Abstract
Combination studies of histone deacetylase inhibitors (HDACi) and proteasome inhibitors are providing preclinical framework to build better strategies against hematologic malignancies. Our previous work found that a novel proteasome inhibitor, NPI-0052, and HDACi synergistically induce apoptosis in leukemia cells in a caspase-8– and oxidant-dependent manner. Here we extend those observations to primary leukemia cells and identify novel mechanisms of synergy. Because the proximal targets of NPI-0052 and HDACi are inhibition of proteasome activity and histone acetylation, we initially examined those biochemical events. Increased acetylation of histone-H3 was detected in Jurkat and CLL primary cells treated with NPI-0052, alone or in combination with various HDACi (MS/SNDX-275 or vorinostat). Hyperacetylation by NPI-0052 occurred to a lesser extent in caspase-8–deficient cells and in cells treated with an antioxidant. These results indicate that NPI-0052 is eliciting caspase-8 and oxidative stress-dependent epigenetic alterations. In addition, real-time PCR revealed that MS/SNDX-275 repressed expression of the proteasomal β5, β2, and β1 subunits, consequently inhibiting respective enzymatic activities. Overall, our results suggest that crosstalk by NPI-0052 and HDACi are contributing, along with caspase-8 activation and oxidative stress, to their synergistic cytotoxic effects in leukemia cells, reinforcing the potential clinical utility of combining these 2 agents.
Introduction
In children and young adults, leukemia is the most commonly occurring type of cancer. Existing therapies for leukemia rely on chemotherapy composed of steroids, anthracyclines and nucleoside analogs, and/or stem cell transplantations.1-3 Despite a relatively high cure rate (upwards of 85%), long-term sequelae are often seen in patients and include cardiac complications and an increased risk of second malignancies.4,5 Therefore, a major challenge in leukemia research is to develop new therapies to decrease toxicity, maintain remission, and prolong survival of patients
Inhibition of the ubiquitin-proteasome pathway has proven to be a fruitful strategy for specific hematologic malignancies. Bortezomib, the first and only Food and Drug Administration (FDA)–approved proteasome inhibitor, is currently in clinical use as a single agent in refractory multiple myeloma and mantle cell lymphoma.6,7 The success of bortezomib has generated interest in the discovery and development of other proteasome inhibitors. NPI-0052 (salinosporamide A) is a novel proteasome inhibitor that is distinct from bortezomib in structure, binding, and potency.8-11 Previous findings from our group and others show that NPI-0052 targets the 20S proteasome by inhibiting the chymotrypsin-, caspase-, and trypsin-like activities with distinct potency and specificity (inhibiting the chymotrypsin- and caspase-like activities more effectively than the trypsin-like activity) in leukemia cells. This profile of proteasome inhibition by NPI-0052 results in apoptosis via a caspase-8– and reactive oxygen species (ROS)–dependent route in leukemia cells.10 These features represent unique aspects of NPI-0052, because bortezomib's cytoxicity relies on both caspase-8 and caspase-9 equivalently,12 and as we show here, NPI-0052 increases intracellular levels of ROS to a greater degree than equimolar doses of bortezomib. Given these differences, which are relevant to apoptosis induction, NPI-0052 may be useful in malignancies, such as leukemia, where bortezomib, as a single agent, failed to have a therapeutic advantage.13
In leukemia, in vitro data indicated strong activity, but early clinical trials of bortezomib did not show significant responses,13 thus combination studies of proteasome inhibitors with other agents are abundant. One group of agents that are currently being tested in combination with proteasome inhibitors are histone deacetylase inhibitors (HDACi), which are a structurally diverse group of epigenetically targeted anticancer agents that inhibit histone deacetylases (HDACs).14 HDACs, together with histone acetyl transferases, primarily regulate the acetylation status of histones, which in turn alters chromatin structure promoting either transcriptional activation or repression. Thus, HDACi can influence gene transcription and expression. HDACi have been reported to synergistically interact with proteasome inhibitors to induce apoptosis in multiple model systems,15-17 and clinical trials examining bortezomib and several HDACi are in progress. We have previously reported that NPI-0052 synergizes with 2 distinct HDACi, MS-275 and valproic acid (VPA), to induce apoptosis in acute lymphocytic leukemia (ALL) cells. This synergy was caspase-8–dependent and was more potent compared with a bortezomib/HDACi regimen.10
Several mechanisms of interaction between proteasome inhibitors, primarily bortezomib and MG132, and HDACi have been described, including ROS generation,10,15,18 Bim up-regulation,16 and JNK activation.15 In this study, we focus on examining the immediate targets of proteasome inhibitors and HDACi: proteasome proteolytic β subunits and their catalytic activity and histone acetylation status. Surprisingly, we found that NPI-0052 and HDACi have overlapping functional effects. NPI-0052 alone strongly promoted the acetylation of histone-H3, resulting in hyperacetylation in cell lines and in lymphocytes isolated from chronic lymphocytic leukemia (CLL) patients. This effect is more pronounced when NPI-0052 is combined with HDACi. Importantly, bortezomib did not promote hyperacetylation, underscoring this biochemical event as unique to NPI-0052. Hyperacetylation by NPI-0052 was reversed by an antioxidant and did not occur in caspase-8–deficient cells, implicating caspase-8 and ROS in this novel effect. In addition, MS-275 was found to target the proteasome by reducing mRNA expression levels of proteolytic β subunits and their corresponding catalytic activities. Because synergy between NPI-0052 and HDACi was observed in peripheral mononuclear cells isolated from acute and chronic leukemia patients at very low subtoxic doses, our results urge the clinical testing of NPI-0052/HDACi regimens in leukemia and suggest that drug-related side effects in patients may be limited.
Methods
Cell lines, patient material, and cell culture
The human leukemia Jurkat cell line was purchased from ATCC (Manassas, VA). Cells were maintained in RPMI media supplemented with 10% heat-inactivated fetal bovine serum (FBS; Hyclone, Logan, UT), 2 mM l-glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin (Cellgro, Mediatech, Herndon, VA). The caspase-8–deficient Jurkat cells, I9.2,19 were kindly provided by Dr Michael Andreeff (University of Texas M. D. Anderson Cancer Center, Houston, TX) and were maintained in RPMI media with N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), sodium pyruvate, and l-glutamine containing 15% FBS and 1.0% penicillin-streptomycin. Cell lines were maintained at 37°C with 5% CO2. Peripheral blood from acute myelogeneous leukemia (AML) and CLL patients was collected after informed consent was obtained in accordance with the Declaration of Helsinki, under the aegis of protocols reviewed by Institutional Review Boards of M. D. Anderson Cancer Center. Mononuclear cells were isolated using Ficoll-Paque PLUS gradient (Amersham Biosciences, Uppsala, Sweden) as previously described.20 Isolated cells were washed with medium and immediately plated for assays.
Reagents
The proteasome inhibitor, NPI-0052, was provided by Nereus Pharmaceuticals (San Diego, CA). Bortezomib (Millennium Pharmaceuticals, Cambridge, MA) was acquired from the M. D. Anderson Cancer Center pharmacy. The HDACi, MS-275, was purchased from Sigma-Aldrich (St Louis, MO), and vorinostat (Merck, Whitehouse Station, NJ) was a gift from Dr David McConkey (University of Texas M. D. Anderson Cancer Center). Fluorogenic substrates, suc-LLVY-amc and z-LLE-amc were obtained from AG Scientific (San Diego, CA). The antioxidant, N-acetyl cysteine (NAC), was purchased from Sigma-Aldrich. The dye, hydroethidium (HEt), was obtained from Molecular Probes (Eugene, OR). The caspase-3 substrate, DEVD-amc, was obtained from Biomol International, LP (Plymouth Meeting, PA). Antibodies were from the following sources: acetyl histone-H3 (Upstate Biotechnology, Temecula, CA), histone-H3 (Abcam, Cambridge, MA), p27 (BD Transduction Laboratories, San Diego, CA), ubiquitin (Santa Cruz Biotechnology, Santa Cruz, CA), and actin (Sigma-Aldrich).
