Abstract
In this study, we demonstrate the suitability of the vertebrate Danio rerio (zebrafish) for functional screening of novel platelet genes in vivo by reverse genetics. Comparative transcript analysis of platelets and their precursor cell, the megakaryocyte, together with nucleated blood cell elements, endothelial cells, and erythroblasts, identified novel platelet membrane proteins with hitherto unknown roles in thrombus formation. We determined the phenotype induced by antisense morpholino oligonucleotide (MO)–based knockdown of 5 of these genes in a laser-induced arterial thrombosis model. To validate the model, the genes for platelet glycoprotein (GP) IIb and the coagulation protein factor VIII were targeted. MO-injected fish showed normal thrombus initiation but severely impaired thrombus growth, consistent with the mouse knockout phenotypes, and concomitant knockdown of both resulted in spontaneous bleeding. Knockdown of 4 of the 5 novel platelet proteins altered arterial thrombosis, as demonstrated by modified kinetics of thrombus initiation and/or development. We identified a putative role for BAMBI and LRRC32 in promotion and DCBLD2 and ESAM in inhibition of thrombus formation. We conclude that phenotypic analysis of MO-injected zebrafish is a fast and powerful method for initial screening of novel platelet proteins for function in thrombosis.
Introduction
Platelets play a central role in hemostasis, adhering to damaged or activated endothelium and aggregating to form a thrombus. Receptor-ligand interactions on the platelet surface regulate these processes. Recent transcriptome and proteome studies of platelets or their precursor cell, the megakaryocyte (MK), have increased our understanding of the complement of proteins present in the human platelet membrane.1-3 Approximately 10 000 genes are transcribed in MKs1,2,4 and less than 1% of these are lineage restricted compared with nucleated blood cell elements and erythroblasts.4 Some of the functionally best-characterized platelet membrane proteins are represented in this small set of transcripts (eg, glycoprotein [GP] IIb), whereas others encode transmembrane proteins whose roles in hemostasis and thrombosis are unknown. Unfortunately, most of the approaches that can be applied to characterize novel genes are of limited use when studying human platelets or investigating large sets of candidate genes. For example, large-scale RNAi screens, commonly used in other cell types, cannot be replicated for platelets because of a combination of factors, including the inability to culture human platelets in vitro or the low efficiency of viral transformation of human hematopoietic stem cells and their progeny.
In this study, we investigated the suitability of zebrafish for functional analysis of novel platelet genes. The zebrafish has recently emerged as an attractive model organism for studying vertebrate hemostasis, as it uniquely combines the advantages of genetic tractability with biologic relevance.5 The coagulation pathways and genetic programs regulating hematopoiesis and vasculogenesis are conserved,6-9 which support the use of zebrafish in the study of human blood diseases. Moreover, zebrafish have nucleated thrombocytes that function in a similar manner to human platelets.10-12 Adhesion and aggregation in response to platelet agonists are comparable,11 and antisera against the human forms of GPIb-α and GPIIb are reactive with zebrafish thrombocytes.11
Zebrafish offer several practical advantages over mice for this type of study. First, external fertilization and transparency of zebrafish embryos allow better visualization of early blood-related phenotypes than mice, where development occurs in utero. Second, zebrafish can survive without blood for the first 7 days of life, allowing the function of genes that give lethal phenotypes when knocked out in mammals to be investigated. Finally, knockdown of gene function by antisense morpholino oligonucleotides (MOs) in zebrafish is more suitable to the screening of a large number of genes than classic gene knockout in mice13 and gives an immediate phenotype.
Using a laser-induced thrombosis model to characterize thrombus formation in arteries of zebrafish where known and novel platelet genes were knocked down by MOs, we show that key proteins involved in platelet aggregation and in coagulation in humans, namely, GPIIb and coagulation factor (F) VIII, are also required for normal hemostasis in the zebrafish. We previously identified a series of genes encoding platelet transmembrane proteins that may represent novel regulators of human platelet function2 and have now used the zebrafish model to conduct the first reverse genetics screen in zebrafish of candidate genes identified in systems biology studies of human blood cell elements.
Methods
Microarray studies and bioinformatics
Comparative whole-genome expression analysis of the major blood cell elements and in vitro–differentiated MKs and erythroblasts (EBs) was described previously.4 In summary, natural killer cells, monocytes, granulocytes, B cells, cytotoxic T cells, and helper T cells were isolated by positive selection using magnetic bead technology from peripheral blood from consenting volunteer blood donors of the Cambridge BioResource. In vitro–differentiated MKs and EBs were derived from CD34+ hematopoietic stem cells obtained from 5 different anonymized cord bloods collected after informed consent in accordance with the Declaration of Helsinki and the guidelines of Eurocord Netherlands.2 The transcriptomes of these and human umbilical vein endothelial cells were profiled by microarray and differentially expressed genes were identified by pairwise comparisons.4 Approval for these studies was obtained from the National Health Service Research Ethics Committee (Huntingdon, United Kingdom). Functional protein domains were predicted using the Eukaryotic Linear Motif resource.14
Confirmation of transcript expression in human platelets
The expression of candidate genes was validated by quantitative TaqMan real-time polymerase chain reaction (RT-PCR). Total RNA (100 ng) was reverse-transcribed using the reverse transcription system (Promega, Madison, WI) and an oligo (dT) primer. After a 1-hour incubation at 42°C, 2 ng cDNA was amplified with template-specific primers using the Mini Opticon real-time PCR system with SYBR green master mix (Bio-Rad, Hercules, CA). Reactions were incubated for 94°C for 10 minutes, and real-time PCR performed more than 40 cycles (94°C for 30 seconds, 52°C for 30 seconds, 72°C for 30 seconds). Data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using a standard curve generated with a dilution series of GAPDH plasmid from which copy number per nanogram of RNA was determined. CD16+ cells had the lowest level of GAPDH; therefore, the ratio of GAPDH copies per nanogram (CD16+) to GAPDH copies per nanogram (other cell type) was used to normalize the remaining data.
