory disease (in the absence of 17p deletion), mono-allelic TP53 mutation, and impaired DNA damage response. Other techniques that may assess a functional DNA damage response, such as FACS analysis of p53 target proteins induced after DNA damage, may be more challenging to standardize than RT-PCR and also require viable cells. In this respect, the finding of reduced miR-34a basal levels is of particular interest. This observation is different from the previous data of indistinguishable mRNA gene expression profiles of TP53 mutated, ATM mutated, and CLL with functional p53/ATM without induction of DNA damage.41
It was interesting to see that most CLL cases harboring a mono-allelic TP53 mutation showed impaired miR-34a level and failure to up-regulate miR-34a and thus exhibit a functional defect similar to the cases with 17p deletion. This finding supports recent evidence that TP53 mutations by themselves may be associated with poor prognosis.42 The demonstration of decreased miR-34a basal levels in cases with TP53 mutation could be interpreted as an oncogenic function of mutated TP53, where the mutated protein acts to decrease miR-34a levels and thus leads to reduced apoptosis.
As p53 is not detectable in unstressed CLL cells (without TP53 mutation), it is unlikely to up-regulate basal levels of miR-34a.43 The fact that miR-34a levels were high in steady state in most cases with good risk features (CD19+ B cells show low expression of miR-34a) suggests that the function of normal levels of miR-34a in CLL may be different from high, induced levels in the context of DNA damage and p53 activation.43 Taken together, these findings suggest that miR-34a levels may have to act in concert with other p53 targets (eg, Bax and p21) after DNA damage, and this may explain why basal levels can be high without induction of apoptosis.
In addition to the down-regulation by impaired p53 as demonstrated in our study, miR-34a expression has very recently been shown to be silenced in several types of cancer due to aberrant CpG methylation of its promoter.44 Prostate carcinomas and melanoma, as well as different epithelial cancer cell lines displayed CpG methylation of the miR-34a promoter and concomitant loss of miR-34a expression. How far promoter hypermethylation contributes to low levels of miR-34a in a fraction of CLL cases remains to be determined.
Cases with 11q deletion showed much variation in the levels of miR-34a. This supports previous reports where only a proportion of cases with 11q deletion show a complete inactivation of ATM (by mutation), and the status of the second allele may be crucial in this respect.7,10,12 Our analysis supports these findings by showing that 11q deletion may be associated with normal response to DNA damage (p53/p21 induction) as well as normal levels of miR-34a with an intact up-regulation upon IR. The detailed study of miR-34a may thus help dissect the heterogeneity of 11q cases and its differential prognostic impact. Because we could not detect any miR-34b/c even in cases without 11q deletion, a role of these miRs, which might have been suggested by their location in the minimally deleted region on 11q23, is unlikely.
Although our study was not designed to identify targets of miR-34a in CLL that mediate its functions within and potentially outside the p53 pathway, the demonstration of down-regulation of cyclin-E2, CDK6, E2F5, and BCL2 protein levels after induction of miR-34a expression is important.26 In addition, it has recently been shown that both mRNA and protein levels of cyclin D1 (CCND1) and cyclin-dependent kinase 6 (CDK6) are reduced by miR-34a.32 Future work should focus on how the different p53 targets orchestrate the differential effects. From our current study the correlation of p21, Bax, and miR-34a suggest that these targets, as opposed to Puma, may act in concert in mediating p53-dependent effects in CLL. It is easy to foresee that some of the known targets of miR-34a will mediate control of apoptosis as well in CLL as shown in the present study, and further studies should be aimed to uncover these connections (Figure 6). It is likely that in other diseases with known prognostic impact of TP53 or 17p deletion, similar mechanisms might be active. In this respect, it is interesting to note that a recent small study investigating miR profiles in follicular and diffuse large B-cell lymphoma, suggested a potential prognostic impact of miR-34a expression.45 It is thus tempting to speculate that miR-34 functions may play an important and broader role in different hematologic malignancies. Once these associations are confirmed and clarified, the focus could shift to a treatment approach using the reactivation of miR-34a, either through the p53 pathway or potentially independently of it, as a therapeutic intervention to reestablish sensitivity to therapy.