DNA fragmentation
Cell death was determined by staining cells with propidium iodide (PI) and analyzed by flow cytometry. Briefly, cells were incubated with indicated drug combinations for 24 hours, followed by centrifugation and resuspending cells with phosphate-buffered saline (PBS) containing 50 μg/mL PI (with 0.1% Triton X-100 and 0.1% sodium citrate). Samples were stored at 4°C for 24 hours and vortex mixed before analysis on the FL-3 channel by flow cytometry (FACSCalibur; Becton Dickinson; Franklin Lakes, NJ). The percentage of apoptotic cells was determined by measuring the percentage of cells in the subdiploid population using CellQuest Software (BD Biosciences, San Jose, CA).
Transient transfection
The overexpression construct for caspase-8 was kindly provided by Dr Scott H. Kaufmann (Mayo Clinic, Rochester, MN). Using EcoRV and NotI, the full-length procaspase-8 open reading frame was cloned downstream of the cytomegalovirus promoter into the multiple cloning site of pCMS-5A, which also contains an internal ribosomal entry site followed by cDNA encoding a histone H2B/enhanced green fluorescent protein (EGFP) fusion to mark successfully transfected cells. This construct was transiently transfected into caspase-8–deficient, I9.2, cells using Nucleofector kit V (Amaxa Biosystems, Cologne, Germany) according to the manufacturer's protocol. After 22 hours, the brightest 10% of histone H2B/EGFP-positive cells of the total population were sorted by flow cytometry. The EGFP-positive cells were immediately treated with 10 nM NPI-0052 or diluent for 6 hours before collecting protein lysates for immunoblotting.
Cell lysates and immunoblotting
Five million cells were treated with different drug concentrations for the times indicated. Cells were lysed with Triton X-100 lysis buffer (PBS with 1% Triton X-100, 25 mM Tris, pH 7.5, and 150 mM sodium chloride [Fisher Scientific, Fair Lawn, NJ]) containing protease inhibitors for 1 hour followed by centrifugation. Lysates (50 μg) were collected and separated on sodium dodecyl sulfate (SDS)–polyacrylamide gels. Protein was transferred to nitrocellulose membranes and blocked for 2 hours at room temperature with 5% milk/Tris-buffered saline with 0.05% Tween-20 (TBST). Membranes were incubated with 1:1 000 concentrations of primary antibodies in 5% milk/TBST overnight at 4°C, followed by corresponding secondary antibodies for 30 minutes at room temperature. Bound antibodies were detected using enhanced chemiluminescence.
Immunoprecipitation
Whole cell lysates were prepared from 10 million cells as described above. Lysates were precleared using 25 μL protein A/G agarose beads (Santa Cruz Biotechnology) and rotating for 1 hour at 4°C. Supernatants were collected and histone-H3 or p27 protein was immunoprecipitated with histone-H3 or p27 antibody overnight at 4°C, and subsequently incubated with 50 μL protein A/G agarose beads for 2 hours. After centrifugation, supernatants were electrophoresed on a 15% SDS-polyacrylamide gel. Protein was transferred to a nitrocellulose membrane, followed by blocking with milk and immunoblotting with 1:500 dilution of ubiquitin antibody overnight. After washing, blot was incubated with secondary antibody and protein was detected using enhanced chemiluminescence.
Real-time polymerase chain reaction
Total RNA extraction was performed from 5 million cells using an RNeasy mini kit (QIAGEN, Valencia, CA) according to the manufacturer's protocol. Reverse transcription reaction was carried out for each sample using 2 μg RNA with Omniscript RT kit (QIAGEN) per the manufacturer's protocol. Quantitative polymerase chain reaction (PCR) was carried out using 200 ng cDNA per reaction with iQ SYBR Green PCR master mix (Bio-Rad Laboratories, Hercules, CA) and specific β5, β2, β1,21 or actin primers. The annealing temperature for all PCR samples was 58°C, and the reaction was carried out for 45 cycles. Actin was amplified using the following primers: 5′ primer, 5′-TGTGCCCATCTACGAGGGGTATGC-3′; and 3′ primer, 5′-GGTACATGGTGGTGCCGCCAGACA-3′. Relative expression of genes of interest were analyzed by calculating the cycle threshold (Ct) values and normalized to actin Ct values.
20S Proteasome activity assays
Fluorogenic peptides, suc-LLVY-amc and z-LLE-amc, were used to determine the chymotrypsin-like and caspase-like activity of the proteasome in leukemia cells as previously described.22 Cells were incubated with 5 μM MS-275 for 24 hours, followed by washing with PBS and centrifugation. Cells were lysed by freezing/thawing with 20 mM Tris, pH 7.5, 0.1 mM ethylenediaminetetraacetic acid (EDTA), pH 8.0, 20% glycerol, 0.05% Nonidet-P40, 1 mM 2-β mercaptoethanol, and 1 mM ATP. Lysates were centrifuged and supernatants were combined with fluorogenic peptides in 50 mM HEPES, pH 7.5, and 5 mM ethyleneglycoltetraacetic acid (EGTA), pH 7.0. Samples were analyzed with a spectrofluorometer (SpectraMax Gemini EM; Molecular Devices, Sunnyvale, CA) using an excitation of 380 nm and an emission of 460 nm. The amount of fluorescence (amc) released correlates with the amount of proteasome activity of the specific proteolytic target. Fluorescence is expressed in relative fluorescent units (RFU).
Intracellular superoxide levels
The cell-permeable dye, HEt, was used to measure intracellular superoxide levels. One million cells were incubated in the presence or absence of NPI-0052, HDACi, or NAC as indicated per experiment for 12 hours. Cells were harvested and washed with PBS. After centrifugation, the cell pellet was resuspended with 1 mL PBS containing 10 μM HEt and incubated for 30 minutes at 37°C in the dark. Samples were harvested, resuspended with 500 μL PBS, and analyzed by measuring the fluorescence intensity by flow cytometry on the FL-3 channel.
Caspase-3 activity assay
Caspase-3 activity was determined as previously described,10 using the fluorogenic substrate DEVD-amc. Briefly, after exposure to NPI-0052 and/or vorinostat for 8 hours, cells were harvested and lysed in PBS by freezing/thawing on dry ice. Samples were aliquoted in triplicates in a 96-well plate with 150 μL DEVD buffer with 50 μM DEVD-amc. Release of fluorescence (amc) was measured using a spectrofluoremeter using an excitation of 355 nm and an emission of 460 nm. Fluorescence generated by the cleavage of fluorogenic peptide is proportional to caspase-3 activity.
Statistical analyses
The data presented are the mean (± standard deviation [SD]) from 3 independent experiments performed in triplicate with similar results. The Student t test was performed to determine statistical significant differences between samples. A P value less than .05 was considered statistically significant. Isobologram analyses using the Chou and Talalay method with Calcusyn (Biosoft, Ferguson, MO) were used to determined synergism.23 A combination index (CI) value less than 1.0 indicates synergistic effects. A CI value equal to 1.0 indicates additive interactions. A CI value greater than 1.0 indicates antagonistic interactions.
Results
Combination of HDACi and proteasome inhibitors results in synergistic cell death in leukemia cells
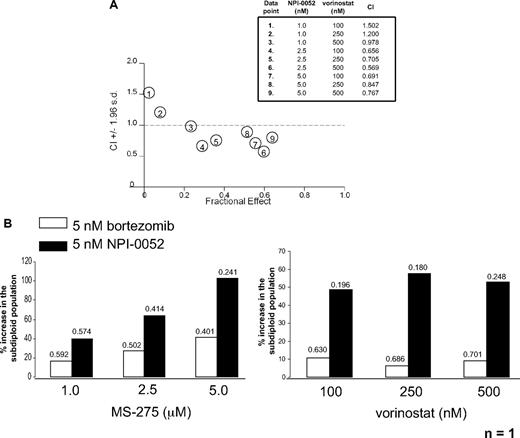
Previous reports have demonstrated synergy between proteasome inhibitors and HDACi in numerous cancer cell models.15,24,25 Our own published work demonstrated higher degrees of synergistic cell death in leukemia cells treated with NPI-0052 and either MS-275 or VPA compared with bortezomib combinations with the same HDACi.10 Here we tested whether NPI-0052 would also synergize with the sole HDACi that is currently FDA approved—vorinostat (SAHA, Zolinza), which is an hydroxamic acid, pan-HDACi, targeting class I, II, III, and IV HDACs.26,27 Cell death was assessed by measuring the subdiploid population of cells stained with PI followed by flow cytometric analysis. To determine synergy, the percentage of apoptotic cells of 3 separate PI experiments were used to determine median dose effects to calculate CI values as described above.23 NPI-0052 (2.5 and 5.0 nM) synergized with all doses of vorinostat (100-500 nM) tested as indicated by CI values less than 1.0 (Figure 1A). The 1.0 nM NPI-0052 dose was additive with the 500 nM vorinostat dose but had antagonistic (CI > 1.0) effects when combined with lower doses of vorinostat (100 and 250 nM).