Antibody staining of human platelets
Polyclonal antisera were generated against Escherichia coli–produced human recombinant proteins in mice as described previously.2 cDNAs corresponding to the extracellular domains of human ANTXR2 (residues 33-318), BAMBI (residues 27-153), DCBLD2 (residues 66-163), ESAM (residues 30-247), and LRRC32 (residues 29-296) were used to produce the immunizing protein. Each antiserum was characterized by immunoblot as described previously.2 For flow cytometry experiments, human blood was collected into trisodium citrate and platelet-rich plasma obtained by centrifugation. For intracellular staining, platelets in platelet-rich plasma were fixed, washed, and then permeabilized using an IntraStain kit (Dako, Ely, United Kingdom) according to the manufacturer's instructions. Samples were incubated with individual mouse sera (1/100), washed, and bound; antibody detected by flow cytometry (EPICS MCL-XL; Beckman Coulter, Buckinghamshire, United Kingdom) using a phycoerythrin-conjugated goat anti–mouse reagent. Platelets were identified by their light scatter properties and phycoerythrin fluorescence measured on a log scale. Preimmune sera from the same mouse were used as negative control.
MO sequences and injections
Zebrafish orthologs were predicted by reciprocal BLAST analysis using the Ensembl utilities at http://www.ensembl.org. The criteria for a probable ortholog was more than or equal to 35% amino acid identity over the entire length of the protein, or more than or equal to 50% if the zebrafish protein sequence was only partial. Antisense MOs (Gene Tools, Philomath, OR) were designed complementary to the 5′ sequence near the start of translation (atg), or at splice junctions (sp)13 (Table 1). Up to 12 ng MO in Danieau solution was microinjected into the yolk of 1- to 4-cell–stage embryos as described previously.13 Dose-response experiments were carried out for all MOs to determine the highest nontoxic dose for use in further experiments. RT-PCR was carried out on the splice MO-injected larvae and their controls as described in supplemental material (available on the Blood website; see the Supplemental Materials link at the top of the online article), with primers listed in Table S1.
Zebrafish embryo collection
General maintenance, collection, and staging of the zebrafish were carried out according to the Zebrafish Book.15 Embryos were maintained in egg water (60 mg/L Red Sea salts, 2 mg/L methylene blue) until the appropriate stage. The approximate stages are given in days post-fertilization (dpf) at 28°C according to morphologic criteria.16
Phenotypic analyses
Morphology.
Gross morphology was analyzed by intravital light-field microscopy. Inspection for spontaneous bleeding was carried out on unmanipulated larvae 2 to 4 dpf.
Laser-induced thrombosis model.
Zebrafish larvae that showed normal blood circulation at 3 or 4 dpf were anesthetized in Tricaine solution (300 μM; Sigma-Aldrich, St Louis, MO) for 20 minutes and placed under the microscope (Leica DMRXA microscope, Wetzlar, Germany; 40× water-immersion objective). Injury of the caudal artery endothelium was by a nitrogen-pulse ablating laser (VSL-337; LSI Laser Science, Newton, MA) and was delivered for 1 to 5 seconds at 3 pulses/second at the maximum intensity setting. Each larva was injured only once. Images of wild-type and CD41–green fluorescent protein (GFP) transgenic zebrafish17 were acquired with a CCD camera (Princeton Scientific Instruments, Monmouth Junction, NJ) using Metamorph software version 6.0 (Molecular Devices, Sunnyvale, CA). Time to attachment (TTA) corresponded to the first cell adhering to the injured vessel after injury (t = 0). Thrombus size was calculated at 1-minute intervals after injury by measuring thrombus surface area (TSA) using ImageJ software.18 Briefly, for each image an outline was drawn around the thrombus (Figure 3B, examples) and the surface area within calculated automatically. For each experiment, typically 10 to 20 MO-injected larvae were screened per MO with a similar number of matched sibling controls tested before and afterward, the total experiment lasting less than 4 hours. Each experiment was repeated at least once using offspring of different parentage.
Statistical analysis of the thrombosis model
For individual experiments, statistical analyses of TTA and TSA were performed using an unpaired Student t test. For the combined analyses, TTA was modeled using Cox regression stratifying by experimental day. The severity of the injury did not significantly affect the model, and so was not adjusted for. Although TSA is highly skewed, log10 (TSA) is approximately normally distributed; therefore, log10 (TSA) was fitted with a linear mixed model with (1) fixed effects for larva type (control or MO-injected fish) and time after injury (1, 2, 3, or 4 minutes) and (2) random effects for experimental day and larva within experimental day. The estimated coefficients were antilogged to transform back to the original scale (square pixels). Using contrasts, we tested whether the effect of the 2 MOs tested differed. In most cases, there was no significant difference (P < .05), and so the MOs were combined.