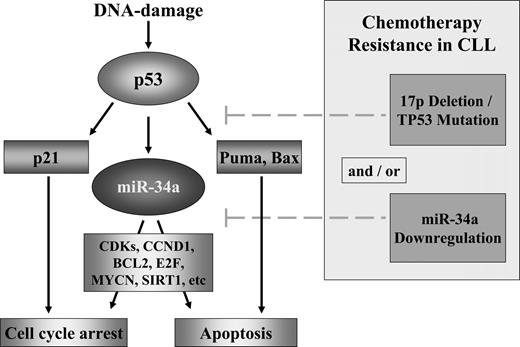
Model of the p53/miR-34a network and its potential impairment in CLL. DNA damage activates p53 through the activation of ATM. p53 induces cell-cycle arrest through p21 and apoptosis through different targets including Puma. miR-34a is a direct transcriptional target of p53 and in case of a functional pathway, miR-34a will be up-regulated. miR-34a induces cell death through apoptosis and cell-cycle arrest through silencing of its potential targets as cyclin-dependent kinases 4 and 6, CCND1, MYCN, BCL2, and Sirtuin1.22,26-33,44,47 In CLL, different modes of interference in this model occur including mutation/deletion of TP53 and down-regulation of miR-34a.
Model of the p53/miR-34a network and its potential impairment in CLL. DNA damage activates p53 through the activation of ATM. p53 induces cell-cycle arrest through p21 and apoptosis through different targets including Puma. miR-34a is a direct transcriptional target of p53 and in case of a functional pathway, miR-34a will be up-regulated. miR-34a induces cell death through apoptosis and cell-cycle arrest through silencing of its potential targets as cyclin-dependent kinases 4 and 6, CCND1, MYCN, BCL2, and Sirtuin1.22,26-33,44,47 In CLL, different modes of interference in this model occur including mutation/deletion of TP53 and down-regulation of miR-34a.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank A. Benner for helpful discussions.
This work was supported by Jose Carreras Foundation (München, Germany; R06/28v, R08/26f), Else Kröner Fresenius Stiftung (Bad Homburg, Germany; P20/07//A11/07), and Global CLL Research Foundation (Houston, TX). The cooperation leading to parts of this work was fostered by the ERIC (European Research Initiative CLL, www.ericll.org).
Authorship
Contribution: T.Z., H.D., and S.S. designed research, analyzed data, and wrote the paper; J.M. performed research, analyzed data, and wrote the paper; J.D. contributed to design of research; and E.E., A.K., A.B., D.K., D.W., J.O., and D.M. performed research and analyzed and discussed data.
Conflict-of-interest disclosure: T.Z. has received reimbursement for the costs of participating in scientific meetings and honoraria from the pharmaceutical industry. S.S. has received reimbursement for costs of participating in scientific meetings, research support, and honoraria from the pharmaceutical industry. The remaining authors declare no competing financial interests.
Correspondence: Dr. Stephan Stilgenbauer, Department of Internal Medicine III, University of Ulm, Albert Einstein Allee 23, 89081 Ulm, Germany; e-mail: stephan.stilgenbauer@uniklinik-ulm.de.

![Figure 1. miR-34a is induced in CLL after IR, and 17p deletion is associated with defective induction. (A) CLL with 17p deletion (n = 3) show impaired miR-34 up-regulation over time. In comparison, CLL with favorable genetics (13q deletion as sole abnormality; n = 3) show miR-34a up-regulation peaking around 32 hours after IR. Values are expression levels relative to the nonirradiated sample (normalized for RNA by U6 control). (B) Relative expression levels of miR-34a were low in cases with 17p deletion with and without IR. The comparison to all other cases showed significantly lower expression of miR-34a in cases with 17p deletion (n = 12) versus all other cases (n = 48; median, 2.5 vs 32.8; P < .001). Similarly, up-regulation of miR-34a was negligible compared with the majority of cases without 17p deletion (median, 5.8 [1.38-12.9] vs 121.8 [1.79-608.9]; P < .001).