Abstract
Decitabine (also referred to as 5-aza-2′-deoxycytidine) is a drug that has recently been approved by the Food and Drug Administration (FDA) for the treatment of myelodysplastic syndrome (MDS). The mechanism of action is believed to be the blocking of DNA methylation and thereby reactivating silenced genes involved in harnessing MDS. When analyzing reactivation of genes involved in Burkitt lymphoma (BL), we discovered that decitabine also sensitizes tumor cells by inducing DNA damage. This sensitization is grossly augmented by the MYC oncogene, which is overexpressed in BL, and occurs in cells lacking a functional p53 tumor suppressor pathway. In p53-deficient BL cells and p53−/− mouse embryo fibroblasts, Myc overrides a transient G2-block exerted by decitabine via activation of Chk1. This triggers aneuploidy and cell death that correlates with, but can occur in the absence of, Epstein-Barr virus (EBV) reactivation, caspase activation, and/or expression of the BH3-only protein Puma. In vivo modeling of Myc-induced lymphoma suggests that decitabine constitutes a potential new drug against lymphoma that would selectively sensitize tumor cells but spare normal tissue.
Introduction
c-Myc is encoded by the founding member of the MYC family of oncogenic transcription factors that also includes MYCN and MYCL. Overexpression of one of these oncogenes is seen in most human cancers and represents the most frequently deregulated genetic event in cancer. The reason for the recurrent selection for Myc activation in cancer is its biologic activities, which include many of the hallmarks of cancer such as stimulation of cell proliferation, inhibition of anti-growth signals and differentiation, induction of angiogenesis and even immortalization. Paradoxically though, Myc also triggers apoptosis,1,2 which is a cellular defense against cancer. For that reason, mutations in genes governing or executing apoptotic pathways are always accompanying Myc mutations in tumors.3
The tumor suppressor p53 is crucial in the defense against activated Myc oncogenes and it can be activated in 2 major ways: one pathway involving the Arf tumor suppressor and one involving the upstream kinases of the DNA damage response (DDR), ATM/ATR and DNA-PK. How Myc induces Arf4 is not completely understood, but when it does, Arf inhibits the E3 ubiquitin ligase Mdm2, which is a transcriptional target of p53 and acts as its negative regulator.5 By inhibiting Mdm2, Arf causes p53 accumulation, resulting in the activation of genes downstream of p53, such as those encoding the cell-cycle inhibitor p21, DNA repair proteins, or proapoptotic proteins of the Bcl-2 family, like Noxa and Puma.5 A similar scenario is the end result of Myc activating a DDR, where unscheduled Myc-induced replication causes strand breaks leading to activation of ATM, ATR or DNA-PK.6 Downstream of these are the Chk1/2 kinases, which regulate the phosphorylation of cyclin:Cdk complexes to transiently arrest cells in the S and G2 phases of the cell cycle. They also phosphorylate p53, which disrupts the interaction with Mdm2, resulting in p53 activation and a more prolonged arrest.7 Importantly, however, because high levels of Myc can repress p21 gene transcription,8,9 the outcome of p53 activation by both the Arf and the DDR pathways, in the context of Myc overexpression, tips the balance toward apoptosis rather than cell-cycle inhibition.8,10 This may explain why oncogene-induced senescence seen in many precancerous lesions is rarely seen in Myc-induced tumors, but may be triggered when Myc itself is inactivated.11
Genetic studies in mice have shown that the pathways triggering p53 described above are essential for keeping Myc in check but they are not mutually exclusive. For instance, inactivation of Arf or Atm in Myc-transgenic mice that normally develop B-cell lymphoma (eg, Eμ-Myc mice) results in a dramatic acceleration of disease, but inactivation of p53 is even more deleterious, suggesting that both Arf and DDR pathways are important.12-15 Interestingly, analyses of B-cell lymphomas that develop in Myc-transgenic mice and in the human disease Burkitt lymphoma (BL), where Myc is overexpressed in B lymphocytes because of translocation of MYC to any of the 3 immunoglobulin-encoding loci, have shown that Arf and p53 mutations occur spontaneously during tumor development.12,13,16,17 Besides p53, other genes, such as members of the Bcl-2 family, are also implicated as regulators of Myc-induced apoptosis.3
Methylation of cytosines in CpG sequences of promoter regions is commonly observed during tumor progression to inactivate genes whose products are important for processes such as DNA repair, cell-cycle regulation, cell adhesion, angiogenesis and apoptosis.18 Thus, strategies to inactivate DNA methyltransferases, causing reactivation of silenced cancer genes, hold promise for use in the clinic.19 5-Aza-2′-deoxycytidine (decitabine, marketed as Dacogen) is one of the most frequently used tools when investigating CpG DNA methylation and since 2006 has been a Food and Drug Administration (FDA)–approved treatment for myelodysplastic syndrome (MDS) and chronic myelomonocytic leukemia.20,21 When using this compound to examine the role of reactivated apoptotic genes in Burkitt lymphoma, we noted that decitabine elicited a DNA damage response leading to cell death of Myc-expressing cells via p53-independent pathways. We propose that decitabine may be used in a wider spectrum of cancer diseases because it can operate both as a classic DNA damaging drug and as a demethylating drug. Clinically relevant mouse models are used here to illustrate the former.
Methods
Materials
Decitabine (5-aza-2′-deoxycytidine) was purchased from Sigma-Aldrich (St Louis, MO) or was generously received from MGI Pharma (Johnson & Johnson Pharmaceutical Research & Development, a division of Janssen Pharmaceutica, Beerse, Belgium). Pan-caspase inhibitor Q-VD-OPH, the Caspase-2 inhibitor ZDVAD-FMK, and the Caspase-2 colorimetric kit were all from Biovision (Mountain View, CA), whereas the in vivo active PFTα prodrug22 was from Calbiochem (#506152; San Diego, CA). Primary antibodies were obtained from Abcam (Puma; Cambridge, MA), Dako (human p53; Glostrup, Denmark), Santa Cruz Biotechnology (mouse p53, p21; Santa Cruz, CA), Sigma-Aldrich (β-actin), Millipore (γ-H2AX; Billerica, MA), Nordic Biosite (Chk1; Täby, Sweden) and Cell Signaling Technology (Caspase-9, NBS, phosphorylated (p-)NBS, p-p53, p-Chk1, Cdc2 and p-Cdc2; Danvers, MA). Horse-radish peroxidase-conjugated antibodies against mouse and rabbit antibodies were from GE Healthcare Life Sciences (Piscataway, NJ), whereas the Cy5-conjugated anti–mouse antibody was from Jackson Immunoresearch (West Grove, PA).