NPI-0052 synergizes with HDAC inhibitors in primary and cultured leukemia cells. (A) Synergistic interactions between NPI-0052 and vorinostat. Jurkat cells were treated with indicated low doses of NPI-0052 and vorinostat for 24 hours. After PI cell staining, DNA fragmentation was assessed by flow cytometry. The averages of 3 independent experiments were taken to determine synergy by calculating the combination index value (CI) by isobologram analysis with Calcusyn software. A CI less than 1.0 indicates synergism. Table defines dose combinations for each data point shown and CI values. (B) HDAC inhibitors synergize more effectively with NPI-0052 than bortezomib in one AML patient sample. Mononuclear cells isolated from peripheral blood from an AML patient were treated with an HDACi/proteasome inhibitor regimen as indicated for 24 hours. DNA fragmentation was assessed after PI staining. Shown is the percent increase in the subdiploid population. CI values, indicating synergy, for each combination are shown above each bar.
NPI-0052 synergizes with HDAC inhibitors in primary and cultured leukemia cells. (A) Synergistic interactions between NPI-0052 and vorinostat. Jurkat cells were treated with indicated low doses of NPI-0052 and vorinostat for 24 hours. After PI cell staining, DNA fragmentation was assessed by flow cytometry. The averages of 3 independent experiments were taken to determine synergy by calculating the combination index value (CI) by isobologram analysis with Calcusyn software. A CI less than 1.0 indicates synergism. Table defines dose combinations for each data point shown and CI values. (B) HDAC inhibitors synergize more effectively with NPI-0052 than bortezomib in one AML patient sample. Mononuclear cells isolated from peripheral blood from an AML patient were treated with an HDACi/proteasome inhibitor regimen as indicated for 24 hours. DNA fragmentation was assessed after PI staining. Shown is the percent increase in the subdiploid population. CI values, indicating synergy, for each combination are shown above each bar.
Synergy was also observed in one AML patient sample treated with proteasome inhibitors and 2 different HDACi, MS-275 and vorinostat. These compounds are structurally distinct and block different classes of HDACs because the benzamide MS-275 inhibits Class I HDACs (HDAC 1, 2, and 3).14,28 Mononuclear cells from an AML patient were exposed to either 5 nM bortezomib or NPI-0052 combined with a range of doses of MS-275 (1.0-5.0 μM) or vorinostat (100-500 nM). The NPI-0052/HDACi regimen had a higher percent increase of the subdiploid population compared with cells treated with a bortezomib/HDACi combination (Figure 1B). Both proteasome inhibitors displayed synergy with MS-275 and vorinostat, however, NPI-0052/HDACi CI values were more potently synergistic than those obtained with equamolar bortezomib/HDACi (Figure 1B values above bars). These differential synergistic effects with NPI-0052 versus bortezomib could be reflecting the fact that NPI-0052 is a more potent inhibitor than bortezomib, because equamolar doses rather than equipotent doses were used. However, these studies were conducted in the interest of showing effects of low doses of proteasome inhibitors and HDACi to minimize possible toxicities. Although only one AML patient sample was tested, they were consistent with the statistically significant results obtained in cell lines. Together these results document synergistic effects of low doses of NPI-0052 with vorinostat and HDACi in both a primary AML specimen and an ALL cell line.
NPI-0052/HDACi regimen induces oxidative stress
Our results from Figure 1A demonstrate that NPI-0052 synergizes with vorinostat to induce apoptosis. Oxidative stress has been implicated in the mechanism of apoptosis induction by proteasome inhibitors and HDACi.15 Furthermore, we have previously shown that the combination of NPI-0052 with MS-275 dramatically potentiates superoxide levels in Jurkat cells than either agent alone.10 Based on these observations, we tested if cytotoxicity with the NPI-0052/vorinostat combination was related to increased oxidative stress. First we examined DNA fragmentation in cells treated with NPI-0052 and/or vorinostat in the presence of the free radical scavenger, NAC. Results show the combination of 5 nM NPI-0052 and 500 nM vorinostat caused a significant increase in cell death compared with either agent alone or control cells (Figure 2A). In addition, NAC protected nearly 50% against apoptosis induced by the NPI-0052/vorinostat combination (Figure 2A), indicating that oxidative stress is in part contributing toward the enhanced cytotoxicity induced by these drugs. Next, we confirmed if NPI-0052 in combination with vorinostat could increase ROS levels. As expected, cells treated with 5 nM NPI-0052 for 12 hours displayed higher superoxide levels compared with control (Figure 2B). A relatively low dose of vorinostat (500 nM) did not alter superoxide levels in Jurkat cells compared with diluent. However the combination of the 2 compounds resulted in a significant increase in intracellular superoxide levels compared with control (P < .01) or 5 nM NPI-0052 (P < .05). Pretreatment with NAC prevented ROS generation as a result of combination treatment (Figure 2B,C). In addition, we found that Jurkat cells treated with NPI-0052 had significantly higher levels of ROS than cells treated with bortezomib (Figure 2D), providing a potential explanation for why stronger synergy measured by apoptosis induction was observed in primary AML cells with NPI-0052/vorinostat compared with bortezomib/vorinostat treatment (Figure 1B).
Oxidative stress is elevated as a consequence of NPI-0052 and vorinostat interactions. (A) NAC partially protects from NPI-0052/vorinostat-induced apoptosis. Jurkat cells were pretreated with 24 mM NAC for 30 minutes, followed by treatment with diluent, 500 nM vorinostat, 5 nM NPI-0052, or combination of drugs for 24 hours. Apoptosis was assessed by PI staining and subsequent flow cytometric analysis. Statistical differences were calculated: *P < .05 for NPI-0052/vorinostat-treated cells compared with control (dimethyl sulfoxide [DMSO]); **P < .01 for NPI-0052/vorinostat compared with NPI-0052/vorinostat/NAC. (B) Intracellular superoxide production in Jurkat cells treated with NPI-0052/vorinostat regimen is decreased with NAC. Cells were treated as indicated for 12 hours, followed by staining with HEt and analyzed by flow cytometry. Shown are the mean fluorescence values of 3 independent experiments. (C) NPI-0052/vorinostat increases superoxide levels, which are decreased by NAC. Shown is a representative histogram of the 3 experiments performed for HEt staining (Figure 4B). Solid line represents control cells, solid bold line is 5 nM NPI-0052/500 nM vorinostat-treated cells and dashed line indicates 5 nM NPI-0052/500 nM vorinostat cells pretreated with 24 mM NAC. (D) NPI-0052 generates more superoxide levels than bortezomib in Jurkat cells. Cells were exposed to 5 nM NPI-0052, bortezomib, or diluent for 12 hours and stained with HEt. Samples were analyzed by flow cytometry. Shown are mean fluorescence values of 3 independent experiments.
Oxidative stress is elevated as a consequence of NPI-0052 and vorinostat interactions. (A) NAC partially protects from NPI-0052/vorinostat-induced apoptosis. Jurkat cells were pretreated with 24 mM NAC for 30 minutes, followed by treatment with diluent, 500 nM vorinostat, 5 nM NPI-0052, or combination of drugs for 24 hours. Apoptosis was assessed by PI staining and subsequent flow cytometric analysis. Statistical differences were calculated: *P < .05 for NPI-0052/vorinostat-treated cells compared with control (dimethyl sulfoxide [DMSO]); **P < .01 for NPI-0052/vorinostat compared with NPI-0052/vorinostat/NAC. (B) Intracellular superoxide production in Jurkat cells treated with NPI-0052/vorinostat regimen is decreased with NAC. Cells were treated as indicated for 12 hours, followed by staining with HEt and analyzed by flow cytometry. Shown are the mean fluorescence values of 3 independent experiments. (C) NPI-0052/vorinostat increases superoxide levels, which are decreased by NAC. Shown is a representative histogram of the 3 experiments performed for HEt staining (Figure 4B). Solid line represents control cells, solid bold line is 5 nM NPI-0052/500 nM vorinostat-treated cells and dashed line indicates 5 nM NPI-0052/500 nM vorinostat cells pretreated with 24 mM NAC. (D) NPI-0052 generates more superoxide levels than bortezomib in Jurkat cells. Cells were exposed to 5 nM NPI-0052, bortezomib, or diluent for 12 hours and stained with HEt. Samples were analyzed by flow cytometry. Shown are mean fluorescence values of 3 independent experiments.