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/16/10.1182_blood-2008-08-172254/7/m_zh80020929210001.jpeg?Expires=1768075018&Signature=C7mgcr6k2fIlmJ4YbSa6yyw3GcITJrnuGIlGG9zlj9-hNUHxpXllGSy5gihS6b2GeH1va17M2OJ6lR-tb9GaqEDgjPvxXOFdS5ph3VT6lEaRSvCzZHQmG4lbvtC5hiCjDN6zih9JMsCbp23whKE7Lr-350xsm8UsB-7-p-nsNriw5ISsWruWVdWb9e~f9f70VRRPEFc80-0fXTCNLGN2dF0BnB6nwxedT-ceYAMUISoTcUGprRKWFi2ohhVjyXJk7gBhbBI0z63~2IEmDr8potWxXLeT~ympfJDvJVc4~~Yc3aStG0LEPH0D3tJWd19u8W2U4RXT1uzne1b~tIJKng__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. CLL exhibits differential miR-34a expression based on genetic background. Relative expression levels of miR-34a are shown in cases stratified by genomic aberrations and TP53 mutation without (■) and with IR (▲). CLL cases with a deletion of 17p show a severely reduced level of miR-34a expression with and without IR. Similarly, cases with TP53 mutation but no 17p deletion showed failure to up-regulate miR-34a after IR. The 2 cases with “normal” up-regulation had a wild-type TP53 allele in 50% to 70% of cells (* case with 70% wild-type allele [c.455C > T], ** case with 50% wild-type allele [c.847C > T]), likely explaining the up-regulation of miR-34a after IR. Cases with 11q deletion did not show different basal expression or induction compared with cells without 17p-, 11q-, or TP53 mutation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/16/10.1182_blood-2008-08-172254/7/m_zh80020929210002.jpeg?Expires=1768075018&Signature=RG92s~L7vNX3wPasprJtxGbkBLzLlKF5~ii-TWAXu~siZAaGs0Ddc393NjjVNlm4Pwu6sitragYQiHVav0WCmKGx1hIbKM-wd6g7VcxTzuIvZp~L3se5fhUE5DSuSPjpdKLMxJPf-wYZZNaurMAmqeeK1gNSMmBqOUhg843j2sUbKEfnE~q9V71ZSrCuGISnBjnJqYyhvfM1cNW8SANNgaoEkMXhSgkgNOHRA8JbHUlabkfCIJA4-zgkPTWOKuqru7cfWBTV0qmVncyzExheGZfoPr5~3drTbHZ87ICSFRYnecjIIk9~OE6xWdKM4Own20S0VtinORfJx5gGIdLlwg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 1. miR-34a is induced in CLL after IR, and 17p deletion is associated with defective induction. (A) CLL with 17p deletion (n = 3) show impaired miR-34 up-regulation over time. In comparison, CLL with favorable genetics (13q deletion as sole abnormality; n = 3) show miR-34a up-regulation peaking around 32 hours after IR. Values are expression levels relative to the nonirradiated sample (normalized for RNA by U6 control). (B) Relative expression levels of miR-34a were low in cases with 17p deletion with and without IR. The comparison to all other cases showed significantly lower expression of miR-34a in cases with 17p deletion (n = 12) versus all other cases (n = 48; median, 2.5 vs 32.8; P < .001). Similarly, up-regulation of miR-34a was negligible compared with the majority of cases without 17p deletion (median, 5.8 [1.38-12.9] vs 121.8 [1.79-608.9]; P < .001).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/16/10.1182_blood-2008-08-172254/7/m_zh80020929210001.jpeg?Expires=1768809754&Signature=oPxVz~DY7KfdpM62Ltpukcc7q75moYyihgxRV7wvC5JCa0HdxRpY43J4YoUlBFG5o5K~jB8a8eQiT2AXJAQwCLpemJat3ayRmc~l2DXkx8w3vBp1QMGLSAOPidOfxXJZIQ6uX~0ZKsiPmLGZKyqPy5tP8k7OUcOC1srE9~SnyYL0J4YLopb4WgWqpqViOc8ptla8n3~-Y6N77hIfspQj26shTCIF4FB3NNR6G1hqIe8qkdHTv5ETiYfUHN6eqU9bFWYMpEc17BwCq3LNOwgUnCMnJ7LnjJ~16xxS57rnhrzin3ZJGjoQN43t~v-BXz1l64iQYtrbeoLA7zauDrLGFw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. CLL exhibits differential miR-34a expression based on genetic background. Relative expression levels of miR-34a are shown in cases stratified by genomic aberrations and TP53 mutation without (■) and with IR (▲). CLL cases with a deletion of 17p show a severely reduced level of miR-34a expression with and without IR. Similarly, cases with TP53 mutation but no 17p deletion showed failure to up-regulate miR-34a after IR. The 2 cases with “normal” up-regulation had a wild-type TP53 allele in 50% to 70% of cells (* case with 70% wild-type allele [c.455C > T], ** case with 50% wild-type allele [c.847C > T]), likely explaining the up-regulation of miR-34a after IR. Cases with 11q deletion did not show different basal expression or induction compared with cells without 17p-, 11q-, or TP53 mutation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/16/10.1182_blood-2008-08-172254/7/m_zh80020929210002.jpeg?Expires=1768809754&Signature=l0Hv6C-MV5k6qwdEFqzHSiDBZ6wXhKApYztEJkJrFi5CvU2B9dkBdKJy6ioZ7FbSiVNf5tVQOyeQt5XT9fhvRKR~bBoIYBlfRi-lXFwcXkAZ~tXbxkCbYt-n6yMxCzols1L-6sbiDxEYR0UMzhoU-35y1M2qkGMsECmBtbLoc5xWe5Ej53RkzyjYc310jJacM8Qzp974B3pWQtB-FUgWaX8Q-EEGFOUuYPbJnQr1bMzcEWFJe-5~Y0QU1U27zlbecFAdZTgvutVwbxykdVY6w8fprpRZuDXMV-v~DuSM3lLLh0tQR3SEx~9j1eTmMydp~D6UBZFOtXK83fKADGEegA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)