Results
ANTXR2, BAMBI, DCBLD2, ESAM, and LRRC32 are present in human platelets and are expressed at higher levels in MKs than in other blood cell lineages
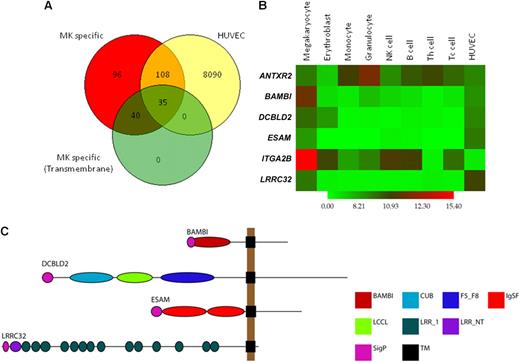
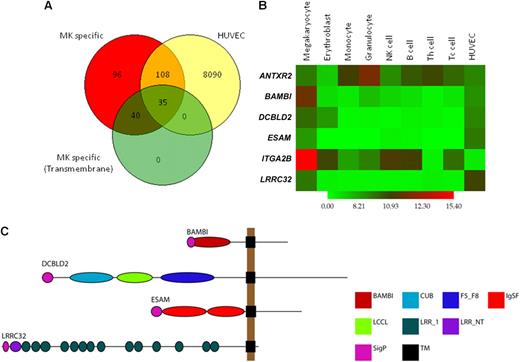
In a previous study, comparative whole-genome expression analysis of helper and cytotoxic T cells, monocytes, B cells, NK cells, granulocytes, EBs, and MKs allowed the identification of genes with up-regulated expression in MKs compared with all other cell lineages.4 Because platelets are derived from MKs, we hypothesized that this subset of genes would provide good candidates for proteins playing a role in thrombus formation. Analysis of the expression data indicated that 10 314 genes are present in MKs, of which 1035 are up-regulated in MKs relative to EBs and 279 are uniquely expressed in MKs relative to all other blood cells (Figure 1A).
Candidate gene selection and validation. Candidate genes were selected from a list of 75 MK-specific genes encoding transmembrane proteins. (A) Overlap analysis of MK-specific, present in HUVECs and transmembrane protein encoding genes. Of the 279 MK-specific genes, 143 are also present in HUVECs, and 35 of these encode transmembrane proteins. (B) Heatmap of log2 transformed, normalized intensity values for the 6 MK genes used in this study, showing increased expression of 4 in MKs and HUVECs. (C) Domain architecture of BAMBI, DCBLD2, ESAM, and LRRC32 as predicted using the Eukaryotic Linear Motif resource. The length of the black line represents the number of amino acids with domain positions to scale. Domain acronyms: CUB indicates complement C1r/C1s, Uegf, Bmp1; F5_F8, coagulation factor 5/8 type; IgSF, immunoglobulin fold; LCCL, Limulus factor C, Coch-5b2 and Lgl1; LRR_1, leucine rich repeats; LRRNT, leucine rich repeat N-terminal domain; SigP, signal peptide; TM, transmembrane domain.
Candidate gene selection and validation. Candidate genes were selected from a list of 75 MK-specific genes encoding transmembrane proteins. (A) Overlap analysis of MK-specific, present in HUVECs and transmembrane protein encoding genes. Of the 279 MK-specific genes, 143 are also present in HUVECs, and 35 of these encode transmembrane proteins. (B) Heatmap of log2 transformed, normalized intensity values for the 6 MK genes used in this study, showing increased expression of 4 in MKs and HUVECs. (C) Domain architecture of BAMBI, DCBLD2, ESAM, and LRRC32 as predicted using the Eukaryotic Linear Motif resource. The length of the black line represents the number of amino acids with domain positions to scale. Domain acronyms: CUB indicates complement C1r/C1s, Uegf, Bmp1; F5_F8, coagulation factor 5/8 type; IgSF, immunoglobulin fold; LCCL, Limulus factor C, Coch-5b2 and Lgl1; LRR_1, leucine rich repeats; LRRNT, leucine rich repeat N-terminal domain; SigP, signal peptide; TM, transmembrane domain.
To narrow down this selection, we used Ensembl and InterPro interrogation of the 279 MK-specific transcripts to identify those encoding putative transmembrane domains because proteins that span the plasma membrane often play crucial roles in modifying cell function and are attractive targets for drug design. Seventy-five genes were identified and are listed in Table S2, and these included genes for well-known MK/platelet-specific receptors (eg, GP6 and GP9), recently reported and confirmed MK/platelet-specific genes (eg, LRRC32, SUCNR1, and C21orf21)2 and novel genes. Finally, we compared this list to the transcriptome of resting human umbilical vein endothelial cells (HUVECs) to determine which of these genes are expressed in both cell lineages because a significant number of known MK/platelet-specific membrane proteins are also expressed in endothelial cells (Figure 1A). Thirty-five genes were identified (Figure S1), and from these we selected 4 for study in the zebrafish thrombosis model, based on (1) their unknown function in thrombosis at time of selection, (2) clear MK and endothelial cell–specific transcript expression (Figure 1B), (3) interesting protein domains (Figure 1C), and (4) reliable zebrafish ortholog identification (Table 2). The integrin gene ITGA2B (encoding GPIIb) and the gene encoding 1 of the 2 known anthrax receptors, ANTXR2, which are more widely expressed, were also included as controls.