Cell culture
293T human kidney cells were purchased from ATCC (Manassas, VA) and cultured in Dulbecco Modified Eagle Medium (DMEM) with 10% fetal calf serum (FCS), 2 mM l-glutamine, 1 mM sodium pyruvate, and antibiotics. Burkitt lymphoma cell lines and mouse lymphoma lines established from tumors that arose in λ-Myc transgenic mice were cultured at a density between 105 and 106 cells/mL in RPMI 1640 medium with 10% FCS, 2 mM l-glutamine, 50 μM β-mercaptoethanol, 0.1875% sodium bicarbonate and antibiotics. Mouse embryo fibroblasts (MEFs) were generated from timed matings between p53 heterozygous mice,23 according to previous methodology.4 All experiments were repeated using multiple MEF isolates.
Viral infections
Retroviruses were made by calcium phosphate-mediated cotransfection of 293T cells with MSCV-IRES-puro (with or without MycER) together with ecotropic helper plasmids expressing gag, pol, and env. Twenty-four hours after transfection, culture supernatants were harvested every 8 hours, pooled, filtered, and used to infect MEFs in the presence of 8 μg/mL polybrene. Cells infected with MSCV-IRES-puro-based retroviruses were selected in the presence of 4 μg puromycin.
Lentiviral infections were made by calcium phosphate-mediated cotransfection of 293T cells with packaging plasmids pCMV-dR8.2 dvpr and pCMV VSV-G (Addgene, Cambridge, MA) plus either one shRNA constructs (Sigma-Aldrich) directed against the human Puma mRNA or against the mouse caspase-2 mRNA. Twenty-four hours after transfection, the different supernatants were harvested 3 times every 8 hours, filtered, and used to infect Akata cells or λ#820 by 3 rounds of spinoculation (20 minutes at 50g) during 24 hours in the presence of 8 μg/mL polybrene. The cells were selected by culturing them in the presence of 1 (λ#820) or 2 (Akata) μg/mL puromycin.
Cell-cycle and apoptosis analyses
For cellular staining with propidium iodine (PI), lymphoma cells were collected by centrifugation and MEFs were trypsinized and collected by centrifugation together with its original culture supernatant. The cells were resuspended in 0.5 mL of phosphate-buffered saline (PBS) containing 50 μg/mL PI and 0.5 mL of Vindelövs reagent (10 mM Tris, 10 mM NaCl, 75 μM propidium iodine, 0.1% Igepal, and 700 U/L RNase adjusted to pH 8.0). The PI-stained cells were kept in the dark at 4°C for 30 to 60 minutes and then analyzed with a FACScalibur flow cytometer (BD Biosciences, San Jose, CA) using the FL3 channel in a linear scale. To distinguish between G2 and M phase, a method based on drop in granularity (side scatter) in M phase was used.24
Apoptosis was determined using DNA histograms on PI-stained cells (as above) and was based on the number of cells that carried one log less than diploid DNA content (sub-G1) in the FL2 channel in a logarithmic scale. Apoptosis was also measured by flow cytometry using annexin V conjugated to R-phycoerythrin (PE; BD Biosciences) that labels phosphatidylserine that flips to the outside of apoptotic cells, and 7-aminoactinomycin D (7-AAD; Sigma-Aldrich) that labels DNA, as per BD Biosciences' recommendation.
Western blot analysis
Cell pellets or tumors crushed in liquid nitrogen were lysed essentially as described before.4 Protein was separated on sodium dodecyl sulfate– polyacrylamide gel electrophoresis (SDS-PAGE) gels and subsequently transferred to nitrocellulose membranes. Antibody staining was carried out in TBS-Tween and antibody-binding was visualized by enhanced chemiluminescence using SuperSignal from Pierce (Rockford, IL).
RNA preparation and analysis
RNA from cultured cells was isolated using Trizol reagent (Invitrogen, Carlsbad, CA). cDNA synthesis was performed on 1 μg RNA using iScript first strand synthesis kit (Bio-Rad, Hercules, CA). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using the SYBRgreen PCR mastermix (Biotools, Madrid, Spain), cDNA and primers directed against Puma (human), caspase-2 (mouse), and Ubiquitin (Ub; human and mouse) on an IQ real-time PCR machine (Bio-Rad). Relative mRNA levels were calculated using the ΔΔCT method, comparing expression of Puma or caspase-2 with that of Ub.
Immunofluorescence
BL cells were treated with 10 Gy of γ-IR or 2 μM decitabine, and 24 or 48 hours after the initiation of treatment, they were harvested by cytospin onto glass slides. The cells were overlaid with a PBS solution containing 4% paraformaldehyde and 0.2% Triton X-100 for 20 minutes to fix and permeabilize and then washed in PBS and blocking solution (5% FCS in PBS). After 1 hour of blocking, primary γ-H2AX antibody in solution was added, and the slides were incubated overnight at 4°C in a moisture chamber. The cells were washed and incubated with a fluorescein isothiocyanate (FITC)–conjugated secondary antibody and subsequently stained with Hoechst and mounted. The slides were analyzed by fluorescence microscopy.
Mouse experiments
All animal experiments were performed in accordance with the Regional Animal Ethic Committee Approval no. A6-08 or no. A18-08. The p53 knockout mice,23 iMycEμ,25 and λ-Myc mice,26 all on a C57/BL6 background, were obtained from The Jackson Laboratory (Bar Harbor, ME), the National Cancer Institute (NCI) mouse depository, and a kind gift from Dr Georg Bornkamm (GSF, Munich, Germany), respectively. C57/BL6 mice used for tumor transplants were from Taconics (Hudson, NY). All transgenic or transplanted mice were observed daily for signs of disease. Sick mice were either killed or treated with decitabine. When tumor-bearing mice were killed, tumors and lymphoid organs were collected for analyses or tissue banking. Tumors were either snap-frozen down as pieces and/or dispersed into single-cell suspensions and slowly frozen alive in freezing medium (B-cell medium, 40% FCS and 10% dimethyl sulfoxide [DMSO]).