HDAC inhibitors target the proteasome
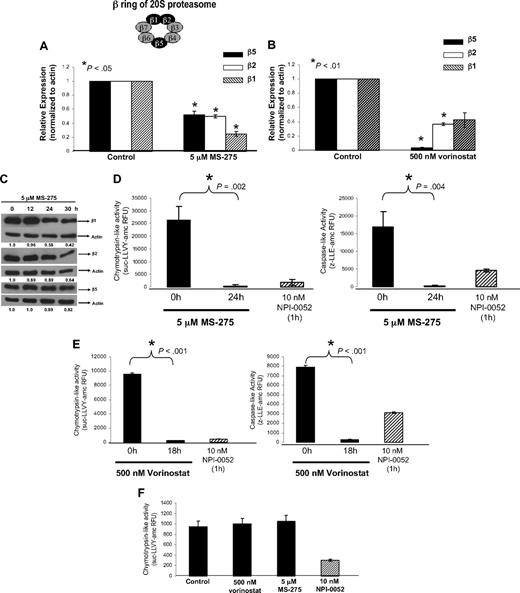
Next we examined proteasome activity to gain insight into the observed synergy between NPI-0052 and HDACi. Decreased enzymatic activity of the 3 active sites, which are localized to the β5, β2, and β1 subunits of the 20S proteasome has been observed in NPI-0052–treated leukemia cells.10,29 To determine if HDACi can also promote proteasome inhibition by altering gene expression of these subunits, RNA was collected from Jurkat cells exposed to 5 μM MS-275 or 500 nM vorinostat for 24 hours and subjected to real-time PCR. As shown in Figure 3A and B, both MS-275 and vorinostat significantly reduced mRNA levels of catalytic β5, β2, and β1 subunits of the proteasome. A time-dependent reduction of protein levels of β5, β2, and β1 subunits with MS-275 was also observed (Figure 3C). These proteasomal subunits contain the chymotrypsin-, trypsin-, and caspase-like activities, respectively.30,31
HDACi, (MS-275 and vorinostat), target the proteasome. (A) MS-275 decreased mRNA expression of catalytic β5, β2, β1 proteasomal subunits. Real-time PCR analyzed expression levels of catalytic β subunits of the proteasome in Jurkat cells exposed to 5 μM MS-275 for 24 hours. Shown is the relative expression, which was calculated by first normalizing all samples to corresponding actin expression, than to control (cells treated with DMSO). *P < .05 compared with 0 hours. (B) Vorinostat decreases mRNA levels of β5, β2, β1 proteasomal subunits. Jurkat cells were treated with 500 nM vorinostat for 18 hours, and RNA was collected to analyze expression of β subunits by real-time PCR. Shown is the relative expression. *P < .01 compared with 0 hours. (C) MS-275 reduces β5, β2, β1 subunit protein expression. Jurkat cells were exposed to 5 μM MS-275, and protein lysates were collected at indicated time points. Western blot analysis and specific antibodies determined expression of β subunits and actin. Numerical values under each panel are the densitometry ratios of β subunit to actin and are normalized to 0 hours. (D) MS-275 inhibits the chymotrypsin-like and caspase-like activity of the proteasome. Jurkat cells were exposed to 5 μM MS-275 for 24 hours. The chymotrypsin-like and caspase-like activity was determined by measuring the fluorescence (amc) intensity released by the cleavage of fluorogenic substrate suc-LLVY-amc or z-LLE-amc, respectively. Proteasome activity was evaluated in RFU. Treatment with proteasome inhibitor, 10 nM NPI-0052, for 1 hour was used as a positive control. Statistical differences between 0 and 24 hours with MS-275 were determined by Student t test, *P = .002 for chymotrypsin-like and P = .004 for caspase-like. (E) Vorinostat diminishes proteasomal chymotrypsin-like and caspase-like activities. After an 18-hour exposure to 500 nM vorinostat, chymotrypsin-like and caspase-like activities in Jurkat cells were measured using fluorogenic substrates as previously described. Student t test determined statistical differences between 0 and 18 hours. (F) HDACi do not inhibit the rate-limiting activity of the proteasome in isolated proteasomes. Purified 20S proteasomes were combined with diluent, 5 μM MS-275, 500 nM vorinostat, or 10 nM NPI-0052 for 30 minutes at room temperature. The chymotrypsin-like activity was analyzed using fluorogenic substrate suc-LLVY–amc and activity was evaluated in RFU. NPI-0052 was used as a positive control.
HDACi, (MS-275 and vorinostat), target the proteasome. (A) MS-275 decreased mRNA expression of catalytic β5, β2, β1 proteasomal subunits. Real-time PCR analyzed expression levels of catalytic β subunits of the proteasome in Jurkat cells exposed to 5 μM MS-275 for 24 hours. Shown is the relative expression, which was calculated by first normalizing all samples to corresponding actin expression, than to control (cells treated with DMSO). *P < .05 compared with 0 hours. (B) Vorinostat decreases mRNA levels of β5, β2, β1 proteasomal subunits. Jurkat cells were treated with 500 nM vorinostat for 18 hours, and RNA was collected to analyze expression of β subunits by real-time PCR. Shown is the relative expression. *P < .01 compared with 0 hours. (C) MS-275 reduces β5, β2, β1 subunit protein expression. Jurkat cells were exposed to 5 μM MS-275, and protein lysates were collected at indicated time points. Western blot analysis and specific antibodies determined expression of β subunits and actin. Numerical values under each panel are the densitometry ratios of β subunit to actin and are normalized to 0 hours. (D) MS-275 inhibits the chymotrypsin-like and caspase-like activity of the proteasome. Jurkat cells were exposed to 5 μM MS-275 for 24 hours. The chymotrypsin-like and caspase-like activity was determined by measuring the fluorescence (amc) intensity released by the cleavage of fluorogenic substrate suc-LLVY-amc or z-LLE-amc, respectively. Proteasome activity was evaluated in RFU. Treatment with proteasome inhibitor, 10 nM NPI-0052, for 1 hour was used as a positive control. Statistical differences between 0 and 24 hours with MS-275 were determined by Student t test, *P = .002 for chymotrypsin-like and P = .004 for caspase-like. (E) Vorinostat diminishes proteasomal chymotrypsin-like and caspase-like activities. After an 18-hour exposure to 500 nM vorinostat, chymotrypsin-like and caspase-like activities in Jurkat cells were measured using fluorogenic substrates as previously described. Student t test determined statistical differences between 0 and 18 hours. (F) HDACi do not inhibit the rate-limiting activity of the proteasome in isolated proteasomes. Purified 20S proteasomes were combined with diluent, 5 μM MS-275, 500 nM vorinostat, or 10 nM NPI-0052 for 30 minutes at room temperature. The chymotrypsin-like activity was analyzed using fluorogenic substrate suc-LLVY–amc and activity was evaluated in RFU. NPI-0052 was used as a positive control.
To confirm if this decreased expression of catalytic mRNA subunit expression had a functional effect, we next measured proteasomal proteolytic activity. Fluorogenic substrates were used to analyze chymotrypsin-like and caspase-like activities as previously described.22 After 24 hours, MS-275 significantly inhibited both chymotrypsin-like and caspase-like activities (Figure 3D) in whole cell lysates. Similarly, vorinostat also reduced chymotrypsin-like and caspase-like activities (Figure 3E). Cells exposed to 10 nM NPI-0052 for 1 hour were used as a positive control for these assays. In addition, experiments using isolated proteasomes in combination with HDACi (Figure 3F) were conducted to determine if inhibition of the rate-limiting chymotrypsin-like activity of the proteasome by HDACi was a result of a direct interaction with proteasomal subunits. Neither HDACi (vorinostat nor MS-275) inhibited chymotrypsin-like activity in isolated proteasomes. In contrast, NPI-0052, which has been shown to bind and inhibit the β5 subunit of yeast proteasome,9 does inhibit the chymotrypsin-like activity of purified rabbit 20S proteasomes. These results suggest that MS-275 targets the proteasome by reducing mRNA and protein levels of catalytic β subunits, thereby diminishing cellular levels of 2 of its proteolytic activities, the chymotrypsin- and caspase-like activities.