The MK-specific nature of the first 4 transcripts and the relative overexpression of the latter 2 in granulocytes and MKs is shown via a heatmap (Figure 1B). Functional domain prediction identified a single BMP and activin membrane-bound inhibitor homolog in BAMBI, a discoidin, CUB, and LCCL domain in DCBLD2, 2 immunoglobulin folds in endothelial cell–selective adhesion molecule (ESAM), and 15 leucine-rich repeats in LRRC3214 (Figure 1C). The restricted expression of LRRC32 has been reported in earlier studies from our laboratory and was confirmed by quantitative PCR for BAMBI, DCBLD2, and ESAM (data not shown).
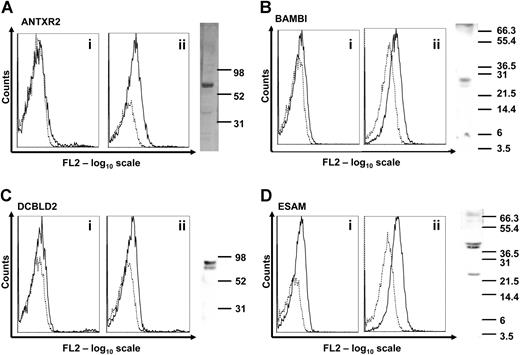
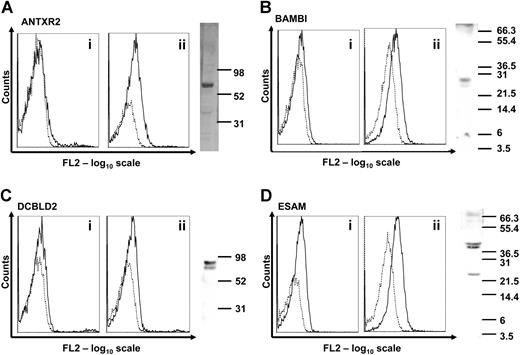
To verify that these genes are expressed in human platelets at the protein level, we used a combination of flow cytometry and Western blot with purpose-generated polyclonal antibodies (Figure 2). Expression of the LRRC32 protein in human platelets was already demonstrated.2 ANTXR2, BAMBI, DCBLD2, and ESAM were detected either on the native platelet surface and/or intracellularly after permeabilization of the platelets. In Western blots with platelet lysates, a band of expected size (27 kDa) was detected for BAMBI, whereas the others were of higher molecular weight, most likely due to glycosylation. ANTXR2 was approximately 67 kDa, DCBLD2 was 85 kDa, and ESAM ran as 2 bands of 40 and 43 kDa (Figure 2 right panels).
Expression of novel transmembrane proteins in human platelets. (A-D) Flow cytometric detection of the novel transmembrane proteins was carried out in platelet-rich plasma (i) or in permeabilized platelets (ii). Binding of individual antibodies to human platelets measured as fluorescence intensity is indicated by solid lines and matched preimmune serum by dotted lines. Expression of each protein in human platelet lysates was analyzed by Western blot. Molecular weight markers are indicated on each blot in kilodaltons. Bands of 67, 27, 85, and 40/43 kDa were detected, corresponding to ANTXR2, BAMBI, DCBLD2, and ESAM proteins, respectively. The presence of LRRC32 in platelets using similar detection procedures has already been reported.2
Expression of novel transmembrane proteins in human platelets. (A-D) Flow cytometric detection of the novel transmembrane proteins was carried out in platelet-rich plasma (i) or in permeabilized platelets (ii). Binding of individual antibodies to human platelets measured as fluorescence intensity is indicated by solid lines and matched preimmune serum by dotted lines. Expression of each protein in human platelet lysates was analyzed by Western blot. Molecular weight markers are indicated on each blot in kilodaltons. Bands of 67, 27, 85, and 40/43 kDa were detected, corresponding to ANTXR2, BAMBI, DCBLD2, and ESAM proteins, respectively. The presence of LRRC32 in platelets using similar detection procedures has already been reported.2
Replication of a Glanzmann thrombasthenia phenotype validates zebrafish as a suitable model organism for platelet functional studies
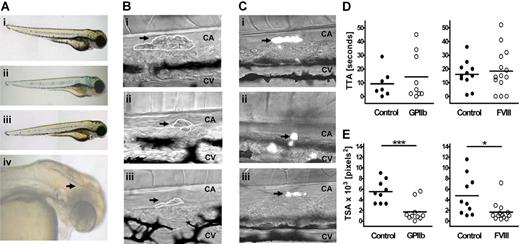
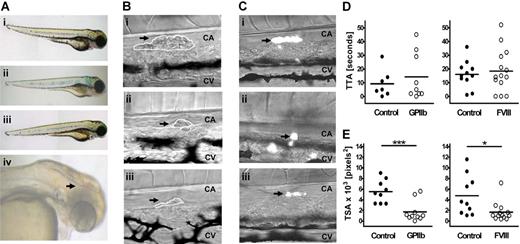
To establish whether zebrafish would be suitable for studying platelet receptor function, we knocked down the gene encoding GPIIb, aiming to replicate the well-characterized phenotype.19 In patients with GT (Glanzmann thrombasthenia), platelet aggregation, but not adhesion, is severely impaired because of loss of function of integrin GPIIbIIIa. As expected, the GPIIb MO-injected fish did not display any gross morphologic malformation, nor was spontaneous bleeding observed (Figure 3Aii). We hypothesized that any possible defect in thrombocyte aggregation in the fish might be accentuated by impairment of the coagulation pathway, and tested this by concurrent knockdown of the gene encoding the procoagulant protein FVIII. A subset of these double MO-injected fish bled spontaneously, most commonly in the tail, but intracranial bleeds were also observed (20% and 5% of larvae, respectively; Figure 3Aiv). Knockdown of the FVIII gene on its own did not produce a spontaneous bleeding phenotype (Figure 3Aiii).