Results
Decitabine causes aneuploidy and cell death in Burkitt lymphoma cells carrying mutations in TP53
Previous studies have shown that loss of the proapoptotic BH3-only protein Puma accelerates Myc-induced B-cell lymphomagenesis in mice and that a selection against Puma expression occurs in BL of humans.27,28 In BL, methylation of the PUMA gene was observed in many primary tumors and cell lines, correlating with low expression of Puma. Treatment of these lines with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (decitabine) caused a reactivation of Puma expression, demonstrating that methylation was needed for PUMA gene silencing.28 We wanted to evaluate if Puma reactivation was the sole cause of the apoptosis that results from decitabine treatment of KemI and Akata BL cells, which both exhibit PUMA methylation. Using lentiviruses carrying shRNA directed against PUMA, we were able to knockdown the expression of Puma RNA and protein in the Akata cell line (Figure S1A,B, available on the Blood website; see the Supplemental Materials link at the top of the online article). Interestingly, Puma knockdown reduced the level of caspase-9 activation and annexin V positivity, hallmarks of the mitochondrial apoptosis regulated by Puma,29 but it did not prevent cell death completely, and in long term experiments, it had no effect at all (Figure S1B,C, and data not shown). Therefore, Puma is in part a mediator of the apoptotic response to decitabine, but Puma-independent death is also involved.
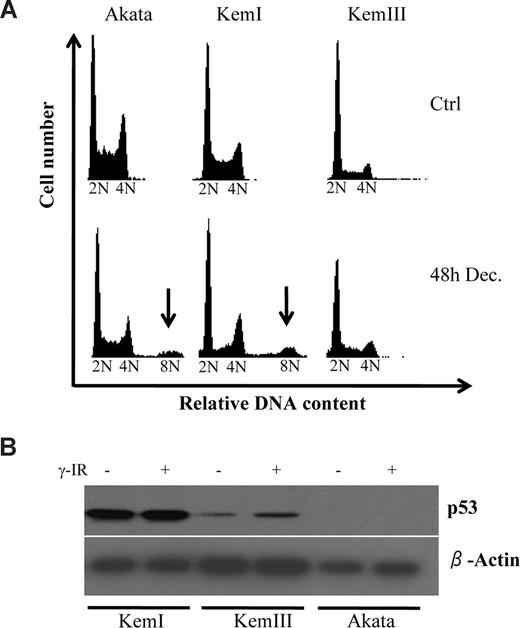
To gain more insight into decitabine-induced cell death in BL cells, we analyzed these cells by fluorescence-activated cell sorting (FACS) to produce DNA histograms. One of the most striking things noted besides the considerable sub-G1 population that had accumulated 48 hours after decitabine treatment, was the appearance of cells carrying much higher than 4N content (Figure 1A), indicative of aneuploidy. This was seen in both Akata and KemI cells, but not in KemIII cells, which originate from the same patient as KemI cells. Because KemIII cells have a latency III Epstein-Barr virus (EBV) program activated, we reasoned that this sub-line had been able to propagate in culture because of the antiapoptotic functions of EBV. This would bypass the selection for mutations of the TP53 tumor suppressor gene that most BL cells undergo after some time in culture.30-32 Indeed, Western blot analysis shown in Figure 1B confirms that KemI carries a mutant TP53,28 because p53 protein level was very high and did not exhibit the expected increase by DNA damage after γ-irradiation (γ-IR). Akata cells did not express any p53 at all, confirming that they have a truncation mutation in the TP53 gene.32 Finally, KemIII cells expressed low amounts of p53, but the levels increased upon DNA damage, characteristic for cells carrying a wild-type TP53 gene.33
Decitabine induces aneuploidy in p53 deficient Burkitt lymphoma (BL) cells. (A) The BL cell lines Akata, KemI, and KemIII were treated with 2 μM decitabine (Dec) for 48 hours. The cell-cycle distribution was determined by flow cytometry of cells stained with propidium iodide. Cells regarded as aneuploid are indicated by arrows. (B) TP53 status correlates with presence of decitabine-induced aneuploidy of BL cells. Cells were irradiated with 10 Gy of ionizing radiation (γ-IR) and harvested 24 hours after treatment for analysis with Western blot using a monoclonal antibody against p53. The KemI cell line exhibits abnormally high levels of p53 irrespective of DNA damage, which is indicative of a TP53 mutant status. KemIII cells induce p53 after γ-IR treatment, as predicted by cells carrying wild-type TP53. Akata cells do not express p53, as previously shown.32
Decitabine induces aneuploidy in p53 deficient Burkitt lymphoma (BL) cells. (A) The BL cell lines Akata, KemI, and KemIII were treated with 2 μM decitabine (Dec) for 48 hours. The cell-cycle distribution was determined by flow cytometry of cells stained with propidium iodide. Cells regarded as aneuploid are indicated by arrows. (B) TP53 status correlates with presence of decitabine-induced aneuploidy of BL cells. Cells were irradiated with 10 Gy of ionizing radiation (γ-IR) and harvested 24 hours after treatment for analysis with Western blot using a monoclonal antibody against p53. The KemI cell line exhibits abnormally high levels of p53 irrespective of DNA damage, which is indicative of a TP53 mutant status. KemIII cells induce p53 after γ-IR treatment, as predicted by cells carrying wild-type TP53. Akata cells do not express p53, as previously shown.32
Decitabine-induced cell death and aneuploidy does not only occur in EBV-positive B-cell lymphoma lines
Previous studies have shown that a switch from latency I into latency III and the lytic EBV program can be achieved by culturing EBV positive BL lines in the presence of decitabine.34 Because both Akata and Kem I cells are EBV positive, we wanted to elucidate if viral proteins were responsible for the observed Puma-independent cell death and/or aneuploidy. To that end, we compared the phenotype of a large panel of B-cell lymphoma lines that varied in their EBV and p53 status to find possible correlations between these parameters. As can be seen in Figure S2, decitabine caused aneuploidy in 3 of the 4 EBV positive lines (Akata, Raji and P3HRI) and in both the mouse lymphoma lines (λ#820 and λ#1061), but only weakly in the EBV positive line Daudi, which can elicit a wild-type p53 response. Cell death was observed in all lines with modest effects observed in Daudi and DG75, which can induce p21 accumulation in response to DNA damage and, therefore, appear arrested in response to decitabine (Figure S2).