Increased histone-H3 expression and hyperacetylation of histone-H3 by NPI-0052
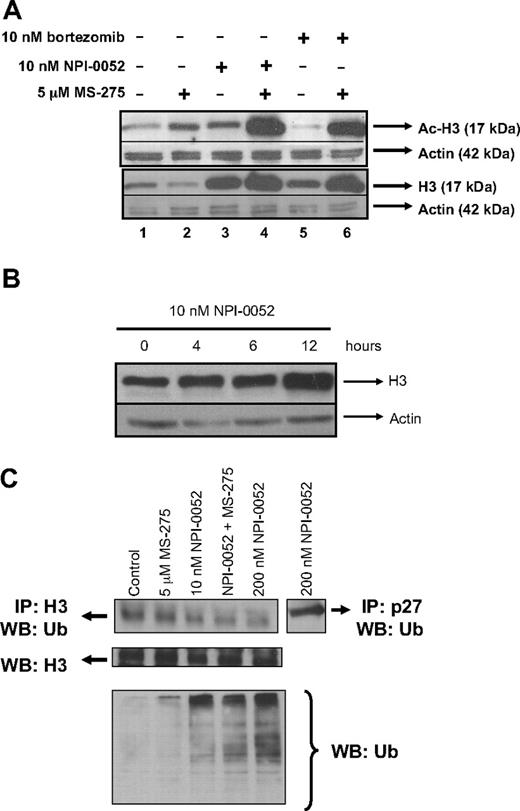
Because HDACi counteract HDACs thereby promoting acetylation, we examined the acetylation status of histone-H3 in Jurkat cells treated with MS-275 in combination with NPI-0052 or bortezomib. As expected, hyperacetylation of histone-H3 was detected in cells exposed to 5 μM MS-275 for 6 hours (Figure 4A lane 2). Interestingly, cells treated with a low dose of NPI-0052 (10 nM) also displayed hyperacetylation of histone-H3 (Figure 4A lane 3); and this effect was more pronounced when NPI-0052 was combined with MS-275 (Figure 4A lane 4). Similar results were obtained in Jurkat cells exposed to NPI-0052 and vorinostat (data not shown). Cells treated with 10 nM bortezomib alone did not increase acetylation of histone-H3 (Figure 4A lane 5) as seen with 10 nM NPI-0052 (Figure 4A lane 3), but hyperacetylation occurred with the bortezomib/MS-275 combination (Figure 4A lane 6). Examination of total histone-H3 levels revealed that cells treated with proteasome inhibitors, either alone or in combination, displayed more histone-H3 protein expression (Figure 4A lanes 3-6) compared with cells treated with diluent (Figure 4A lane 1) or HDACi (Figure 4A lane 2). Again, cells exposed to 10 nM NPI-0052 (Figure 4A lane 3) had elevated histone-H3 protein expression compared with cells treated with an equimolar dose of bortezomib (Figure 4A lane 5). Further examination of histone-H3 protein expression revealed that levels were increased by 10 nM NPI-0052 in a time-dependent manner (Figure 4B) beginning at 4 to 6 hours after exposure.
NPI-0052 increases histone-H3 expression and its hyperacetylation. (A) NPI-0052 increases histone-H3 levels and hyperacetylation. Jurkat cells were exposed to 10 nM bortezomib, 10 nM NPI-0052, 5 μM MS-275, or a combination for 6 hours. Cell lysates were prepared and subjected to SDS polyacrylamide gel electrophoresis (SDS-PAGE) and immunobloted for acetylated (Ac)–H3, histone-H3 (H3), and actin. (B) NPI-0052 increases histone-H3 protein expression. Lysates from Jurkat cells treated with 10 nM NPI-0052 were collected at the indicated times. Western blot analyzed histone-H3 and actin expression. (C) NPI-0052 does not induce accumulation of ubiquitinated-histone-H3. Immunoprecipitation (IP) of histone H3 or p27 was performed in protein lysates from Jurkat cells that were treated with 5 μM MS-275, 10 nM NPI-0052, 200 nM NPI-0052, or a combination with MS-275 and NPI-0052 for 6 hours. After IP, samples were run on SDS-PAGE and blotted for ubiquitin. Bottom panel, protein lysates from Jurkat cells treated with the indicated compounds for 6 hours were immunoblotted for total ubiquitin protein expression.
NPI-0052 increases histone-H3 expression and its hyperacetylation. (A) NPI-0052 increases histone-H3 levels and hyperacetylation. Jurkat cells were exposed to 10 nM bortezomib, 10 nM NPI-0052, 5 μM MS-275, or a combination for 6 hours. Cell lysates were prepared and subjected to SDS polyacrylamide gel electrophoresis (SDS-PAGE) and immunobloted for acetylated (Ac)–H3, histone-H3 (H3), and actin. (B) NPI-0052 increases histone-H3 protein expression. Lysates from Jurkat cells treated with 10 nM NPI-0052 were collected at the indicated times. Western blot analyzed histone-H3 and actin expression. (C) NPI-0052 does not induce accumulation of ubiquitinated-histone-H3. Immunoprecipitation (IP) of histone H3 or p27 was performed in protein lysates from Jurkat cells that were treated with 5 μM MS-275, 10 nM NPI-0052, 200 nM NPI-0052, or a combination with MS-275 and NPI-0052 for 6 hours. After IP, samples were run on SDS-PAGE and blotted for ubiquitin. Bottom panel, protein lysates from Jurkat cells treated with the indicated compounds for 6 hours were immunoblotted for total ubiquitin protein expression.
To determine if increased histone-H3 levels were a direct consequence of proteasome inhibition, immunoprecipitation with a polyclonal histone-H3 antibody was performed to detect ubiquitinated histone-H3. Immunoprecipitation was followed by immunoblotting with an ubiquitin antibody to detect ubiquitinated histone-H3, because proteins destined to be degraded by proteasome are tagged by a polyubiquitin chain.32 Neither NPI-0052 alone nor in combination with MS-275 induced increased ubiquitination of histone H3 compared with controls (Figure 4C). However, NPI-0052 (10 and 200 nM) alone or together with MS-275 caused accumulation of total polyubiquitinated proteins, confirming that, at these doses, NPI-0052 is inhibiting the proteasome (Figure 4C bottom panel). To prove that NPI-0052 caused accumulation of known proteasome substrates, we detected ubiquitinated p27 by immunoprecipitation (Figure 4C last lane). Because this was successful, our data suggest that the increased protein expression of histone-H3 levels in response to NPI-0052 is not a result of accumulation of ubiquitinated histone-H3.
Antioxidant reduces increased histone-H3 expression and hyperacetylation of histone-H3 by NPI-0052
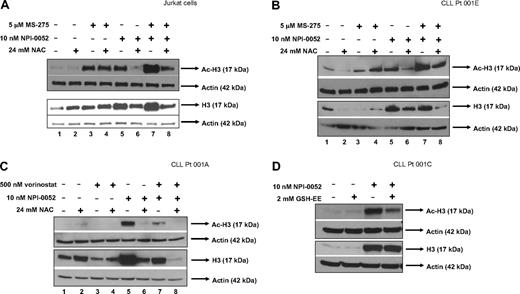
We next addressed the possibility that the oxidative stress generated by NPI-0052 alone or in combination with HDACi could be contributing toward the acetylation status of histone-H3. Protein lysates were collected from Jurkat cells pretreated with NAC followed by exposure to NPI-0052 and MS-275 or either agent alone for 6 hours. Western blot analysis showed similar levels of increased acetylation in cells treated with 5 μM MS-275 (Figure 5A top panel lane 3) and 10 nM NPI-0052 (Figure 5A top panel lane 5), whereas the combination resulted in more hyperacetylation (Figure 5A top panel lane 7) compared with control (Figure 5A top panel lane 1) or either compound by itself (Figure 5A top panel lanes 3,5). As seen in Figure 4A and B, histone-H3 expression was increased in lysates that were treated with the proteasome inhibitor (bottom panel lanes 5,7). The addition of the antioxidant reduced both histone-H3 levels and hyperacetylation by NPI-0052 alone (Figure 4A,B bottom panel lane 6) or in combination with MS-275 (Figure 4A,B bottom panel lane 8), suggesting that: (1) oxidative stress is involved in NPI-0052's effect on histone-H3 expression and acetylation and (2) the increased acetylation might correlate with increased histone-H3 expression. Similar results were also obtained in 2 separate CLL patient samples that were treated with NPI-0052 in combination with either MS-275 (Figure 5B) or vorinostat (Figure 5C). NPI-0052 alone or in combination MS-275 or vorinostat caused hyperacetylation and increased histone-H3 expression (Figure 5B,C lanes 5,7) in CLL primary cells. Hyperacetylation by NPI-0052/vorinostat (Figure 5C lane 7) was not as striking as NPI-0052 by itself (Figure 5C lane 5) or NPI-0052/MS-275 (Figure 5B lane 7) in CLL patients' samples. These differences in acetylation status could be in part due to sample heterogeneity in the patient cells or perhaps a lack of sensitivity to vorinostat. Nevertheless, our finding that NPI-0052 alone or with HDACi promoted acetylation and heightened expression of histone-H3 was consistent in both cell lines and primary cells. These effects were attenuated in the presence of NAC (Figure 5B,C lanes 6,8) in 2 different CLL specimens. In addition, as demonstrated in Figure 5D, pretreatment with glutathione ethyl ester reversed hyperacetylation of histone-H3 by NPI-0052 in primary CLL cells. These findings demonstrate for the first time that NPI-0052, alone or in combination with HDACi, affects the expression levels of histone H3 and its acetylation in both cell lines and patient samples in a manner regulated by ROS.