Thrombus formation in GPIIb and FVIII MO-injected fish in a laser-induced arterial injury model. (A) Representative phenotypes of 3 dpf control siblings (i) and fish injected with 2 ng GPIIb MO (itga2b atg1, ii), 4 ng FVIII MO (f8 sp2, iii), or coinjected with both MOs at 2 ng and 8 ng, respectively (iv). An intracranial hemorrhage, as observed in 5% of larvae, is indicated by arrowhead (iv). (B) Representative images of thrombus formation (arrows) 2 minutes after injury in 3-dpf wild-type larvae (i) or larvae injected with 2 ng GPIIb (ii) or 4 ng FVIII MOs (iii). CA indicates caudal artery; CV, caudal vein. White lines represent each thrombus as carried out in ImageJ for determining TSA. (C) Representative images of thrombus formation in 4-dpf CD41-GFP fish controls (i), and siblings injected with 2 ng GPIIb (ii) or 4 ng FVIII (iii) MOs showing thrombocyte-rich thrombi. (D) TTA and (E) TSA (2 minutes after injury) were measured and compared with uninjected sibling controls. Statistical analyses were performed using an unpaired Student t test (*P < .05; ***P < .001).
Thrombus formation in GPIIb and FVIII MO-injected fish in a laser-induced arterial injury model. (A) Representative phenotypes of 3 dpf control siblings (i) and fish injected with 2 ng GPIIb MO (itga2b atg1, ii), 4 ng FVIII MO (f8 sp2, iii), or coinjected with both MOs at 2 ng and 8 ng, respectively (iv). An intracranial hemorrhage, as observed in 5% of larvae, is indicated by arrowhead (iv). (B) Representative images of thrombus formation (arrows) 2 minutes after injury in 3-dpf wild-type larvae (i) or larvae injected with 2 ng GPIIb (ii) or 4 ng FVIII MOs (iii). CA indicates caudal artery; CV, caudal vein. White lines represent each thrombus as carried out in ImageJ for determining TSA. (C) Representative images of thrombus formation in 4-dpf CD41-GFP fish controls (i), and siblings injected with 2 ng GPIIb (ii) or 4 ng FVIII (iii) MOs showing thrombocyte-rich thrombi. (D) TTA and (E) TSA (2 minutes after injury) were measured and compared with uninjected sibling controls. Statistical analyses were performed using an unpaired Student t test (*P < .05; ***P < .001).
To analyze the phenotype of the GPIIb MO-injected fish in more detail, a thrombosis model was developed. This method is similar to those described in earlier studies using laser injury to initiate thrombus formation in zebrafish20,21 and mice22 and allows qualitative and quantitative differences in thrombus formation between MO-injected fish and controls to be measured. The transparency of zebrafish larvae at this stage allows blood cell attachment to the site of injury and subsequent thrombus formation to be visualized by intravital light microscopy, without intervention. In addition, we used a well-characterized transgenic zebrafish line with fluorescently labeled thrombocytes, where GFP expression is under the control of the CD41 promoter,17 to examine the specific contribution of thrombocytes to thrombus formation in control and MO-injected fish.
An ablating laser was used to injure the endothelial cell wall of the caudal artery of 3- to 4-dpf larvae and the TTA and TSA was measured in each case. These endpoints were chosen because they allow distinction between 2 stages of primary hemostasis: thrombus initiation (including endothelial cell activation, thrombocyte tethering, and activation) and thrombus growth (including aggregation and firm adhesion). In control larvae, thrombi could be reproducibly induced (Figure 3Bi), with the first blood cell attaching within 5 to 20 seconds (Figure 3D). Thrombi were occlusive in approximately 10% of cases, and partial but incomplete lysis was observed after 2 to 10 minutes (data not shown). As expected, TTA was not significantly delayed in GPIIb MO-injected fish, but a significant reduction (> 50%) in TSA compared with controls was observed (P < .001) (Figure 3D,E left panels). Similarly, loss of FVIII should have little effect on TTA while decreasing thrombus size23 ; and, as anticipated, the TSA in FVIII MO-injected fish was only 33% that of controls (P = .016) accompanied by a normal TTA (Figure 3D,E right panels). Expression of itga2b and f8 genes in 3- to 4-dpf control embryos and their successful knockdown in GPIIb and FVIII MO-injected larvae were confirmed by RT-PCR (Figure S2).
Finally, analysis of thrombus formation in the CD41-GFP transgenic zebrafish (Figure 3C; Movie S1) showed that thrombi were initiated by thrombocytes, although in a few cases (< 10%) the first cell to attach was not fluorescent (data not shown). The phenotype of GPIIb and FVIII MO-injected transgenic fish was the same as in MO-injected wild-types; and in all cases, the thrombi were composed primarily of thrombocytes (Figure 3C).