Although the mouse lymphoma lines also exhibited aneuploidy, indicating that this feature can occur independently of the human-specific EBV, we reasoned that the virus may still play a role in potentiating aneuploidy in the human cells that were devoid of p53 function. We therefore analyzed the expression of some of the EBV genes that have been shown to be induced by decitabine and whose products may exert a function in regulating apoptosis or aneuploidy. The mRNAs encoding the master transcriptional regulator EBNA-2 and its downstream target genes LMP-1, LMP-2, and ZEBRA (BZLF-1) were all induced by decitabine at the time point when aneuploidy was observed. However, only ZEBRA was induced to protein levels similar to Raji and KemIII, which express a EBV latency III program (Figure S3A,B).
Decitabine causes a transient G2 arrest in BL cells with mutations in TP53
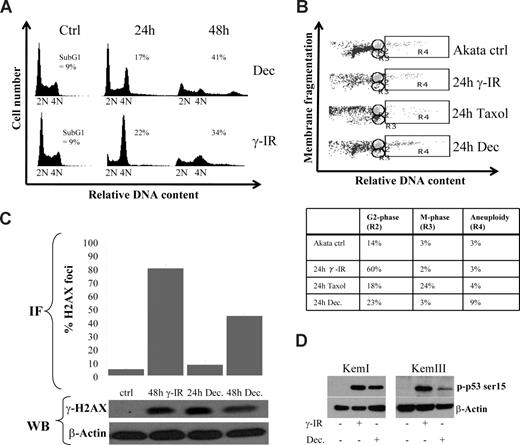
To set the stage for aneuploidy, we reasoned that one or several checkpoints had been overridden. Indeed, by analyzing Akata cells 24 hours after decitabine administration, an enrichment of cells arrested in the G2/M phase was observed that was dissolved at 48 hours, correlating with aneuploidy or cell death (Figure 2A). This is an event that very often can be seen in p53-deficient cells subjected to DNA damage, because in normal cells p53 and its downstream target p21 are dispensable for the rapid Chk1 and Chk2-mediated arrest but responsible for the prolonged G2 arrest.35-37 We therefore investigated if DNA damage by γ-IR treatment of Akata cells would result in a similar cell-cycle profile as decitabine treatment. The kinetics were far from identical, which is not surprising because γ-IR treatment results in an immediate DNA damage, whereas decitabine requires incorporation into DNA, but the basic events were similar, both treatments caused an arrest followed by aneuploidy or cell death. Using a flow cytometry method that is based on the fact that the nuclear envelope is disintegrated at mitosis causing a drop in granularity (side-scatter) of the cells,24 we were able to show that the transient arrest of Akata cells upon γ-IR or decitabine treatment was in G2 and not in M (Figure 2B).
Decitabine-induced aneuploidy and cell death is preceded by a DNA damage-induced transient G2 block. (A) Cell-cycle distribution of Akata cells 24 and 48 hours after 10 Gy of γ-IR or start of culture in the presence of 2 μM decitabine (Dec). The amount of cells with a less than diploid DNA content was regarded as apoptotic (ie, sub-G1, only number shown). (B) Decitabine and γ-IR treatment transiently arrests Akata cells in the G2 phase. To distinguish between the G2 and M phase of the cell cycle, loss of granularity due to nuclear envelope breakdown in mitosis was used during FACS analyses. To be able to set the M-phase gate correctly, Akata cells were treated with 1 μg/mL Taxol, which resulted in an accumulation of cells in gate R3. The remaining cells with the same DNA content but with higher granularity were collected in gate R2, depicted as the G2-phase. Quantifications of the cell-cycle distribution are shown in the table. (C) Decitabine treatment induces a DNA damage response (DDR). Akata cells were treated with either 5 μM decitabine or 10 Gy of γ-IR and analyzed for the presence of phosphorylated H2AX (γ-H2AX) in foci by immunofluorescence. The graph shows quantification of the percentage of cells with a signal that exceeds that of an arbitrarily chosen background staining. The results are based on quantifications of at least 100 cells counted from at least 3 different slides. A representative staining of a cell subjected to DNA damage is shown in Figure S4C. The bottom panel shows a Western blot analysis of lysates from cells treated with decitabine or γ-IR. The loading order is the same as the label on the graph. (D) Decitabine treatment leads to phosphorylation of p53 on serine 15. KemI and KemIII cells were treated with either decitabine or γ-IR and analyzed for phosphorylation of p53 serine 15. Both mutant (KemI) and wild-type (KemIII) p53 showed serine 15 phosphorylation, which is indicative of a DNA damage response.
Decitabine-induced aneuploidy and cell death is preceded by a DNA damage-induced transient G2 block. (A) Cell-cycle distribution of Akata cells 24 and 48 hours after 10 Gy of γ-IR or start of culture in the presence of 2 μM decitabine (Dec). The amount of cells with a less than diploid DNA content was regarded as apoptotic (ie, sub-G1, only number shown). (B) Decitabine and γ-IR treatment transiently arrests Akata cells in the G2 phase. To distinguish between the G2 and M phase of the cell cycle, loss of granularity due to nuclear envelope breakdown in mitosis was used during FACS analyses. To be able to set the M-phase gate correctly, Akata cells were treated with 1 μg/mL Taxol, which resulted in an accumulation of cells in gate R3. The remaining cells with the same DNA content but with higher granularity were collected in gate R2, depicted as the G2-phase. Quantifications of the cell-cycle distribution are shown in the table. (C) Decitabine treatment induces a DNA damage response (DDR). Akata cells were treated with either 5 μM decitabine or 10 Gy of γ-IR and analyzed for the presence of phosphorylated H2AX (γ-H2AX) in foci by immunofluorescence. The graph shows quantification of the percentage of cells with a signal that exceeds that of an arbitrarily chosen background staining. The results are based on quantifications of at least 100 cells counted from at least 3 different slides. A representative staining of a cell subjected to DNA damage is shown in Figure S4C. The bottom panel shows a Western blot analysis of lysates from cells treated with decitabine or γ-IR. The loading order is the same as the label on the graph. (D) Decitabine treatment leads to phosphorylation of p53 on serine 15. KemI and KemIII cells were treated with either decitabine or γ-IR and analyzed for phosphorylation of p53 serine 15. Both mutant (KemI) and wild-type (KemIII) p53 showed serine 15 phosphorylation, which is indicative of a DNA damage response.
Decitabine elicits a DNA damage response
Previous studies have shown that decitabine can induce effects other than DNA hypomethylation, including DNA damage.38,39 Thus, we sought to determine if a DNA damage response (DDR) was triggered in KemI and Akata cells that lack functional p53. Phosphorylation of histone H2AX (γ-H2AX) is one of the earliest and most reliable markers for an activated DDR40 and it is carried out by the proximal kinases ATM and ATR. γ-H2AX can be detected by immunofluorescence as foci at the site of lesion in the nucleus of damaged cells. We observed elevated levels of γ-H2AX in Akata (Figure 2C and Figure S4C) and KemI (not shown) cells using immunofluorescence, which was confirmed by Western blot analysis. Moreover, to further confirm that a DDR was elicited by decitabine, we also analyzed the phosphorylation status of p53 as a surrogate marker for an activated DDR. As seen in Figure 2D, serine 15 of p53 was strongly phosphorylated in KemI and KemIII cells treated with γ-IR and decitabine. Furthermore, other key mediators of the DDR like NBS1 and Chk1 were activated as measured by their phosphorylation status in cells treated with γ-IR and decitabine (Figure S4A-B). Taken together with the results from the cell-cycle analyses, we postulate that decitabine induces a DDR causing a transient Chk1- (or Chk2)–mediated growth arrest in p53-deficient BL cells, which is overridden, resulting in aneuploidy and cell death.
Myc is a sensitizer to decitabine
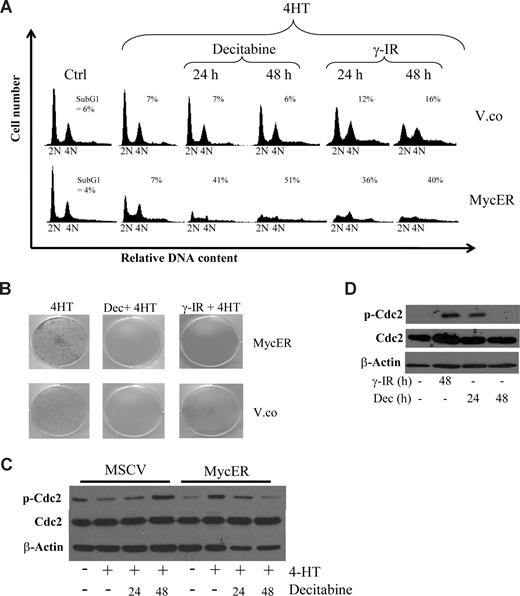
The KemI and Akata cells used in this study have been in culture for many decades so it was of interest to set up a system of primary cultures to see if we could mimic the effects with a minimal amount of genetic lesions. We used early-passage p53-deficient mouse embryo fibroblasts (MEFs) that were found to be sensitive to decitabine, but not as sensitive as the BL lines. We therefore transduced p53−/− MEFs with a control retrovirus (MSCV-IRES-puro) or one engineered to express a conditionally active form of c-Myc, the MycERTAM protein,41 to be able to assess the role of Myc in the sensitization of p53-deficient cells. MycERTAM encodes a fusion protein between c-Myc and the ligand binding domain of the estrogen receptor (ER). Addition of the estrogen analog 4-hydroxytamoxifen (4-HT) to the growth medium releases the MycERTAM protein from binding to heat-shock proteins (Hsp) in the cytoplasm and exposes the nuclear localization signal of Myc to induce nuclear translocation and Myc activation. Using this system, we activated Myc in cells that were untreated or had been treated with decitabine or γ-IR. As expected, activation of Myc resulted in an increase in the proportion of cells in S-phase (Figure 3A). However, concomitant treatment with decitabine or γ-IR resulted in an induction of apoptosis to a much higher degree in p53−/− MEFs carrying an activated Myc oncogene than in cells transduced with the MSCV-IRES-puro vector control retrovirus. Moreover, clonogenic survival assays over 10 days showed that p53−/− MEFs are sensitive to DNA damage by decitabine and γ-IR and that Myc overexpression kills the remaining cells (Figure 3B). Thus, the results show that it is not only loss of p53 function that sensitizes BL cells to DNA damage by decitabine, but also the active MYC oncogene.
Myc provokes G2 checkpoint override that sensitizes p53 knockout mouse embryo fibroblasts (MEFs) to decitabine-induced cell death. (A) Low passage p53−/− MEFs were infected with retroviruses made either with MSCV-IRES-puro (vector control) or with MSCV-MycER-IRES-puro vectors. After selection, the cells were cultured in the presence or absence of 4-HT, to induce nuclear translocation and activation of MycER, and either treated with 10 Gy γ-IR or 2 μM decitabine (Dec) for the indicated time points. The cells were harvested and stained with propidium iodide and their cell-cycle distribution was analyzed by FACS. (B) The same cells as in panel A were also seeded into 6-well plates and cultured for 10 days after γ-IR or decitabine. Cultures with freshly added decitabine daily for 3 or 10 days produced the same results and a representative experiment of 3 independent clonogenic survival assays is shown. (C) Western blot analysis showing levels of total and phosphorylated cyclin-dependent kinase 1 (Cdc2) in p53−/− MEFs subjected to decitabine and Myc activation for the indicated times. Less phosphorylated Cdc2 suggests checkpoint override. (D) Akata cells were treated with either decitabine or γ-IR and analyzed for phosphorylation of Cdc2 by Western blot analysis.