An antioxidant attenuates hyperacetylation of histone-H3 by NPI-0052 and HDACi in cell lines and primary cells. (A) NAC decreases histone-H3 expression and hyperacetylation of histone-H3 in Jurkat cells treated with NPI-0052 and MS-275. Cells were pretreated with 24 mM NAC for 30 minutes followed with 5 μM MS-275, 10 nM NPI-0052, or combination treatment for 6 hours. Cells were harvested, lysed, and analyzed by Western blot analysis for expression of Ac-H3, histone-H3, and actin. (B) Increased histone-H3 expression and hyperacetylation as a result of NPI-0052/MS-275 interaction is decreased by NAC in CLL patient sample. Peripheral blood from patient material (patient no. 1) was separated by centrifugation with a Ficoll gradient. Isolated cells were exposed to indicated doses of NPI-0052 and MS-275 for 6 hours after exposure to 24 mM NAC. Cell lysates were analyzed for Ac-H3, histone-H3, and actin expression by Western blot analysis. (C) Hyperacetylation of histone-H3 and increased histone-H3 expression by NPI-0052/vorinostat in CLL primary cells were attenuated with NAC. Mononuclear cells were isolated from CLL patient peripheral blood (patient no. 2). Cells were treated for 6 hours with 500 nM vorinostat, 10 nM NPI-0052, or a combination after exposure to 24 mM NAC for 30 minutes. Protein lysates were prepared and Western blot analyzed expression for Ac-H3, histone-H3, and actin. (D) Supplementation with glutathione ethyl ester (GSH-EE) reduces hyperacetylation of histone-H3 by NPI-0052 in primary cells. Isolated CLL primary cells (patient no. 3) were pretreated with 2 mM GSH-EE for 1 hour, followed by exposure to 10 nM NPI-0052 or diluent for 6 hours. Protein lysates were prepared and analyzed by Western blot analysis for Ac-H3, histone-H3, and actin expression.
An antioxidant attenuates hyperacetylation of histone-H3 by NPI-0052 and HDACi in cell lines and primary cells. (A) NAC decreases histone-H3 expression and hyperacetylation of histone-H3 in Jurkat cells treated with NPI-0052 and MS-275. Cells were pretreated with 24 mM NAC for 30 minutes followed with 5 μM MS-275, 10 nM NPI-0052, or combination treatment for 6 hours. Cells were harvested, lysed, and analyzed by Western blot analysis for expression of Ac-H3, histone-H3, and actin. (B) Increased histone-H3 expression and hyperacetylation as a result of NPI-0052/MS-275 interaction is decreased by NAC in CLL patient sample. Peripheral blood from patient material (patient no. 1) was separated by centrifugation with a Ficoll gradient. Isolated cells were exposed to indicated doses of NPI-0052 and MS-275 for 6 hours after exposure to 24 mM NAC. Cell lysates were analyzed for Ac-H3, histone-H3, and actin expression by Western blot analysis. (C) Hyperacetylation of histone-H3 and increased histone-H3 expression by NPI-0052/vorinostat in CLL primary cells were attenuated with NAC. Mononuclear cells were isolated from CLL patient peripheral blood (patient no. 2). Cells were treated for 6 hours with 500 nM vorinostat, 10 nM NPI-0052, or a combination after exposure to 24 mM NAC for 30 minutes. Protein lysates were prepared and Western blot analyzed expression for Ac-H3, histone-H3, and actin. (D) Supplementation with glutathione ethyl ester (GSH-EE) reduces hyperacetylation of histone-H3 by NPI-0052 in primary cells. Isolated CLL primary cells (patient no. 3) were pretreated with 2 mM GSH-EE for 1 hour, followed by exposure to 10 nM NPI-0052 or diluent for 6 hours. Protein lysates were prepared and analyzed by Western blot analysis for Ac-H3, histone-H3, and actin expression.
Synergistic effects are mediated by caspase-8
Earlier research in leukemia and multiple myeloma showed a reliance on caspase-8 for NPI-0052 action.8,10 Furthermore, we found that the antioxidant NAC did not prevent caspase-8 activation by NPI-0052, indicating that both oxidative stress and caspase-8 activation contribute to toxicity of this proteasome inhibitor, albeit by parallel rather than linear pathways.10 To further explore whether caspase-8 is required for ROS production by NPI-0052, we used a Jurkat variant cell line that lacked caspase-8 (I9.2)19 and its normal counterpart (Jurkat) to compare intracellular superoxide levels. Cells were treated with 5 μM MS-275, 10 nM NPI-0052, or the combination for 12 hours, followed by HEt staining and flow cytometric analysis. Figure 6A shows graphical representative of 4 separate experiments and indicates that more superoxide was detected in Jurkat cells compared with cells that lacked caspase-8. Jurkat cells treated with 5 μM MS-275 or 10 nM NPI-0052 had moderately increased superoxide levels, but when cells were treated with both agents together these superoxide levels were heightened. In contrast, caspase-8–deficient cells treated only with NPI-0052 alone or in combination with MS-275 produced ROS, but these levels were significantly less than those observed with normal Jurkat cells. Similar results were obtained with vorinostat (Figure 6B). These results suggest that ROS levels produced by NPI-0052 as a single agent or in combination with either MS-275 or vorinostat partially rely on caspase-8 activation.
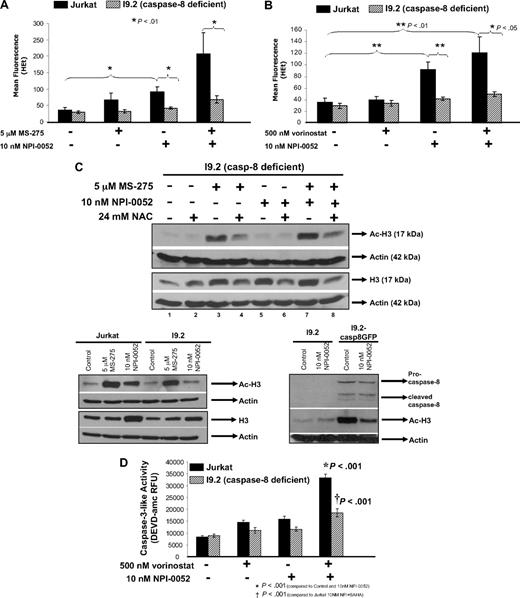
Caspase-8 mediates ROS production, hyperacetylation of histone-H3, and caspase-3 activation by NPI-0052/HDACi. (A) Decreased superoxide levels in a caspase-8–deficient cell line. Jurkat cells and caspase-8–deficient Jurkat cells (I9.2) were treated with 5 μM MS-275, 10 nM NPI-0052, or a combination for 12 hours. Superoxide levels were measured by staining cells with HEt and subsequent flow cytometric analysis on the FL-3 channel. Shown are mean fluorescence values for 4 independent experiments. (B) Lower superoxide levels are detected in caspase-8–deficient cells with vorinostat/NPI-0052 treatment. I9.2 and Jurkat cells were exposed to 500 nM vorinostat, 10 nM NPI-0052, or a combination for 12 hours. Dihydroethidium staining was used to detect superoxide levels. Shown are mean fluorescence values for 4 independent experiments. (C) Caspase-8 plays a role in the hyperacetylation of histone-H3 by NPI-0052. Top panel, the I9.2 (caspase-8–deficient) cells were incubated with 24 mM NAC for 30 minutes, before 5 μM MS-275, 10 nM NPI-0052, or a combination for a total of 6 hours. Cell lysates were analyzed by Western blot analysis for Ac-H3 and actin. Bottom left panel, protein lysates from Jurkat and I9.2 cells treated with 10 nM NPI-0052, 5 mM MS-275, or diluent for 6 hours were analyzed for Ac-H3 and histone H3 protein expression by Western blot analysis. Bottom right panel, protein lysates from I9.2 cells and I9.2/caspase-8/EGFP-transfected cells exposed to DMSO or 10 nM NPI-0052 for 6 hours were evaluated for Ac-H3, caspase-8, and actin protein expression by Western blot analysis. (D) Caspase-8 mediates caspase-3 activation by NPI-0052/vorinostat regimen. Caspase-3 activity was measured in Jurkat and I9.2 cells exposed to DMSO, 500 nM vorinostat, 10 nM NPI-0052, or the combination for 8 hours. *P < .001 compared control or either agent alone; †P < .001 compared with Jurkat cells incubated with NPI-0052/vorinostat.