Screening by reverse genetics in zebrafish identifies putative roles for BAMBI, DCBLD2, ESAM, and LRRC32 in thrombus formation
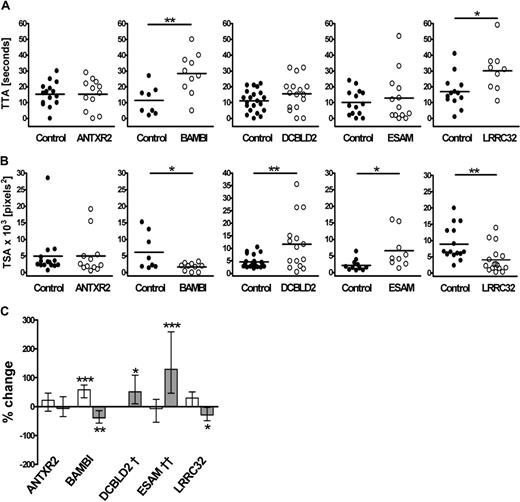
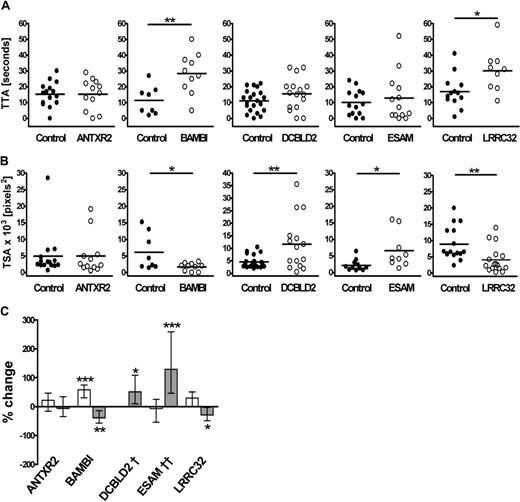
To investigate the potential roles of the 4 candidate genes in thrombus formation, we identified the zebrafish ortholog for each gene (Table 2) and designed 2 MOs per gene to act as mutual controls for assessment of the knockdown phenotype in the thrombosis model. The zebrafish ortholog of the control gene, ANTXR2, was similarly assessed. As shown for itga2b and f8 genes, antxr2, bambi, dcbld2, esam, and lrrc32 genes are expressed in 3- to 4-dpf embryos (Figure S2). No transcript was detected in embryos injected with the splice blocking MOs, confirming successful knockdown of each gene in the zebrafish (Figure S2). Representative results for TTA and TSA, obtained in MO-injected larvae compared with siblings, from a single experiment are presented in Figure 4A and B. Overall, 42 to 70 larvae were tested for each gene, and the effect of MOs directed at 2 different sites was similar (Table 3). The one exception was ESAM, where just one of the 2 MOs (esam sp1) resulted in degradation of the esam transcript (Figure S2); thus, we lack a corroborating result for this protein. However, the concordance of the ESAM phenotype with mouse knockout data, discussed later, suggests that the result with esam sp1 is specific.
Effects of antisense knockdown of ANTXR2, BAMBI, DCBLD2, ESAM, and LRRC32 zebrafish orthologs on thrombus formation. TTA (A) and TSA (B) in the MO-injected fish were compared with that of their matched sibling controls. Each experiment is representative of more than 4 experiments, including similar numbers for both groups, using 2 nonoverlapping MOs. Zebrafish larvae were injected with the following MOs: 2 ng ANTXR2 atg1, 2 ng ANTXR2 sp1, 2 ng BAMBI atg1, 1 ng BAMBI sp1, 4 ng DCBLD2 atg1, 0.5 ng DCBLD2 sp1, 1 ng ESAM sp1, 12 ng ESAM sp2, 4 ng LRRC32 atg1, or 2 ng LRRC32 sp1. Statistical analyses were performed using an unpaired Student t test (*P < .05; **P < .005). (C) Results from all experiments and for each of the 2 MOs tested per gene were combined for TTA (□) and TSA ( ) using Cox regression and linear mixed modeling, respectively, as described in “Statistical analysis of the thrombosis model” (*P < .05; **P < .005; ***P < .001). †TTA data for each DCBLD2 MO were significantly different and therefore were not combined. ††Results for TSA are applicable for esam sp1 only. Detailed P values for the combined and individual MO analysis are presented in Table 3.
) using Cox regression and linear mixed modeling, respectively, as described in “Statistical analysis of the thrombosis model” (*P < .05; **P < .005; ***P < .001). †TTA data for each DCBLD2 MO were significantly different and therefore were not combined. ††Results for TSA are applicable for esam sp1 only. Detailed P values for the combined and individual MO analysis are presented in Table 3.
Effects of antisense knockdown of ANTXR2, BAMBI, DCBLD2, ESAM, and LRRC32 zebrafish orthologs on thrombus formation. TTA (A) and TSA (B) in the MO-injected fish were compared with that of their matched sibling controls. Each experiment is representative of more than 4 experiments, including similar numbers for both groups, using 2 nonoverlapping MOs. Zebrafish larvae were injected with the following MOs: 2 ng ANTXR2 atg1, 2 ng ANTXR2 sp1, 2 ng BAMBI atg1, 1 ng BAMBI sp1, 4 ng DCBLD2 atg1, 0.5 ng DCBLD2 sp1, 1 ng ESAM sp1, 12 ng ESAM sp2, 4 ng LRRC32 atg1, or 2 ng LRRC32 sp1. Statistical analyses were performed using an unpaired Student t test (*P < .05; **P < .005). (C) Results from all experiments and for each of the 2 MOs tested per gene were combined for TTA (□) and TSA ( ) using Cox regression and linear mixed modeling, respectively, as described in “Statistical analysis of the thrombosis model” (*P < .05; **P < .005; ***P < .001). †TTA data for each DCBLD2 MO were significantly different and therefore were not combined. ††Results for TSA are applicable for esam sp1 only. Detailed P values for the combined and individual MO analysis are presented in Table 3.
) using Cox regression and linear mixed modeling, respectively, as described in “Statistical analysis of the thrombosis model” (*P < .05; **P < .005; ***P < .001). †TTA data for each DCBLD2 MO were significantly different and therefore were not combined. ††Results for TSA are applicable for esam sp1 only. Detailed P values for the combined and individual MO analysis are presented in Table 3.
Combined analysis of data from both MOs for each gene revealed significant alterations in cell attachment and/or thrombus growth for all 4 candidate genes but not for the ANTXR2 MO-injected fish (Figure 4C). BAMBI MO-injected fish had a longer TTA with an average increase of 58% (P < .001) and impaired thrombus development where TSA was reduced by 39% (P = .006). The effect of the DCBLD2 knockdown on TTA was ambiguous but resulted in a clear 51% increase in TSA compared with the controls (P = .013). The ESAM MO-injected fish showed normal TTA but increased thrombus size (P < .001). In the LRRC32 MO-injected fish, there was a marginally significant increase in TTA (30% increase, P = .051), whereas TSA was reduced by 29% compared with controls (P = .034). The P values for individual MOs and combined values are summarized in Table 3.
Discussion
The zebrafish has been proposed as a relevant model for hemostasis and thrombosis5,24,25 and presents several attractive features over the standard mouse model, the most practical being the transparency of zebrafish embryos, amenability to gene knockdown, and the short time needed to generate and study the MO-injected fish. In the present study, we report on the use of zebrafish for the initial rapid functional analysis of genes derived from postgenomic studies, with a hitherto unknown role in thrombosis.
We chose to validate this approach by knockdown of the zebrafish gene encoding the integrin subunit αIIb (GPIIb), a well-characterized platelet receptor in humans that is deficient in GT patients. The GPIIb MO-injected fish were morphologically identical to controls, concordant with GT patients where only a small proportion of patients display severe spontaneous bleeding.19 Moreover, when we accentuated the GT phenotype by concurrent knockdown of the gene encoding FVIII,7 the coagulation factor perturbed in hemophilia A patients, we observed spontaneous tail and intracranial bleeds, which correlate with increased severity of bleeding observed in rare cases of coexistence of GT and other inherited bleeding diseases19 or in hemophilia patients who have inadvertently used aspirin.26
We next used a laser-induced thrombosis model for more detailed analysis of the GT phenotype. This method allows the kinetics of thrombus formation to be measured in vivo, by observing thrombus initiation and development in real time. We observed that thrombus formation was severely impaired in GPIIb MO-injected fish compared with controls, although there was no difference in initial cell recruitment to the site of vascular injury, in accordance with observations made using GT mouse models27-29 and the well-characterized role of GPIIbIIIa in the later aggregation, but not initial adhesion, stages of thrombus formation.30 These observations highlighted the utility of the system to separate the processes of thrombus initiation from growth, not possible by analysis of gross morphology. Moreover, because knockdown of a critical receptor such as GPIIb did not produce a spontaneous bleeding phenotype, it is clear that the laser-induced thrombosis model is a more sensitive and informative tool to test for loss and gain of function.
A potential drawback with zebrafish knockdown experiments is that all cells and tissues are affected; MOs injected at the 1- to 4-cell stage will be present in every cell in the developing fish. Because we therefore cannot assume that the induced phenotype is purely the result of a defect in the function of thrombocytes alone, we selected genes encoding transmembrane proteins, which in humans have up-regulated expression in platelets or MKs compared with other blood cell types, to optimize our chances of identifying novel genes involved in thrombus formation. The zebrafish orthologs of BAMBI, DCBLD2, ESAM, LRRC32, and the control ANTXR2 were knocked down and morphologic and hemostatic phenotypes assessed 3 dpf. In total, more than 900 larvae were tested and power was gained by combining between 4 and 6 sets of experiments of each gene, using 2 nonoverlapping MOs. Observation in most of the cases (90%) of the same phenotype with both MOs provided strong evidence that the phenotype is specific to the gene of interest. In addition, for splice blocking morpholinos, we performed RT-PCR and showed that splicing was altered for all genes.
The BAMBI gene, which encodes a protein related to type I receptors of the transforming growth factor-β,31 was the most highly expressed in human MKs. A recent study of BAMBI-deficient mice showed no developmental defects, consistent with our observations in the zebrafish.32 However, here we found that BAMBI is required for both cell attachment to injured endothelium and for thrombus growth, suggesting that it plays a fundamental role in both processes. Analysis of thrombus formation in tissue-specific Bambi−/− mice would be a good next course of action and may unpick whether our observed phenotype is the result of a defect in thrombocyte or endothelial cell function, or both.
The gene DCBLD2 is transcribed in human MKs and, to a lesser degree, in endothelial cells and erythroblasts, but not in any other of the blood cell types tested. DCBLD2 MO-injected zebrafish show normal TTA but form larger thrombi than controls, suggesting that the receptor negatively regulates thrombus formation. The structure of the DCBLD2 protein is similar to the semaphorin-binding neuropilins33 ; thus, the receptor may play a role in signaling. We previously showed at least 7 semaphorin transcripts present in MKs,4 although only 2 have been characterized in platelets34,35 ; hence, this may be an interesting avenue of research.
We previously showed that the expression of LRRC32 is restricted to platelets and megakaryocytes.2 As a structural homolog of GPIbα and GPV, we predicted that the protein may operate as part of a multiprotein complex, perhaps playing a role in adhesion.2 Here, our combined analysis of LRRC32 MO-injected fish shows that the TTA is normal, although when the effects of each MO were examined individually, the TTA was increased in both cases. For one of the MOs, this increase reached significance; therefore, we do not wish to exclude the possibility of LRRC32 playing a role in thrombus initiation. The TSA result was more straightforward; both MOs produced a defect in thrombus growth. We suggest that LRRC32 could play a role in platelet-platelet and platelet-endothelial cell interactions, which would tie in with the propensity of LRR-rich receptors to form homotypic interactions.36 Alternatively, LRRC32 might have an as yet unidentified ligand in plasma, endothelial cells, or both, in line with what is known for von Willebrand factor and its interaction with the platelet-specific receptor GPIbα.
For all of these proteins, our work represents the first study on their putative role in thrombus formation. A recent study, however, has shown that Esam−/− mice form larger thrombi in an in vivo thrombosis model.37 The ESAM protein is expressed on platelet alpha granules38 and plays a role in increasing vascular permeability at endothelial cell tight junctions.39 Thus, it is quite possible that in platelets ESAM functions to destabilize platelet-platelet cell contacts. In agreement with this hypothesis, we found that fish injected with esam sp1 had normal thrombus initiation but dramatically increased thrombus growth and size. Replication of the phenotype observed in mice lends further support to the validity of this zebrafish model.
Knockdown of the zebrafish ANTXR2 ortholog did not significantly alter TTA, nor did it affect thrombus formation in MO-injected fish compared with controls. Because this gene is ubiquitously expressed compared with the other 4, these results show that, as predicted, MK/endothelial-specific gene expression is a strong indicator of a putative function in thrombosis.
The identification of 4 new genes that play a role in thrombus formation, together with the evidence that the proteins are expressed in human platelets, provides enough arguments for these genes to be studied further. On the basis of the functional data gained from this MO-based screen in the zebrafish, we are increasing screening throughput by combining Targeted Induced Local Lesions IN Genomes (TILLING) with automated image capture coupled with computer-based pattern recognition of the processes, leading to thrombus formation. This will allow many more genes to be characterized and put us one step closer to understanding the complex networks of interactions that take place within and between platelets and endothelial cells after vessel wall damage. In addition, the generation of genetically modified platelets and endothelial cells, for example, by Cre/Lox-based tissue-specific gene knockout in mice, will allow a more detailed study of the function of these genes in either tissue.
In conclusion, we have demonstrated the suitability of the zebrafish thrombosis model for the functional characterization of novel platelet proteins. This model will facilitate our understanding of the role of novel genes being identified in transcriptional studies of MKs and platelets and also in genome-wide association studies in patients with myocardial infarction40,41 and has the potential to contribute significantly to systems biology studies of platelets in health and disease.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr H. F. Lin and Dr R. I. Handin (Boston, MA) for their generous gift of the CD41-GFP transgenic zebrafish line.
This work was supported by grants from the European Union 6th Framework Program (LSHM-CT-2004-503485), the Wellcome Trust (grant no. WT082597/Z/07/Z: M.N.O., A.C.; grant no. WT077037/Z/05/Z: D.L.S.), and the National Institute for Health Research in England to National Health Service Blood and Transplant (N.A.W., S.F.G.). Funding for the Cambridge BioResource was from the National Institute for Health Research Biomedical Research grant for Cambridge University Hospitals National Health Service Foundation Trust.
Wellcome Trust
National Institutes of Health
Authorship
Contribution: M.N.O. and I.I.S. designed and performed research, analyzed data, and wrote the paper; N.A.W. and W.H.O. designed research, analyzed data, and wrote the paper; A.W., S.F.G., C.I.J., and A.C. designed and performed research; I.C.M. designed and performed research and analyzed data; J.-J.Z. performed research and analyzed data; S.L.B. and F.D. performed statistical analysis of data; M.S. analyzed data and provided critical reagents; B.d.B. provided bioinformatic support; A.H.G. designed research; D.L.S. designed research and provided laboratory space and reagents; and H.D. designed research and wrote the paper.
A complete list of participating Bloodomics Consortium members appears in the online Supplemental Appendix.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hans Deckmyn, Laboratory for Thrombosis Research, Interdisciplinary Research Centre, KU Leuven Campus Kortrijk, E. Sabbelaan 53, B-8500 Kortrijk, Belgium; e-mail: hans.deckmyn@kuleuven-kortrijk.be.
References
Author notes
*M.N.O. and I.I.S. contributed equally to this study.


 ) using Cox regression and linear mixed modeling, respectively, as described in “Statistical analysis of the thrombosis model” (*P < .05; **P < .005; ***P < .001). †TTA data for each DCBLD2 MO were significantly different and therefore were not combined. ††Results for TSA are applicable for esam sp1 only. Detailed P values for the combined and individual MO analysis are presented in
) using Cox regression and linear mixed modeling, respectively, as described in “Statistical analysis of the thrombosis model” (*P < .05; **P < .005; ***P < .001). †TTA data for each DCBLD2 MO were significantly different and therefore were not combined. ††Results for TSA are applicable for esam sp1 only. Detailed P values for the combined and individual MO analysis are presented in