Myc provokes G2 checkpoint override that sensitizes p53 knockout mouse embryo fibroblasts (MEFs) to decitabine-induced cell death. (A) Low passage p53−/− MEFs were infected with retroviruses made either with MSCV-IRES-puro (vector control) or with MSCV-MycER-IRES-puro vectors. After selection, the cells were cultured in the presence or absence of 4-HT, to induce nuclear translocation and activation of MycER, and either treated with 10 Gy γ-IR or 2 μM decitabine (Dec) for the indicated time points. The cells were harvested and stained with propidium iodide and their cell-cycle distribution was analyzed by FACS. (B) The same cells as in panel A were also seeded into 6-well plates and cultured for 10 days after γ-IR or decitabine. Cultures with freshly added decitabine daily for 3 or 10 days produced the same results and a representative experiment of 3 independent clonogenic survival assays is shown. (C) Western blot analysis showing levels of total and phosphorylated cyclin-dependent kinase 1 (Cdc2) in p53−/− MEFs subjected to decitabine and Myc activation for the indicated times. Less phosphorylated Cdc2 suggests checkpoint override. (D) Akata cells were treated with either decitabine or γ-IR and analyzed for phosphorylation of Cdc2 by Western blot analysis.
To gain insight into the mechanism behind the checkpoint override by Myc and the ensuing apoptosis, we entertained the following option: Chk1 is activated by DNA damage causing a G2 arrest,42,43 which is overridden because of Myc's transcriptional induction of the G2-M regulators Cdc25a-c and M-phase promoting factor (MPF; cyclin B and Cdc2). To get an indication if this was the case, we queried the Myc Target Gene database and the genomic sequences of these regulators and found that many of them appeared to be direct Myc targets (www.myccancergene.org).44 As a further validation of this hypothesis, decitabine caused an inactivating phosphorylation of Cdc2 in p53−/− MEFs, which is known to involve Chk1 and the phosphatase Cdc25c.42,43 In cells overexpressing Myc, on the other hand, Cdc2 phosphorylation was transient, correlating with the premature G2/M transition (Figure 3C). This transient endurance of phosphorylated Cdc2 upon decitabine-induced DDR was also confirmed in Akata cells (Figure 3D).
Myc sensitizes cells to decitabine by inducing cell death pathways that involves caspases
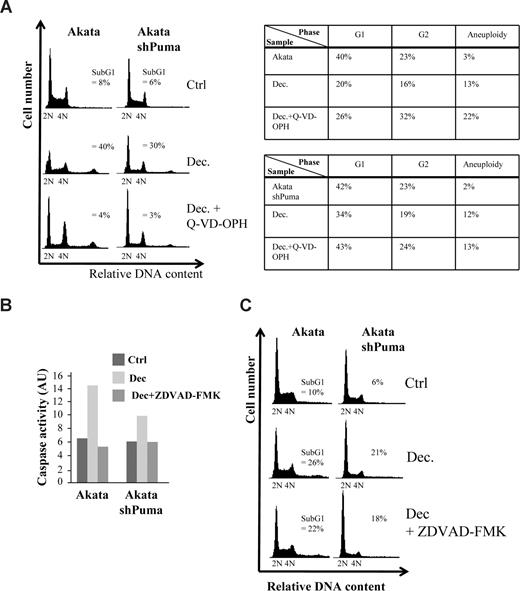
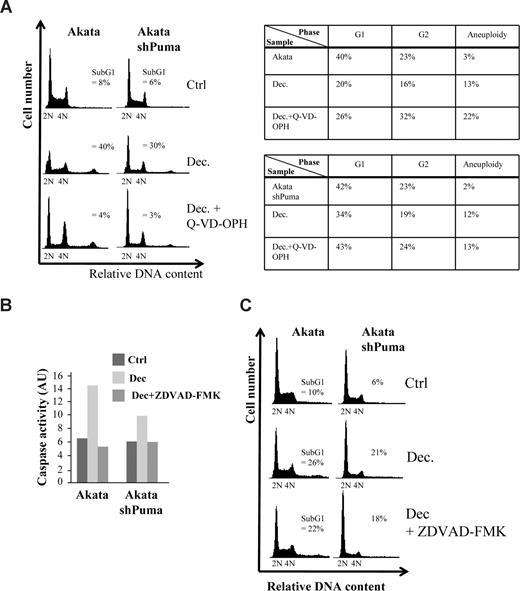
To narrow down the search for the mechanism of cell death, we decided to investigate if the cell death was caspase-dependent. When pretreating Akata cells or Akata cells expressing a shRNA against Puma with the pan-caspase inhibitor Q-VD-OPH, decitabine-induced caspase-9 cleavage and cell death was completely abolished (Figure 4A and data not shown). Interestingly, most of the cells that survived either became aneuploid or were arrested in G2 (Figure 4A table inset), suggesting that checkpoint override into lethal mitosis is the cause of part of the decitabine-induced cell death. Because knockdown of Puma did not result in a G2-arrest or aneuploidy of the surviving cells from decitabine treatment, we reason that Puma is not involved in the mitotic death resulting from checkpoint override and that several forms of cell death are induced by decitabine. Indeed, when using another Myc-induced lymphoma line, λ#820 cells that originated from a p53-deficient primary tumor that arose in λ-Myc transgenic mice, Q-VD-OPH did not completely block cell death, suggesting that even caspase-independent death can occur in response to decitabine (Figure S5A).
Decitabine-induced cell death in Akata cells is caspase-dependent but can occur in the absence of induced caspase-2 activity. (A) Akata cells, or Akata cells expressing an shRNA against Puma (Figure S1) and/or were pretreated with 5 μM of the pan-caspase inhibitor Q-VD-OPH, were grown in the presence or absence of 2 μM decitabine (Dec). Apoptosis (= sub-G1), G1, G2 and aneuploidy (> 4N) were measured by FACS analysis of PI-stained cells and plotted in the table (inset) or DNA histograms. (B) Caspase-2 activity of Akata cells or Akata cells expressing an shRNA against Puma treated or untreated with decitabine was measured by a colorimetric method as measured by the cleavage of the caspase-2 substrate VDVAD-pNA into the chromophore p-nitroanilide (pNA). The specificity of the assay was ascertained by pretreating (1 hour) and culturing the cells in the presence of 20 μM of the caspase-2 inhibitor Z-VDVAD-FMK. The arbitrary units (AU) were calculated by multiplying the absorbance minus background measured at 400 nm with 100. (C) DNA histogram showing that blocking Puma and/or caspase-2 (with Z-VDVAD-FMK) results in a very small decrease in the amount of apoptosis (= sub-G1).
Decitabine-induced cell death in Akata cells is caspase-dependent but can occur in the absence of induced caspase-2 activity. (A) Akata cells, or Akata cells expressing an shRNA against Puma (Figure S1) and/or were pretreated with 5 μM of the pan-caspase inhibitor Q-VD-OPH, were grown in the presence or absence of 2 μM decitabine (Dec). Apoptosis (= sub-G1), G1, G2 and aneuploidy (> 4N) were measured by FACS analysis of PI-stained cells and plotted in the table (inset) or DNA histograms. (B) Caspase-2 activity of Akata cells or Akata cells expressing an shRNA against Puma treated or untreated with decitabine was measured by a colorimetric method as measured by the cleavage of the caspase-2 substrate VDVAD-pNA into the chromophore p-nitroanilide (pNA). The specificity of the assay was ascertained by pretreating (1 hour) and culturing the cells in the presence of 20 μM of the caspase-2 inhibitor Z-VDVAD-FMK. The arbitrary units (AU) were calculated by multiplying the absorbance minus background measured at 400 nm with 100. (C) DNA histogram showing that blocking Puma and/or caspase-2 (with Z-VDVAD-FMK) results in a very small decrease in the amount of apoptosis (= sub-G1).
Besides stimulating M-phase entrance by inducing G2/M regulators (see above), Myc was recently shown to have an inhibitory role in prometaphase by transactivating the mitotic regulators BubR1 and MAD2, which was linked to Myc's propensity to trigger aneuploidy.45 Because Caspase-2 is believed to be one of the initiator caspases responsible for triggering apoptosis upon “mitotic catastrophe”,46 we measured the activity of this caspase in decitabine-treated cells. As seen in Figure 4B, decitabine was able to induce Caspase-2 activity in Akata cells by 48 hours. Interestingly, Akata cells expressing a shRNA directed against Puma mRNA (Figure S1) also activated Caspase-2 when treated with decitabine, but to a slightly lesser extent. We then pretreated the cells with the Caspase-2 inhibitor Z-VDVAD-fmk, which accordingly blocked Caspase-2 activation (Figure 4B). The inhibitor, however, only blocked a very small proportion of cell death in Akata cells (5% ± 2%, P < .05), suggesting that although Caspase-2 was induced, cell death can occur in its absence (Figure 4C). Similarly, lentiviral-mediated shRNA knockdown of caspase-2 mRNA or pretreatment with Z-VDVAD-fmk suggested that Caspase-2 is only partly responsible for the caspase-dependent cell death of λ#820 cells treated with decitabine (Figure S5B-D). Taken together, our data suggest that Puma and/or caspase-2 both participate in caspase-dependent and decitabine-induced cell death but other mechanisms of cell death must be involved.
Clinically relevant mouse models of Myc-induced lymphoma respond to decitabine
We sought to analyze if decitabine would be effective against primary tumor cells in mouse models of Myc-induced lymphomas. We turned to the iMycEμ mouse model that carries a Myc gene targeted to the immunoglobulin heavy chain locus via homologous recombination.25 To create a p53-deficient model, we inter-crossed iMycEμ mice with p53 knockout mice,23 thus generating iMycEμ; p53+/− mice that were followed until they developed tumors. As expected, p53 heterozygous iMycEμ mice developed disease at a grossly accelerated pace (from median survival 198 days to 48 days, P = .001) compared with regular iMycEμ mice housed at our facility (Figure 5A), which has also been seen in another Myc-induced cancer model, the Eμ-Myc mouse.12,13 The tumors that developed were Myc-expressing B-cell lymphomas (B220+) that had infiltrated many lymphoid organs such as spleen, bone marrow, and lymph nodes, and they were either IgM positive or negative, as previously reported for the parental iMycEμ mouse model25 (Figure 5D and Figure S6A,B). Western blot analysis showed that the p53 protein was either lost (LOH) or elevated, indicative of a de novo mutation (Figure 5B). Tumors with compromised p53 function were transplanted into recipient C57/BL6 mice to create colonies of mice that developed the same disease as the donor mouse within 3 to 4 weeks. The colonies were divided into groups that were either left untreated or treated 1 week after transplantation with decitabine. Interestingly, decitabine showed very potent efficacy with just 3 injections during 24 hours (Figure 5C), delaying tumor onset from 23 to 34 days (P = .001). We were also able to cause remission in mice that were injected with decitabine on the day the mice showed signs of palpable lymphoma (data not shown), but the tumors relapsed eventually in all mice. The tumors that arose were not secondary tumors arising because of decitabine treatment, but were Myc-expressing B-cell lymphomas like the original tumors (Figure 5D).
Decitabine treatment delays tumor formation in vivo. (A) iMycEμ mice were bred to p53 knockout mice to produce offspring that were Myc-transgenic and heterozygous for p53. These mice exhibited an accelerated course of disease (median survival 48 days) compared with the iMycEμ mice previously born in the laboratory (dashed line, median survival 198 days). The survival curves and the statistical analyses were performed using the Survival curve function of the GraphPad Prism software (San Diego, CA). (B) Tumors arising in iMycEμ; p53+/− mice were frozen down as live cells, but pieces were also snap frozen for analyses. The snap frozen tumor pieces were analyzed for their p53 status by PCR genotyping and Western blot analysis (WB). In the PCR, intensities between the top mutant and the bottom wild-type bands were compared with assess loss of heterozygosity (LOH). High amount of p53 protein on the WB indicates mutant protein (*). (C) Vials of live cells were thawed, and the tumor cells were transplanted into the tail veins of 12 recipient C57/BL6 mice. One week after transplant, 6 mice were treated with 3 injections of 5 mg/kg decitabine with 8-hour intervals. The survival curve was generated and analyzed using GraphPad Prism and is a representative of transplantation experiments of 3 different tumors. (D) Tumors arising in transplanted mice were analyzed for the expression of c-Myc protein levels by Western blot analysis.
Decitabine treatment delays tumor formation in vivo. (A) iMycEμ mice were bred to p53 knockout mice to produce offspring that were Myc-transgenic and heterozygous for p53. These mice exhibited an accelerated course of disease (median survival 48 days) compared with the iMycEμ mice previously born in the laboratory (dashed line, median survival 198 days). The survival curves and the statistical analyses were performed using the Survival curve function of the GraphPad Prism software (San Diego, CA). (B) Tumors arising in iMycEμ; p53+/− mice were frozen down as live cells, but pieces were also snap frozen for analyses. The snap frozen tumor pieces were analyzed for their p53 status by PCR genotyping and Western blot analysis (WB). In the PCR, intensities between the top mutant and the bottom wild-type bands were compared with assess loss of heterozygosity (LOH). High amount of p53 protein on the WB indicates mutant protein (*). (C) Vials of live cells were thawed, and the tumor cells were transplanted into the tail veins of 12 recipient C57/BL6 mice. One week after transplant, 6 mice were treated with 3 injections of 5 mg/kg decitabine with 8-hour intervals. The survival curve was generated and analyzed using GraphPad Prism and is a representative of transplantation experiments of 3 different tumors. (D) Tumors arising in transplanted mice were analyzed for the expression of c-Myc protein levels by Western blot analysis.
As shown above, p53-deficient mouse and human tumor cells are sensitive to decitabine, but are they more sensitive than cells with functional p53? To test this, we decided to inhibit p53 function with the p53 inhibitor pifithrin-α (PFTα)47 in p53 proficient Myc-induced mouse lymphomas to see if this sensitized them to decitabine. As seen in Figure S7A and B, treating p53 proficient lymphomas with decitabine resulted in remission. Strikingly though, inhibition of p53 with PFTα resulted in a statistically significant protracted onset of tumor development (P = .002). The tumors that arose were not of the same size or predominant localization as in the mice treated only with decitabine, which was not due to changes in expression of c-Myc or p53 (Figure S7C,D).
Discussion
Patients suffering from BL are often responsive to chemotherapy, despite the fact that these tumors frequently carry TP53 mutations. Based on our previous study,10 and data presented herein, we would argue that this responsiveness is due to the combined over-activity of the MYC oncogene, p53 loss, and DNA damage that leads to several different types of cell death. Because decitabine has potential to both demethylate genes and induce DNA damage, perhaps introduction of decitabine and thereby replacing some DNA damaging drugs in anti-cancer regimes will result in improved treatment responses of Myc-induced cancers, such as leukemia/lymphoma, colon, prostate and breast cancer.
The in vitro findings presented here extend and confirm c-Myc's cell-cycle effects to include not only S-phase progression, but also the transition through the G2 and M phases. This is a less appreciated role of c-Myc, which in the case of cancer cells, may play dual and opposite roles; to enhance aneuploidy leading to transformation and to sensitize the transformed cells to DNA-damaging drugs that enforce an arrest in these cell-cycle phases. Myc was recently shown to have an inhibitory role in prometaphase by transactivating the mitotic regulators BubR1 and MAD2, which was linked to Myc's propensity to trigger aneuploidy.45 Concomitant activation of G2/M regulators such as Cdc2:cyclin B may make matters worse by forcing cells into a poorly functional mitosis. If the cells survive, they may end up in a tetraploid G1 phase, which is sensed by a poorly characterized p53-dependent checkpoint consisting of mitotic regulators and the tumor suppressor Lats2.48 Here, in cells lacking p53, this checkpoint is abolished, resulting in a small fraction of cells surviving up to 8N content, making this system a tractable model for studying pathways that govern polyploidization. Because both p53-deficient EBV-positive BL cells and mouse lymphoma cells exhibit the aneuploid cell population, it is possible that EBV may disturb a pathway that is also selected against during Myc-induced lymphomagenesis in the mouse.
EBV is tightly connected to the endemic form of BL (eBL), but the role of the virus in the genesis or maintenance of the BL phenotype is still not completely understood. Once formed, most eBL tumors express only a small number of EBV genes known as the latency type I program.34 It is believed that this program enables the cancer cell to escape immune recognition, but conversion into a program that expresses more genes (latency III) can readily occur in vitro. Because in vitro culture adds other stresses to the cell, it appears as if BL lines either turn on EBV latency III or mutate TP53. A switch from latency I into III and lytic replication can, however, also be accomplished by compounds that affect silencing, such as decitabine and sodium butyrate, which target DNA methylation and histone acetylation, respectively.34,49 We observed a correlation between induced expression of the lytic regulator ZEBRA in response to decitabine, which could exert a function either in the observed aneuploidy or cell death. Intriguingly, however, ZEBRA induction may also be mediated via the DNA-damaging potential of decitabine, because other chemotherapeutic agents such as doxorubicin, 5-fluorouracil, and cisplatin have been shown to induce a switch to lytic EBV infection.49 More studies are needed to elucidate what effects of decitabine on cells and EBV reactivation are mediated by DNA damage or demethylation.
DNA damage sensitizes p53-deficient cells to many types of cell death including poly(ADP-ribose) polymerase (PARP)–mediated necrosis,50 mitotic catastrophe, and caspase-independent mitotic death (CIMD). The latter was recently shown to involve mitochondrial release of Apoptosis-inducing factor and Endonuclease G, which resulted in DNA fragmentation independent of caspases,51 similar to what we observed in the mouse lymphoma cells. If decitabine triggers CIMD or other types of caspase-independent deaths, then Akata cells must have developed resistance against this alternative form of cell death during the decades it has been in culture world-wide, because the pan-caspase inhibitor protected these cells from decitabine. We, thus, favor an expanded investigation of pathways involved in decitabine-induced death, because this is likely to be important in clinically related issues, such as prediction and prevention of relapses. These studies will be important to optimize future usage of decitabine in the clinic.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Gerard Zambetti and Erik Lundgren for cell lines, Per Holmfeldt and Kristina Ruuth for suggestions on flow cytometry techniques, Göran Roos and Erik Lundgren for help with pathology, and the personnel at Umeå Transgene Core Facility.
This work was supported by Cancerfonden, the Association of International Cancer Research, Vetenskapsrådet, Kempestiftelsen, and Umeå University.
Authorship
Contribution: A.H., L.M.N., and L.P.F. performed research and analyzed data; K.H.M. designed research; and J.A.N. designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: K.H.M was employed by Thermo Scientific after having contributed to the submitted work. The remaining authors declare no competing financial interests.
Correspondence: Jonas Nilsson, PhD, Department of Molecular Biology, By 6K, Umeå University, SE-901 87 Umeå, Sweden; e-mail: jonas.nilsson@molbiol.umu.se.