Caspase-8 mediates ROS production, hyperacetylation of histone-H3, and caspase-3 activation by NPI-0052/HDACi. (A) Decreased superoxide levels in a caspase-8–deficient cell line. Jurkat cells and caspase-8–deficient Jurkat cells (I9.2) were treated with 5 μM MS-275, 10 nM NPI-0052, or a combination for 12 hours. Superoxide levels were measured by staining cells with HEt and subsequent flow cytometric analysis on the FL-3 channel. Shown are mean fluorescence values for 4 independent experiments. (B) Lower superoxide levels are detected in caspase-8–deficient cells with vorinostat/NPI-0052 treatment. I9.2 and Jurkat cells were exposed to 500 nM vorinostat, 10 nM NPI-0052, or a combination for 12 hours. Dihydroethidium staining was used to detect superoxide levels. Shown are mean fluorescence values for 4 independent experiments. (C) Caspase-8 plays a role in the hyperacetylation of histone-H3 by NPI-0052. Top panel, the I9.2 (caspase-8–deficient) cells were incubated with 24 mM NAC for 30 minutes, before 5 μM MS-275, 10 nM NPI-0052, or a combination for a total of 6 hours. Cell lysates were analyzed by Western blot analysis for Ac-H3 and actin. Bottom left panel, protein lysates from Jurkat and I9.2 cells treated with 10 nM NPI-0052, 5 mM MS-275, or diluent for 6 hours were analyzed for Ac-H3 and histone H3 protein expression by Western blot analysis. Bottom right panel, protein lysates from I9.2 cells and I9.2/caspase-8/EGFP-transfected cells exposed to DMSO or 10 nM NPI-0052 for 6 hours were evaluated for Ac-H3, caspase-8, and actin protein expression by Western blot analysis. (D) Caspase-8 mediates caspase-3 activation by NPI-0052/vorinostat regimen. Caspase-3 activity was measured in Jurkat and I9.2 cells exposed to DMSO, 500 nM vorinostat, 10 nM NPI-0052, or the combination for 8 hours. *P < .001 compared control or either agent alone; †P < .001 compared with Jurkat cells incubated with NPI-0052/vorinostat.
We next examined if caspase-8 could be playing a role in the hyperacetylation status of histone-H3 and total histone-H3 expression. For this, the caspase-8–deficient Jurkat cells were treated with 5 μM MS-275, 10 nM NPI-0052, or the combination for 6 hours. Immunoblot results show that MS-275 is able to cause hyperacetylation of histone-H3 (Figure 6C top panel lane 3), however, NPI-0052 treatment did not cause histone-H3 acetylation (Figure 6C top panel lane 5). Hyperacetylation of histone-H3 is still observed in cells treated with the combination (Figure 6C top panel lane 7) compared with control (Figure 6C top panel lane 1), however it is not as robust as previously observed with combination treatment in normal Jurkat cells (Figure 5A lane 7). In this experiment, NAC had no effect on hyperacetylation by NPI-0052 (Figure 5A lane 6), suggesting that caspase-8 activation is upstream of ROS involvement in hyperacetylation of histone-H3 as a consequence of proteasome inhibition.
Because NPI-0052 alone is not able to induce hyperacetylation of histone-H3 in I9.2 cells, the acetylation observed in the combination treatment is most likely reflecting the effects of MS-275. To show this, Jurkat cells and I9.2 cells (caspase-8–deficient) were compared side-by-side (Figure 6C bottom panel). Increased total histone-H3 expression levels were detected in both Jurkat and I9.2 cells exposed to 10 nM NPI-0052 compared with cells treated with diluent. However, increased acetylation was only detected in Jurkat cells, whereas the caspase-8–lacking cells did not exhibit hyperacetylation. These results indicate that caspase-8 is playing a role in NPI-0052 induced hyperacetylation of histone-H3, but is not involved in increased histone-H3 levels as a consequence of proteasome inhibition.
To further test the requirement for caspase-8 in NPI-0052–induced hyperacetylation of histone-H3, wild-type caspase-8/GFP was reintroduced into caspase-8–deficient cells (I9.2) followed by a 6-hour incubation with 10 nM NPI-0052. In comparison to parental I9.2 cells, the caspase-8/GFP-transfected cells had more acetylation in the presence of NPI-0052 (Figure 6C bottom right panel lanes 2,4). However, caspase-8/GFP-transfected cells without treatment also displayed hyperacetylation (Figure 6C bottom right panel lane 3) compared with parental cells (Figure 6C bottom right panel lane 1) suggesting that overexpression and activation of caspase-8 is sufficient to alter acetylation status of histone-H3. Surprisingly, NPI-0052 treatment in caspase-8 overexpression I9.2 cells did not result in more acetylation (Figure 6C bottom right panel lane 4) than I9.2 caspase-8/GFP-transfected cells exposed to diluent (Figure 6C bottom right panel lane 3). Caspase-8 expression was confirmed by Western blot analysis.
We next wanted to explore if caspase-8 could also be involved in NPI-0052/vorinostat cytotoxicity, because we had previously observed caspase-8 dependence for NPI-0052/MS-275 cytotoxicity.10 Thus, we measured caspase-3 activation in Jurkat and I9.2 cells treated with 500 nM vorinostat, 10 nM NPI-0052, or the combination. Significantly increased caspase-3 activation (P < .001) was detected in Jurkat cells treated with the combination compared with diluent or either compound by itself (Figure 6D). Furthermore, caspase-3 activity was significantly decreased (P < .001) in cells that lacked caspase-8 in contrast to normal Jurkat cells.
Discussion
Several recent reports have focused on describing the interactions of HDACi and proteasome inhibitors as a therapeutic strategy for both solid and liquid tumors.15,16,18 Promising results from these studies have led to the development of early clinical trials in myeloma, lymphoma, prostate cancer, and leukemia. However, all of these efforts have focused on bortezomib, the sole FDA-approved proteasome inhibitor, or MG132, which shares structural features with bortezomib but is not suitable for clinical use. Published results from our laboratory indicate that a unique proteasome inhibitor, NPI-0052, demonstrates more potent synergy with HDACi than that observed with bortezomib in leukemia cells.10 Our current work provides insight into the mechanism of the observed synergy between NPI-0052 and the HDACi. First, HDACi inhibit the proteasome by repressing gene expression of the β subunits that are responsible for the enzymatic activities of the 20S proteasome. Second and more surprisingly, NPI-0052 promotes histone acetylation in a caspase 8-dependent manner.
The ability of HDACi to inhibit the β subunits of the proteasome has been reported for specific compounds. Trichostatin A and butyrate (both not suitable for clinical use) were found to cause down-regulation of β5, β1, and β2 genes and inhibition of chymotrypsin-like activity in a colonic epithelial cell line.21 In addition, in a gene microarray study examining multiple myeloma cells treated with vorinostat, decreased expression of the β5 proteasomal subunit gene was observed and corresponded to a 50% decrease in chymotrypsin-like activity.33 In leukemia cells, we also detected reduced mRNA expression of β subunits of the proteasome and their corresponding catalytic activities with exposure to vorinostat. Our current study extends those findings to also include the benzamide HDACi, MS-275, which was able to significantly suppress mRNA expression of catalytic β5, β2, and β1 proteasomal subunits in leukemia cells. This reduction had functional consequences: even though MS-275 had no direct effect on proteasome activity (Figure 3F), it diminished the chymotrypsin-like (located in β5 subunit) and caspase-like (found in β1 subunit) activities of the proteasome by more than 90%. Given the fact that MS-275 inhibits Class I HDACs whereas vorinostat is a pan-HDAC inhibitor, our data point toward HDACs 1, 2, and 3 in being responsible for gene expression of the β subunits. Generally, activation of gene transcription is associated with HDACi, however gene repression has also been reported. This likely occurs through modulation of acetylation status of nonhistone proteins such as transcription factors.34 Thus, acetylation and subsequent inactivation of a transcription factor that promotes expression of the proteolytic β subunits could be a potential explanation for our observations. Further experiments need to be done to address this possibility. Little is known regarding the identity of transcription factors that might drive expression of the proteasomal β subunits in human cells. In yeast, the transcription factor Rpn4 promotes expression of the 3 evolutionarily conserved β subunits,35 however, to date a functional mammalian homolog of Rpn4 has yet to be identified.
Our finding that NPI-0052 alone can hyperacetylate histone-H3 was a surprise, particularly because bortezomib at equimolar doses does not. Strikingly, it was observed in both cell lines and primary CLL cells that hyperacetylation of histone-H3 occurs in the presence of NPI-0052 alone or when either proteasome inhibitor (bortezomib or NPI-0052) is combined with HDACi (MS-275 and vorinostat). From previous work, we know that NPI-0052 relies more heavily on caspase-8 activation than bortezomib, because caspase-8 inhibition by pharmacologic or genetic means has a more profound effect on NPI-0052's cytotoxicity.8,10 In addition, ROS (primarily superoxide) are detected at higher levels in NPI-0052–treated cells compared with bortezomib-treated cells (Figure 2D). Therefore, we investigated whether abrogating ROS up-regulation or diminishing caspase-8 would affect histone acetylation by NPI-0052. Pretreatment with NAC prevented hyperacteylation of histone-H3 by NPI-0052, as did loss of caspase-8. Therefore both caspase-8 and oxidative stress appear important in the regulation of hyperacetylation of histone-H3 by NPI-0052. These effects of NPI-0052 on histone acetylation may reflect histone solubility, cell cycle-dependent histone variations, or subcellular localization of histones, because our data also show that total expression of histone-H3 was increased by NPI-0052 in cell lines and primary patient samples. Immunoprecipitation experiments (Figure 4C) indicated that this accumulation of histone-H3 in NPI-0052–treated cells is not due to ubiquitination, and quantitative PCR shows heightened histone-H3 mRNA levels (data not shown). While posttranslational modification of histone H3 is still being investigated, our preliminary findings suggest that NPI-0052's effects are likely via stabilization of a transcription factor that promotes histone-H3 expression.
The mechanism by which caspase-8 is playing a role in NPI-0052 induced hyperacetylation of histone-H3 is unclear. Cleavage of HDAC-3 resulting in cytoplasmic relocalization has been shown to be caspase-8–dependent.36 This process was associated with increased acetylation in human osteosarcoma cells. HDAC-7 has also been identified as a caspase-8 substrate, influencing its localization and activity.37 Based on our previous observations that caspase-8 is activated shortly after proteasome inhibition by NPI-0052,10 it is conceivable that the mode by which NPI-0052 is inducing hyperacetylation of histone-H3 is by first activating caspase-8, which in turn cleaves an HDAC, ultimately affecting acetylation status. Consistent with this hypothesis, reintroduction of caspase-8 in caspase-8–deficient I9.2 cells, which was accompanied by activation of caspase-8 as demonstrated by the appearance of cleaved fragments, also caused hyperacetylation of histone-H3.
The concept of proteasome inhibitors promoting biochemical events traditionally associated with epigenetically targeted drugs is gaining momentum. A recent study in AML cells found that bortezomib can cause DNA hypomethylation thereby preventing gene transcription.38 Interestingly, the authors implicated a Sp1/NF-κB–dependent DNA methyltransferase activity in this effect. Relevant to our study, cursory analysis of transcription factors that drive histone-H3 expression revealed that Sp1 may be involved. Experiments directed toward this possibility are currently under way.
Induction of cell death by proteasome inhibitor/HDAC inhibitor regimens has been well described in terms of apoptotic mechanisms. For the NPI-0052/HDACi combination, caspase-8 and oxidative stress appear to be key proapoptotic events, particularly because they also control histone acetylation status. This raises the question of how caspase-8 is activated by NPI-0052 alone or in combination with HDACi and how ROS become elevated. Answers to these questions, which we hope to provide in future studies, will offer insight into using these agents more effectively, particularly in the setting of acute and chronic leukemias.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors gratefully acknowledge grant support from the National Institutes of Health (NIH, Bethesda, MD; F31 CA123645 to C.M. and NIH R01 CA115811 to J.C.). The authors also greatly appreciate valuable advice and guidance from Scott Kaufmann, MD, PhD, on the manuscript and the caspase-8 overexpression construct generated by his associate, Xue Wei Meng, PhD.
National Institutes of Health
Authorship
Contribution: C.M. performed research, analyzed data, and wrote a draft of the paper; S.R. performed research; M.J.K., W.W., and M.P. contributed vital reagents; and J.C. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: M.P. is an employee of Nereus Pharmaceuticals, the company that manufactures NPI-0052. The remaining authors declare no competing financial interests.
Correspondence: Joya Chandra, Box 853, Pediatrics Research, University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX; e-mail: jchandra@mdanderson.org.

![Figure 2. Oxidative stress is elevated as a consequence of NPI-0052 and vorinostat interactions. (A) NAC partially protects from NPI-0052/vorinostat-induced apoptosis. Jurkat cells were pretreated with 24 mM NAC for 30 minutes, followed by treatment with diluent, 500 nM vorinostat, 5 nM NPI-0052, or combination of drugs for 24 hours. Apoptosis was assessed by PI staining and subsequent flow cytometric analysis. Statistical differences were calculated: *P < .05 for NPI-0052/vorinostat-treated cells compared with control (dimethyl sulfoxide [DMSO]); **P < .01 for NPI-0052/vorinostat compared with NPI-0052/vorinostat/NAC. (B) Intracellular superoxide production in Jurkat cells treated with NPI-0052/vorinostat regimen is decreased with NAC. Cells were treated as indicated for 12 hours, followed by staining with HEt and analyzed by flow cytometry. Shown are the mean fluorescence values of 3 independent experiments. (C) NPI-0052/vorinostat increases superoxide levels, which are decreased by NAC. Shown is a representative histogram of the 3 experiments performed for HEt staining (Figure 4B). Solid line represents control cells, solid bold line is 5 nM NPI-0052/500 nM vorinostat-treated cells and dashed line indicates 5 nM NPI-0052/500 nM vorinostat cells pretreated with 24 mM NAC. (D) NPI-0052 generates more superoxide levels than bortezomib in Jurkat cells. Cells were exposed to 5 nM NPI-0052, bortezomib, or diluent for 12 hours and stained with HEt. Samples were analyzed by flow cytometry. Shown are mean fluorescence values of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/18/10.1182_blood-2008-08-174797/6/m_zh80170933890002.jpeg?Expires=1769099365&Signature=h1RA2E2wrNMAS~GXlChCZVliOckyFyYP0uVOqA64IHSlHF6eUQSV69e~kJ-gKOxXCFR40gEatfNgiJb914ylce9p6Lr4MlJ4bC4yyugDbryxA1psG1tsmxxZWTZ0RQu3aQQcDh5wOxa0vUtbL3PW0azTkhX4DiMqam~6suz4CsEMqwjewihaEF99BJtbfeAal0nspF6RpOrMkYEs2-mav2P~Aet9VvJTKJXzOb9Ob563l2dwJJZzXByjh~KY-WZol~snrdbav6pEzWxQwBeP8oExd0M3eoWyGN0dEXeEtpMHBA1avdmeFR-q~Kvo7Y9uXwnqlySkyWyjPfr95Q8QrQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal