Abstract
Although numerous studies have shown a higher risk of acquiring HIV infection in the presence of other sexually transmitted diseases, the biologic mechanisms responsible for enhanced HIV acquisition are unclear. Because Langerhans cells (LCs) are suspected to be the initial HIV targets after sexual exposure, we studied whether microbial components augment HIV infection in LCs by activating Toll-like receptor (TLR) and nucleotide-binding oligomerization domain (NOD) pattern recognition receptors. We found that TLR1/2 and TLR2/6 agonists dramatically enhanced both HIV susceptibility and replication in immature monocyte-derived LCs, whereas TLR3-5, TLR7-9, and NOD1,2 agonists did not significantly affect HIV infection. The same infection-enhancing effects were observed when LCs were incubated with other related bacterial components as well as with whole Gram+ bacteria. In resident LCs in human skin, TLR2 agonists also significantly increased HIV susceptibility. By contrast, TLR2 agonists and related bacterial components decreased HIV susceptibility in monocyte-derived dendritic cells (DCs). We found that TLR2 activation of LCs, but not DCs, resulted in a significant down-regulation of APOBEC3G, which is a cellular restriction factor for HIV. Given these data, we hypothesize that ligation of TLR2 by Gram+ bacterial products may underlie enhanced sexual transmission of HIV that occurs with concomitant bacterial sexually transmitted disease infections.
Introduction
Epidemiologic studies have suggested a strong association between the acquisition of HIV and other sexually transmitted diseases (STDs).1 The risk ratio for HIV acquisition for a person with genital ulcer disease ranges from 2.2 to 11.3, whereas for nonulcerative STDs, risk ratios range from 3 to 4.2 Additional studies have reported that bacterial vaginosis (BV) is associated with an increase in HIV acquisition.3,4 Genital ulcer disease, such as syphilis or herpes simplex infections, are thought to enhance HIV transmission, because genital ulcerations associated with these infections reduce epithelial barriers to HIV.1 Two nonulcerative diseases, Chlamydia and gonorrhea, are responsible for up to 80% of the cases of all notifiable diseases in the United States,5 whereas nonulcerative STDs, such as BV, Candida, and Trichomonas, are common in Africa.6 Mechanisms responsible for the enhanced HIV transmission by nonulcerative STDs are as of yet unknown, but they may be related to breaching of physical barriers to infection, disturbances in normal vaginal flora, or biologic effects on potential target cells for HIV infection.1
During sexual transmission of HIV, virus crosses mucosal epithelium and is eventually disseminated to proximally located lymphoid organs, where it establishes a permanent infection. Most evidence has indicated that Langerhans cells (LCs) are the initial cellular targets for HIV, and that these cells play a crucial role in disseminating HIV.7-10 LCs are abundantly present within genital mucosal epithelium, and after contact with pathogens, they readily emigrate from tissue to draining lymph nodes. Immature resident LCs express surface CD4 and C-C chemokine receptor (CCR) 5, but not surface CXC chemokine receptor 4. These LCs are readily infected ex vivo with R5 HIV, but not with X4 HIV.11-14 This is consistent with previous epidemiologic observation, which have found that the majority of HIV strains isolated from patients are R5 HIV after initial infection.15 It has been reported that persons with CCR5 homozygous defects are largely protected from sexually acquiring HIV.16 In rhesus macaques, within an hour after intravaginal inoculation of simian immunodeficiency virus (SIV), up to 90% of the SIV-infected cells were LCs, which also supports the role of LCs as initial target cells.17
Studies on dendritic cell–specific intercellular adhesion molecule-3–grabbing non-integrin (DC-SIGN), a C-type lectin receptor (CLR) that is expressed on dermal macrophages and monocyte-derived dendritic cells (mDCs),18,19 have shown that it can bind to HIV glycoprotein (gp) 120 and facilitate HIV infection of T cells in trans.18 Although results from other studies indicate a minor contribution by DC-SIGN in the transmission of HIV from mDCs to T cells,19,20 DC-SIGN may be involved in viral dissemination. In addition, langerin, a LC-specific CLR, has been shown to bind HIV gp120, suggesting that it also participates in viral dissemination.21 However, a recent study revealed that langerin impairs both infection of LCs by HIV and its subsequent viral dissemination.22 This study also showed that langerin was involved in capture of HIV and subsequent internalization within Birbeck granules, where it was degraded. Nevertheless, when LCs were exposed to high viral concentrations of HIV, there was significant infection of LCs by R5 virus, followed by viral transmission to T cells. This suggests that there is a saturation of langerin at higher virus concentrations that overwhelms the protective mechanism of action, so that langerin is ultimately unable to completely prevent LC infection. This might explain the recent in vivo findings that demonstrated that molecules targeting CCR5 were able to protect against mucosal transmission of SIV,23,24 whereas the CLR inhibitor, mannan, could not prevent SIV mucosal transmission in female macaques.24 When taken together with the human epidemiologic data cited above, the results suggest that CD4/CCR5-mediated de novo infection of LCs is the major pathway involved in sexual transmission of HIV.
When encountering pathogens, LCs and dendritic cells (DCs) recognize pathogen-associated molecular patterns (PAMP) by pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs). Subsequently, this leads to production of inflammatory cytokines that ultimately promote innate responses.25 In addition, 2 members of the nucleotide-binding oligomerization domain (NOD) family, NOD1 and NOD2, mediate recognition of special bacterial components, and function as cytosolic sensors for innate recognition of these microorganisms.26 Many studies have shown that ligands of the NOD family can synergize with many of the TLR ligands, including TLR2 ligands, and cause induction of cytokine production.25 For example, lipopolysaccharide (LPS) is recognized by TLR4; lipoproteins and peptidoglycans (PGNs) are recognized by TLR1/TLR2, TLR2/TLR6, NOD1, or NOD2; and flagellin is recognized by TLR5. Alternatively, viral compounds can trigger signaling through endosome-associated receptors, such as TLR3 by dsRNA, TLR7 and TLR8 by ssRNA, and TLR9 by unmethylated cytosine guanine dinucleotide DNA.
Recent studies have suggested that there is involvement of PRRs in STD and BV immune responses, and among the specific PRRs, TLR2 signaling is considered as a critical stimulus for enhanced HIV acquisition during STD and BV infections, because it has been shown to promote HIV replication in various immune cells.27-31 Thus, we hypothesized that when STDs or BV occur, stimulation of PRRs on LCs by bacterial components may augment HIV susceptibility in LCs, thereby leading to enhanced sexual transmission of HIV. In the current study, we examined whether specific agonists for PRRs, microbial components, or whole bacteria could modulate HIV infection levels in LCs and DCs.
Methods
Reagents
Cells were stimulated with synthetic or bacteria-derived TLR and NOD agonists for 24 hours at the following concentrations: Pam3CysSerLys4 (Pam3CSK4, TLR1/2; 0.2-5 μg/mL), heat-killed Listeria monocytogenes (HKLM, TLR2; 0.2-5 × 108/mL), poly(I:C) (TLR3; 20 μg/mL), Escherichia coli K12 LPS (TLR4; 10 ng/mL), flagellin from Salmonella typhimurium (TLR5; 2.5 μg/mL), Pam2CGDPKHPKSF (FSL1, TLR2/6; 2-50 μg/mL), loxoribine (TLR7; 500 μM), ssRNA40/LyoVec (TLR8; 10 μg/mL), CpG oligonucleotide type B (OPN2006, TLR9; 5 μM), lipoteichoic acid (LTA; TLR2; 0.4-10 μg/mL), PGN (TLR2 and NODs; 0.2-5 μg/mL), γ-d-Glu-mDAP (iE-DAP, NOD1; 100 μg/mL), and muramyl dipeptide (MDP, NOD2; 10 μg/mL; all from InvivoGen, San Diego, CA, except for PGN from Sigma-Aldrich, St Louis, MO, and MDP from Calbiochem, Madison, WI). For the blocking experiment, the following monoclonal antibody (mAb) and reagent were used: anti-TLR2 (eBioscience, San Diego, CA) and myeloid differentiating factor 88 (MyD88) homodimerization inhibitory peptide (Imgenex, San Diego, CA). Both Staphylococcus aureus and S typhimurium were gifts from Naoki Yamamoto (University of Osaka, Osaka, Japan). Lactobacillus spp, group B streptococcus, Prevotella bivia, and Bacteroides fragilis were collected from HIV-seronegative women after informed consent was obtained in accordance with the Declaration of Helsinki, using an Institutional Review Board–approved protocol from the University of Yamanashi. All the bacteria were heat-inactivated for 1 hour at 57°C.
Preparation of monocyte-derived LCs and mDCs
Monocyte-derived LCs (mLCs) and mDCs were cultured from peripheral blood mononuclear cells (PBMCs), as described previously.32 Briefly, monocytes were isolated by depletion of magnetically labeled nonmonocytes (Monocyte Isolation Kit II; Miltenyi Biotec, Auburn, CA) from plastic-adherent PBMC obtained from healthy blood donors. Monocytes were cultured in RPMI 1640 (Invitrogen Life Technologies, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (Cell Culture Technologies, Gravesano, Switzerland), 100 U/mL penicillin (Invitrogen Life Technologies), 100 μg/mL streptomycin (Invitrogen Life Technologies), 2 mM l-glutamine (Invitrogen Life Technologies; complete medium) supplemented with 1000 U/mL recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF; R&D Systems, Minneapolis, MN), 1000 U/mL recombinant human interleukin (IL)–4 (R&D Systems), and with mLCs or without mDCs and 10 ng/mL human platelet-derived transforming growth factor (TGF)–β1 (R&D Systems) for 7 days.
HIV infection of mLCs and mDCs in vitro
Purified, pelleted, and titered HIV-1Ba-L, an R5 HIV laboratory isolate (stock at 50% tissue culture–infective dose of 107.17/mL and 1.8 × 1010 virus particles/mL), was purchased from Advanced Biotechnologies (Columbia, MD). For some experiments, 2 × 105 mLCs and mDCs were preincubated with various agonists or inhibitors, and then HIVBa-L at a 1/100 final dilution was added for 2 hours at 37°C. After incubation, cells were harvested, washed 3 times, resuspended in complete medium supplemented with GM-CSF and IL-4 and/or TGF-β1, and cultured for an additional 7 days at the same cellular concentration. Half of the total volume of the medium was replaced with fresh complete medium and GM-CSF and IL-4 every other day. HIV-infected cells were assessed by HIV p24 intracellular staining. HIV infection levels were expressed as a normalized percentage of the positive cells for HIV p24 using a calculated fold difference compared with the mean percentage of the positive cells for HIV p24 in untreated cells.
HIV infection of skin explants ex vivo
Epithelial sheets were obtained from suction blister roofs from HIV-negative healthy donors. Droplets (50 μL) containing HIVBa-L at a 1/100 final dilution were placed on the inside surfaces of sterile plastic culture dish covers. Explants were draped over droplets with the basal epithelial cell surface facing downward. Virus and explants were incubated together in this manner at 37°C for 2 hours. Explants were washed and then floated with the basal epithelial cell sides down. The emigrating cells from the epidermal sheets were collected 3 days after the HIV exposure. HIV-infected LCs were assessed using HIV p24 staining. In some experiments, 1 × 104 emigrated LCs were cocultured with 2 × 106 allogeneic CD4+ T cells for 12 days. Supernatants were harvested every third day and examined for HIV p24 protein content by enzyme-linked immunosorbent assay (ELISA).
Flow cytometry
Single-cell suspensions were stained using the following anti–human mAbs: anti-CD83 (BD Biosciences, San Jose, CA), anti-CD86 (BD Biosciences), anti-CD4 (Beckman Coulter, Fullerton, CA), anti-CCR5 (R&D Systems), anti–DC-SIGN (R&D Systems) directly conjugated to fluorescein isothiocyanate (FITC), antilangerin (Immunotech, Fullerton, CA), anti–TLR-1, -2, -4, -3, -9 (eBioscience) directly conjugated to phycoerythrin (PE), and anti-CD11c (BD Biosciences) and anti-CD1a (BD Biosciences) directly conjugated to allophycocyanin. Biotinylated anti–TLR-6 (eBioscience), and in some experiments, biotinylated antilangerin (R&D Systems) were stained by streptavidin-FITC or allophycocyanin conjugate (BD Biosciences), respectively, and purified anti–E-cadherin (R&D Systems) and anti–TLR-7, -8 (eBioscience) were detected by FITC-conjugated goat anti–mouse or FITC-conjugated rabbit F(ab′)2, respectively. Cells were incubated with Abs for 30 minutes at 4°C, washed 3 times in staining buffer, and examined by FACSCalibur. For measurement of intracellular TLR-3, -9, -7, and -8, a fixation and permeabilization procedure was performed (Cytofix/Cytoperm; BD Biosciences). For HIV p24 intracellular staining, cells were incubated with 10 μg/mL PE-conjugated mouse anti–human langerin mAb and allophycocyanin-conjugated mouse anti–human CD11c mAb for 30 minutes at 4°C. Cells were then fixed and permeabilized with Cytofix/Cytoperm reagents for 20 minutes at 4°C. Cells were then incubated with 10 μg/mL FITC-conjugated mouse anti-HIV p24 mAb (Beckman Coulter) diluted for 30 minutes at 4°C, with the quantified numbers of HIV-infected cells determined by FACSCalibur. mLCs were gated on langerin+ CD11c+ cells, and mDCs were gated on CD11c+ cells.
Real-time quantitative reverse transcription–polymerase chain reaction analysis
Relative mRNA expression was determined by real-time polymerase chain reaction (PCR) using an ABI PRISM 5500 Sequence Detection System (Applied Biosystems, Foster City, CA) with SYBR Green I dye (QIAGEN, Valencia, CA), according to the manufacturer's instructions. Total RNA was isolated using TRIzol (Invitrogen Life Technologies), and cDNA was synthesized using the SuperScript system (Invitrogen Life Technologies). Primers corresponding to human TLR-3, TLR-9, NOD1, NOD2, APOBEC3G, and glyceraldehyde-3-phosphate dehydrogenase (G3PDH) were designed by Takara Bio (Shiga, Japan). Cycle threshold numbers (Ct) were derived from the exponential phase of the PCR amplification. Fold differences in the expression of gene x in the cell populations y and z were derived by 2k, where k = (Ctx − CtG3PDH)y − (Ctx − CtG3PDH)z.
ELISA
mLCs and mDCs were (106 cells/mL) stimulated for 24 hours with TLR and NOD ligands. The culture supernatants were collected after centrifugation, and stored at −80°C for cytokine measurements. The concentrations of cytokines (IL-6, IL-8, tumor necrosis factor (TNF)–α, and IL-10) in the culture supernatants were measured using cytometric bead array (CBA) assays (BD Biosciences). After acquisition of sample data by flow cytometry, results were analyzed using the BD Biosciences CBA analysis software. For measurement of HIV p24 protein levels, supernatants were collected, inactivated with Triton X-100 (Sigma-Aldrich; 2% final concentration), and kept frozen until measurements of HIV p24 protein levels were performed by ELISA (ZeptoMetrix, Buffalo, NY).
Western blot analysis
Proteins of the cells were extracted using a 15-minute incubation in complete lysis buffer containing a protease inhibitor. Equal amounts of protein were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto a transfer membrane (Daiichikagaku, Tokyo, Japan). Western blot was performed to detect human APOBEC3G and used 2.5 μg/mL rabbit immunoglobulin (Ig) G anti-APOBEC3G polyclonal antibody (Abcam, Cambridge, MA). Blots were incubated with the horseradish peroxidase (HRP)–linked secondary antibody, goat anti–rabbit IgG (Pierce, Rockford, IL). Analyses were performed using the HRP Western blot detection system (Pierce). To test for the A3G subcellular localization in some experiments, we separated the pellet (P) fraction, as has been described previously.33 Briefly, cells were lysed with ice-cold lysis buffer (Sigma-Aldrich) for 30 minutes and centrifuged at 35 000 rpm (TLA-120.2 rotor in Optima TLX, Beckman Coulter) for 1.5 hours. Equal volumes of P and supernatant (SN) were loaded on gel and analyzed by Western blotting.
Results
Because ex vivo purification of epidermal LCs from human skin results in a significant spontaneous maturation, it is difficult to monitor PAMP-induced maturation of fresh isolated LCs.34,35 Most of the phenotypic characteristics of LCs depend upon the presence of TGF-β1 in the epidermal microenvironment,36,37 because TGF-β1 promotes the development of LC-like DCs from peripheral blood monocytes.32,38 In general, epidermal LCs can be discriminated from DCs by the expression of E-cadherin and langerin. In 5 experiments, we generated monocyte-derived LCs (mLC) and found expression levels of E-cadherin+ cells and langerin+ cells to be 90.7 + 9.5% (mean + SD) and 35.1 + 14.5%, respectively (data not shown). Based on these findings, we used fluorescence-activated cell sorter (FACS) to isolate highly purified langerin-positive mLCs that could be used as surrogate cells for epidermal LCs. Alternatively, mLCs were identified by gating langerin-positive cells in FACS analyses. To test activation of LCs and DCs after TLR stimulation, mLCs and mDCs were challenged with Pam3CSK4 for TLR1/2, heat-killed L monocytogenes (HKLM) for TLR2, FSL1 for TLR2/6, poly(I:C) for TLR3, LPS for TLR4, flagellin for TLR5, loxoribine for TLR7, ssRNA40 for TLR8, and ODN2006 for TLR9 for 24 hours. Subsequently, we examined expression of CD86, a marker of LC/DC activation. Similar to previously reported findings,35,39,40 there was significant up-regulation of CD86 on langerin+ mLCs by Pam3CSK4, HKLM, FSL1, and poly(I:C), respectively (Figure 1A). These ligands also induced up-regulation of another activation marker, CD83, and stimulated IL-6 and IL-8 production by langerin+ mLCs (data not shown). Bacteria-derived TLR2 agonists, LTA and PGN, also up-regulated expression levels of these activation markers in a dose-dependent manner, whereas the TLR4, 5, 7, 8, and 9 agonists did not (Figures 1A, 2A, and data not shown). These results indicated that mLCs express functional TLR1, TLR2, TLR3, and TLR6 proteins. We also used anti-TLR–specific mAb labeling, which made it possible to confirm the expression of these TLRs on mLCs (data not shown).
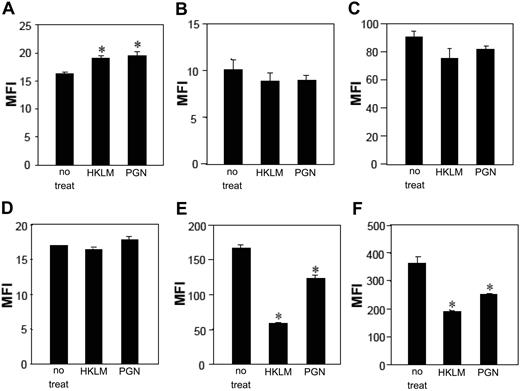
TLR stimuli modulate CD86 expression, HIV susceptibility, and HIV replication in mLCs and mDCs. mLCs and mDCs were cultured in medium alone or with 5 μg/mL Pam3CSK4, 5 × 108/mL HKLM, 20 μg/mL poly(I:C), 10 ng/mL LPS, 2.5 μg/mL flagellin, 50 μg/mL FSL1, 500 μM loxoribine, 10 μg/mL ssRNA40, or 5 μM ODN2006 for 24 hours. The expression of CD86 was examined (A, mLC; D, mDC; MFI, mean fluorescence intensity). Results are shown as means plus or minus SD (*P < .05). mLCs (B,C) or mDCs (E,F) were stimulated with the indicated TLR agonists for 24 hours before (B,E) and after (C,F) HIV exposure. To identify HIV infection levels, mLCs or mDCs were collected 7 days after the HIV exposure, and HIV p24+ cells were assessed in langerin+ CD11c+ mLCs or CD11c+ mDCs (expressed as normalized percentage of positive cells for HIV p24, as has been described in “HIV infection of mLCs and mDCs in vitro”). Mean values in mDCs and mLCs obtained from different donors are shown as horizontal marks. Representative FACS analyses of CD11c and p24 mAb double-stained cells after TLR2 stimulations are shown.
TLR stimuli modulate CD86 expression, HIV susceptibility, and HIV replication in mLCs and mDCs. mLCs and mDCs were cultured in medium alone or with 5 μg/mL Pam3CSK4, 5 × 108/mL HKLM, 20 μg/mL poly(I:C), 10 ng/mL LPS, 2.5 μg/mL flagellin, 50 μg/mL FSL1, 500 μM loxoribine, 10 μg/mL ssRNA40, or 5 μM ODN2006 for 24 hours. The expression of CD86 was examined (A, mLC; D, mDC; MFI, mean fluorescence intensity). Results are shown as means plus or minus SD (*P < .05). mLCs (B,C) or mDCs (E,F) were stimulated with the indicated TLR agonists for 24 hours before (B,E) and after (C,F) HIV exposure. To identify HIV infection levels, mLCs or mDCs were collected 7 days after the HIV exposure, and HIV p24+ cells were assessed in langerin+ CD11c+ mLCs or CD11c+ mDCs (expressed as normalized percentage of positive cells for HIV p24, as has been described in “HIV infection of mLCs and mDCs in vitro”). Mean values in mDCs and mLCs obtained from different donors are shown as horizontal marks. Representative FACS analyses of CD11c and p24 mAb double-stained cells after TLR2 stimulations are shown.
TLR2 stimuli enhance HIV susceptibility in mLCs and resident epidermal LCs. mLCs were stimulated via TLR2 using heat-killed Gram+ bacteria (HKLM; 0.2, 1, 5 × 108/mL), synthetic agonists (Pam3CSK4, 0.2, 1, 5 μg/mL; FSL1, 2, 10, 50 μg/mL), or Gram+ bacterial components (LTA, 0.4, 2, 10 μg/mL; PGN, 0.2, 1, 5 μg/mL) for 24 hours at various concentrations, as has been described in “Methods” (A-C). Cells were stimulated via TLR2 before (B) and after (C) HIV exposure. The expression of CD86 (A) and percentage of positive cells for HIV p24 (B,C) were assessed in langerin+ CD11c+ mLCs or CD11c+ mDCs (MFI, mean fluorescence intensity). Epithelial sheets obtained from suction blister roofs were preincubated with 5 × 108/mL HKLM or 20 μg/mL poly(I:C) for 2 hours, and then exposed to R5 HIV (D). The emigrating cells from the epidermal sheets were collected 3 days after HIV exposure, and HIV-infected epidermal LCs were assessed by HIV p24 intracellular staining (D). Representative FACS analyses of CD11c and p24 mAb double-stained cells are shown. For the assessment of HIV transmission from LCs to CD4+ T cells, emigrated LCs were collected 3 days after HIV exposure and washed, and then 104 LCs were cocultured with 2 × 106 allogeneic CD4+ T cells for 12 days. p24 protein levels in culture supernatants were assessed by ELISA on the indicated days (E). Results are shown as means plus or minus SD (n = 3). *P < .05; **P < .01. All data shown represent at least 2 separate experiments.
TLR2 stimuli enhance HIV susceptibility in mLCs and resident epidermal LCs. mLCs were stimulated via TLR2 using heat-killed Gram+ bacteria (HKLM; 0.2, 1, 5 × 108/mL), synthetic agonists (Pam3CSK4, 0.2, 1, 5 μg/mL; FSL1, 2, 10, 50 μg/mL), or Gram+ bacterial components (LTA, 0.4, 2, 10 μg/mL; PGN, 0.2, 1, 5 μg/mL) for 24 hours at various concentrations, as has been described in “Methods” (A-C). Cells were stimulated via TLR2 before (B) and after (C) HIV exposure. The expression of CD86 (A) and percentage of positive cells for HIV p24 (B,C) were assessed in langerin+ CD11c+ mLCs or CD11c+ mDCs (MFI, mean fluorescence intensity). Epithelial sheets obtained from suction blister roofs were preincubated with 5 × 108/mL HKLM or 20 μg/mL poly(I:C) for 2 hours, and then exposed to R5 HIV (D). The emigrating cells from the epidermal sheets were collected 3 days after HIV exposure, and HIV-infected epidermal LCs were assessed by HIV p24 intracellular staining (D). Representative FACS analyses of CD11c and p24 mAb double-stained cells are shown. For the assessment of HIV transmission from LCs to CD4+ T cells, emigrated LCs were collected 3 days after HIV exposure and washed, and then 104 LCs were cocultured with 2 × 106 allogeneic CD4+ T cells for 12 days. p24 protein levels in culture supernatants were assessed by ELISA on the indicated days (E). Results are shown as means plus or minus SD (n = 3). *P < .05; **P < .01. All data shown represent at least 2 separate experiments.
To determine the effects of TLR signaling on HIV susceptibility or HIV replication of mLCs or mDCs, cells were stimulated with TLR agonists for 24 hours before and after exposure to HIV. To specifically identify HIV-infected cells on a single-cell level, mLCs or mDCs were collected 7 days after HIV exposure and then double-stained with anti-HIV p24 mAb along with other antibodies directed against surface markers for mLCs or mDCs. This methodology made it possible to determine the number of HIV p24+ cells within langerin+/CD11c+ mLCs and within CD11c+ mDCs. Uninfected cells stained with anti-p24 mAb and HIV-infected cells stained with isotype control antibody always showed less than 0.10% positivity, thereby confirming the specificity of the p24 staining (data not shown). Surprisingly, incubation with the TLR2 agonists, including HKLM, Pam3CSK4, and FSL1, before and after HIV exposure dramatically enhanced HIV infection levels in mLCs, whereas incubation with the other agonists did not (Figure 1B,C). Similar effects were observed with the bacteria-derived TLR2 agonists, LTA and PGN, in a dose-dependent manner (Figure 2B,C). We also used ELISA to determine HIV p24 protein levels in mLC culture supernatants. ELISA results correlated with the single-cell flow cytometric data (data not shown). As was seen with LCs, preincubation with TLR2 or TLR3 agonists induced maturation of mDCs (Figure 1D). By contrast to the mLC findings, however, there was partial inhibition of HIV infection in mDCs when these cells were preincubated with TLR2 agonists (Figure 1E,F). Furthermore, TLR3 ligation by poly(I:C) before and after HIV exposures dramatically decreased HIV infection levels in mDCs. Thus, these data suggest that TLR signaling effects on HIV susceptibility and productive infection are differently regulated in LCs and DCs.
To understand how HIV traverses skin and genital mucosa, we recently developed an ex vivo model whereby resident LCs within epithelial tissue explants are exposed to HIV and then allowed to emigrate from tissue, thus mimicking conditions that occur after mucosal exposure to HIV.11,41 Using this model, we examined whether TLR2 stimulation enhanced HIV infection of epidermal LCs, and whether TLR2 stimulation enhanced subsequent HIV transmission from LCs to CD4+ T cells. Similar to the findings observed within mLCs, preincubation of epithelial sheets with HKLM significantly enhanced HIV infection levels in epidermal LCs as well as HIV transmission from emigrated LCs to CD4+ T cells (Figure 2D,E). As controls, epithelial sheets preincubated with poly(I:C) failed to enhance HIV infection levels in LCs, and led to decreased HIV transmission from LCs to CD4+ T cells.
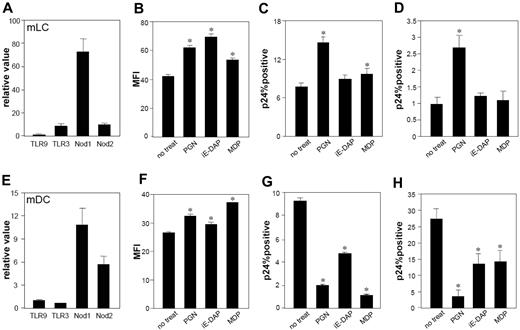
To test whether LCs or DCs express NOD1 and NOD2, mLCs and mDCs were challenged with PGN, the NOD1-specific agonist iE-DAP, and the NOD2-specific agonist MDP. These stimuli caused significant up-regulation of CD86 on mLCs and mDCs (Figure 3B,F). Quantitative reverse transcription (RT)–PCR confirmed the expression of NOD1 and NOD2 by these cells (Figure 3A,E). To determine effects of these NOD stimulators on HIV infection, cells were incubated with PGN, iE-DAP, or MDP for 24 hours before and after HIV exposure. Incubation with iE-DAP or MDP before and after HIV exposure significantly reduced HIV infection levels in mDCs (Figure 3G,H), but not in mLCs. Instead, these stimuli caused slight increases in HIV infection in mLCs (Figure 3C,D). Compared with iE-DAP and MDP, PGN significantly increased HIV infection levels in mLCs, suggesting that the infection-enhancing effects of PGN are primarily mediated by a TLR2 signaling pathway. This assertion is also supported by the fact that anti-TLR2 mAb significantly blocked the ability of PGN to enhance HIV infection in mLCs (Figure 4A).
mLCs and mDCs express functional NOD1 and NOD2. Expression of NOD1 and NOD2, and TLR3 and TLR9, which were used as controls, in mLCs and mDCs was assessed using real-time quantitative RT-PCR analysis (qPCR; A, mLCs; E, mDCs). mLCs and mDCs were stimulated by PGN (5 μg/mL), iE-DAP (for NOD1, 100 μg/mL), and MDP (for NOD2, 10 μg/mL) for 24 hours. mLCs (B-D) or mDCs (F-H) were stimulated via NOD receptors before (C,G) and after (D,H) HIV exposure. The expression of CD86 (B,F) and percentage of positive cells for HIV p24 (C,D,G,H) was assessed in langerin+ CD11c+ mLCs or CD11c+ mDCs (MFI, mean fluorescence intensity). Results are shown as means plus or minus SD (*P < .05). All data shown represent at least 2 separate experiments.
mLCs and mDCs express functional NOD1 and NOD2. Expression of NOD1 and NOD2, and TLR3 and TLR9, which were used as controls, in mLCs and mDCs was assessed using real-time quantitative RT-PCR analysis (qPCR; A, mLCs; E, mDCs). mLCs and mDCs were stimulated by PGN (5 μg/mL), iE-DAP (for NOD1, 100 μg/mL), and MDP (for NOD2, 10 μg/mL) for 24 hours. mLCs (B-D) or mDCs (F-H) were stimulated via NOD receptors before (C,G) and after (D,H) HIV exposure. The expression of CD86 (B,F) and percentage of positive cells for HIV p24 (C,D,G,H) was assessed in langerin+ CD11c+ mLCs or CD11c+ mDCs (MFI, mean fluorescence intensity). Results are shown as means plus or minus SD (*P < .05). All data shown represent at least 2 separate experiments.
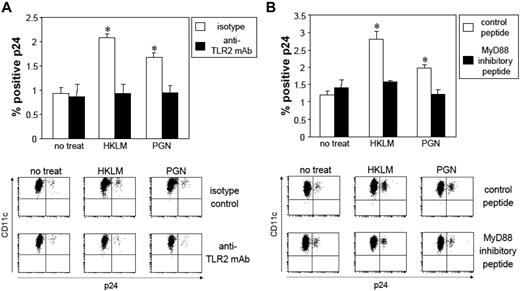
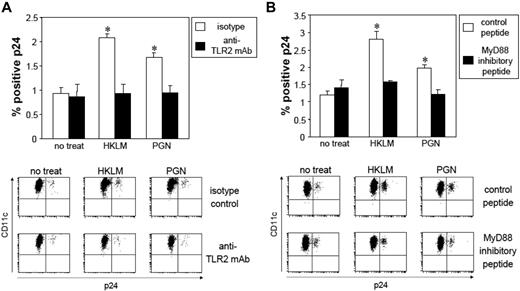
Enhancement of HIV replication by TLR2 ligation in mLCs is dependent on the TLR2-MyD88 signal pathway. After HIV exposure, mLC were preincubated with anti-TLR2 mAb (10 μg/mL) for 30 minutes or MyD88 inhibitory peptide (100 μM) for 24 hours, followed by coculturing with 5 × 108/mL HKLM, or 5 μg/mL PGN for 24 hours. Isotype control or control peptide experiments were performed using the same conditions. HIV-infected mLCs were assessed 7 days later by HIV p24 intracellular staining. (A) Anti-TLR2 mAb; (B) MyD88 inhibitory peptide. Summary of experiments and representative FACS analyses of CD11c and p24 mAb double-stained cells are shown. Results are shown as means plus or minus SD (n = 3). *P < .05. All data shown represent at least 2 separate experiments.
Enhancement of HIV replication by TLR2 ligation in mLCs is dependent on the TLR2-MyD88 signal pathway. After HIV exposure, mLC were preincubated with anti-TLR2 mAb (10 μg/mL) for 30 minutes or MyD88 inhibitory peptide (100 μM) for 24 hours, followed by coculturing with 5 × 108/mL HKLM, or 5 μg/mL PGN for 24 hours. Isotype control or control peptide experiments were performed using the same conditions. HIV-infected mLCs were assessed 7 days later by HIV p24 intracellular staining. (A) Anti-TLR2 mAb; (B) MyD88 inhibitory peptide. Summary of experiments and representative FACS analyses of CD11c and p24 mAb double-stained cells are shown. Results are shown as means plus or minus SD (n = 3). *P < .05. All data shown represent at least 2 separate experiments.
In contrast to the findings observed for Gram+ bacteria-derived or synthetic agonists for TLR2, LPS failed to enhance HIV infection levels in mLCs (Figure 1). This is not surprising, because LCs have been shown to lack expression of the functional TLR4 that is involved in the recognition of Gram− bacterial components.35,39,42 We were also unable to detect expression of TLR4 on mLCs (data not shown). The distinct responsiveness to TLR2 and TLR4 agonists suggests that Gram+ or Gram− bacteria may have differential effects on HIV infection levels in LCs. Therefore, we tested effects of whole Gram+ and Gram– bacteria on HIV susceptibility and HIV replication in LCs. As expected, incubation with S aureus before and after the HIV exposures dramatically enhanced HIV infection levels in langerin+/CD11c+ mLCs, whereas S typhimurium showed little or no effect (Figure 5A,B). Similarly, preincubation with group B streptococcus significantly enhanced HIV infection levels in mLCs, whereas Listeria spp did not (Figure 5C). In contrast, incubation with S aureus or S typhimurium decreased HIV infection levels in mDCs (Figure 5D,E).
Gram+ bacteria enhance both HIV susceptibility and replication in the mLCs. mLCs and mDCs were incubated with heat-killed whole bacteria, including S aureus (105∼7 CFU/mL), S typhimurium (104∼6 CFU/mL), L spp (106 CFU/mL), group B streptococcus (106 CFU/mL), P bivia (106 CFU/mL), or B fragilis (106 CFU/mL) for 24 hours before (A,C,D) and after (B,E) HIV exposure. The percentage of positive cells for HIV p24 was assessed in langerin+ CD11c+ mLCs (A-C) or CD11c+ mDCs (D,E). Results are shown as means plus or minus SD (*P < .05). All data shown represent at least 2 separate experiments.
Gram+ bacteria enhance both HIV susceptibility and replication in the mLCs. mLCs and mDCs were incubated with heat-killed whole bacteria, including S aureus (105∼7 CFU/mL), S typhimurium (104∼6 CFU/mL), L spp (106 CFU/mL), group B streptococcus (106 CFU/mL), P bivia (106 CFU/mL), or B fragilis (106 CFU/mL) for 24 hours before (A,C,D) and after (B,E) HIV exposure. The percentage of positive cells for HIV p24 was assessed in langerin+ CD11c+ mLCs (A-C) or CD11c+ mDCs (D,E). Results are shown as means plus or minus SD (*P < .05). All data shown represent at least 2 separate experiments.
We next tested the capacity of mAbs against TLR2 and a MyD88 inhibitor for their ability to inhibit TLR2-enhanced HIV replication in mLCs. Anti-TLR2 mAb demonstrated a significant blocking capacity for HKLM- and PGN-enhanced HIV replication in mLCs, whereas isotype control IgG did not (Figure 4A). Furthermore, when mLCs were stimulated by HKLM or PGN in the presence of a MyD88 inhibitor peptide, we did not observe TLR2-enhanced infection (Figure 4B), confirming that increased HIV replication that occurs after exposure of mLC to Gram+ bacteria is mediated by TLR2/MyD88 signaling.
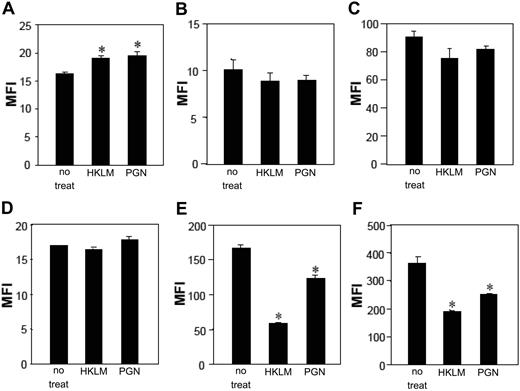
We have previously reported that mLCs and mDCs express CD4 and CCR5 with expression levels for these HIV receptors lower on mLCs compared with mDCs.32 Similar to previous DC studies,32,43 mDCs matured in the presence of HKLM or PGN decreased CCR5 surface expression, whereas CCR5 levels on mLCs were not significantly down-regulated after exposure to these stimuli (Figure 6B,E). Interestingly, these stimuli significantly up-regulated CD4 expression on mLCs, whereas CD4 levels on mDCs were unchanged (Figure 6A,D). Further investigation of the expression levels for the surface CLRs determined that langerin expression on mLCs remained unchanged, whereas DC-SIGN levels on mDCs were markedly down-regulated after HKLM- or PGN-induced maturation (Figure 6C,F). Thus, there was a partial correlation between cell surface expression levels of the HIV receptors and CLRs with the HIV susceptibility phenotype observed in mLCs and mDCs stimulated by bacterial components via TLR2 (Figure 1).
HKLM and PGN modulate surface expression of HIV receptors on mLCs and mDCs. mLCs and mDCs were cultured in medium alone or with 5 × 108/mL HKLM or 5 μg/mL PGN for 24 hours, and the expression of CD4 (A,D), CCR5 (B,E), langerin (C), or DC-SIGN (F) was then examined (A-C, mLCs; D-F, mDCs; MFI). Results are shown as means plus or minus SD (*P < .05). All data shown represent at least 2 separate experiments.
HKLM and PGN modulate surface expression of HIV receptors on mLCs and mDCs. mLCs and mDCs were cultured in medium alone or with 5 × 108/mL HKLM or 5 μg/mL PGN for 24 hours, and the expression of CD4 (A,D), CCR5 (B,E), langerin (C), or DC-SIGN (F) was then examined (A-C, mLCs; D-F, mDCs; MFI). Results are shown as means plus or minus SD (*P < .05). All data shown represent at least 2 separate experiments.
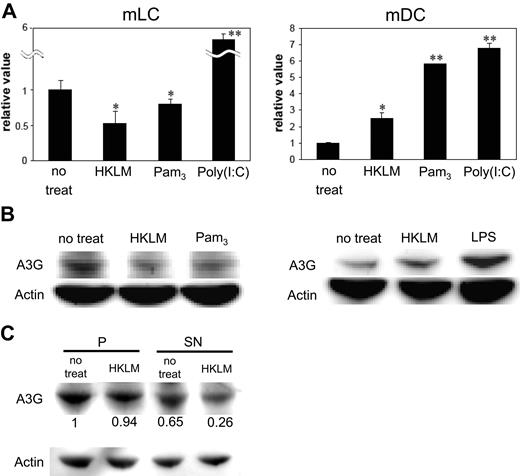
We found that TLR2 stimulation enhanced HIV infection levels in mLCs, even when mLCs were incubated with TLR2 agonists after the HIV exposures. We therefore considered postentry cellular restriction factors for HIV. Recently, Pion et al have reported that between the 2 families that have been shown to restrict HIV infection (tripartite motif [TRIM]/APOBEC),33 T5α and A3F were not strong restriction factors in mDCs.44 However, A3G did function as a potent postentry cellular restriction factor for HIV in mDCs. To analyze whether TLR stimulation modulated A3G activity in mLCs and mDCs, we activated these cells with TLR agonists and then quantified A3G expression. Interestingly, higher levels of A3G were expressed by mLCs versus mDCs (Figure 7). Consistent with a previous report,33 A3G levels were significantly increased in mDCs by LPS (data not shown). In addition, A3G levels were increased by HKLM, Pam3CSK4, and poly(I:C) in mDCs (Figure 7A,B). Surprisingly, the same TLR2 ligation by HKLM and Pam3CSK4 resulted in a significant reduction of A3G levels in mLCs. Poly(I:C) increased A3G levels in mLCs, but not in mDCs.
TLR2, but not TLR3, stimulation negatively modulates A3G expression levels in mLCs. mLCs and mDCs were cultured in medium alone or with 5 × 108/mL HKLM, 5 μg/mL Pam3CSK4, or 20 μg/mL poly(I:C) for 24 hours before HIV exposure. Expression of A3G in mLCs and mDCs was assessed using qPCR (A) or Western blotting (B). Results are shown as means plus or minus SD (*P < .05; **P < .01). Alternatively, cells were lysed and centrifuged at high speed, with equal volumes of the P fraction and the SN fraction, and then analyzed for A3G expression by Western blotting. A3G expression was quantified, with a value of 1 assigned to the pellet fraction in nontreated cells (C). All data shown represent at least 2 separate experiments.
TLR2, but not TLR3, stimulation negatively modulates A3G expression levels in mLCs. mLCs and mDCs were cultured in medium alone or with 5 × 108/mL HKLM, 5 μg/mL Pam3CSK4, or 20 μg/mL poly(I:C) for 24 hours before HIV exposure. Expression of A3G in mLCs and mDCs was assessed using qPCR (A) or Western blotting (B). Results are shown as means plus or minus SD (*P < .05; **P < .01). Alternatively, cells were lysed and centrifuged at high speed, with equal volumes of the P fraction and the SN fraction, and then analyzed for A3G expression by Western blotting. A3G expression was quantified, with a value of 1 assigned to the pellet fraction in nontreated cells (C). All data shown represent at least 2 separate experiments.
Cellular A3G, which resides as a low molecular mass (LMM) active form, can function as a postentry restriction factor for HIV during reverse transcription in mDCs. To evaluate whether A3G was present in a LMM active form in mLCs, we used a previously reported method for mDCs that used high-speed centrifugation to separate the cytosolic proteins from the pellet (P) fraction (which contains heavy membranes, endosomes, and nuclei).33 Using this method, cytosolic A3G was extracted in the SN fraction (A3G SN), whereas the higher molecular forms of A3G were left in the P fraction. Our results indicated that A3G SN was present in mLCs, although it was significantly down-regulated after the HKLM stimulation (Figure 7C). When taken together, these results suggest that the proportion of the A3G SN is well correlated with the HIV restriction phenotype in mLCs (Figures 1, 7A,B).
Discussion
LCs are thought to be the initial cellular targets that play a pivotal role in the viral dissemination during sexual transmission of HIV. To understand the biologic mechanisms by which STDs and BV contribute to increase in HIV acquisition, we tested the hypothesis that bacterial recognition pathways are responsible for modulating LC susceptibility to HIV. We demonstrated that Gram+ bacteria (eg, HKLM) enhance HIV susceptibility in langerin+ mLCs as well as in epidermal LCs. Among the TLR1-9-specific agonists, only TLR2 and TLR3 agonists were able to induce significant maturation and cytokine production in mLCs. Interestingly, TLR2 agonists enhanced HIV susceptibility in mLCs, whereas other TLR-specific agonists, including TLR3, did not. Thus, our findings suggest that TLR2 plays a central role in Gram+ bacteria-enhanced infection that occurs in LCs. This statement is also supported by our findings that TLR2 mAb exhibited a significant ability to block the enhancement of HIV infection in LCs by Gram+ bacteria. It should be noted that the stimulation of TLR2 and TLR3 decreased HIV susceptibility in mDCs. Although it is unlikely that submucosal DCs are directly infected with HIV in the sexual transmission of HIV, this may occur in the presence of ulcerating STD infections. In these instances, our data suggest that TLR2 stimulation by bacterial components may act to protect DC from HIV infection. Overall, the current findings suggest that TLR2 stimulation might induce a cell type–specific pathway in LCs that enhances HIV susceptibility of these cells.
There are several possible mechanisms that could account for enhancement of HIV infection in LCs via Gram+ bacteria/TLR2. We demonstrated that HKLM and PGN significantly increased surface expression of CD4 in mLCs, which may be involved in enhanced HIV susceptibility of LCs. In addition, TLR2 ligation may also affect HIV infection in mLC postviral entry, because the enhancement of HIV infection was observed even if the cells were stimulated after HIV exposure. Recent studies have revealed a crucial contribution for A3G in the susceptibility of various cell types to HIV. In mDCs, it has been shown that A3G functions as an especially strong postentry restriction factor, and its expression level is significantly increased by TLR4 ligation, which further restricts HIV infection.33 Our results demonstrated that TLR2 ligation significantly reduced A3G levels in mLCs, and the proportion of A3G SN was inversely correlated with HIV infection levels in these cells (Figures 1, 7), suggesting that Gram+ bacteria enhance HIV infection in LCs by decreasing A3G expression. Interestingly, there was no significant difference between HIV infection levels in mLCs treated by TLR2 ligands before infection compared with treatment after infection (Figure 1B,C). In contrast, mDC treated by HKLM or Pam3CSK4 demonstrated significantly higher infection levels with pretreatment compared with posttreatment (Figure 1E,F, and data not shown). These results suggest that TLR2-mediated regulation of cell surface HIV receptor expression may contribute to decreased HIV infection in mDCs, but that regulation of cell surface HIV receptor expression contributes relatively little to TLR2-mediated enhancement of HIV infection in mLCs.
We found that A3G expression is strongly induced in mDCs upon TLR2 stimulation; however, it failed to limit HIV replication even if cells were stimulated after infection (Figures 1F, 7A). Because other factors (eg, APOBEC family other than A3G, TRIM family, chemokines, or cytokines) can affect HIV infection levels in macrophages,45 these conditions may supercede restriction effects controlled by A3G in HIV-infected TLR2-stimulated DCs. Recent studies have shown that interferons are strong inducers of A3G, and interferon-β expression is induced in mLCs by activation with TLR3, but not TLR2, agonists.45,46 In our study, TLR3, but not TLR2, agonists significantly increased A3G levels in mLCs (Figure 7A), suggesting that interferons produced by LCs may be involved in the regulation of A3G by TLRs. Thus, mucosal application of TLR3 agonists or interferons may be considered as alternative approaches to decrease sexual transmission of HIV.
Recently, de Jong et al reported that TNF-α stimulated HIV transmission from LCs to T cells by increasing HIV replication in LCs, whereas Pam3CSK4 increased LC capture of HIV and subsequent trans infection of T cells.47 In contrast to our findings, they showed Pam3CSK4 alone did not increase HIV infection in LCs. Considering the indirect effect of TNF-α, we found that TLR2 (but not TLR3) agonists induced abundant TNF-α production by mLCs (data not shown), consistent with previous findings.40,46 It is possible that the enhancement of HIV infection in LCs by TLR2 stimuli observed in our study might be due, at least in part, to increased production of TNF-α by LCs.
Bacterial components are either liberated from live bacteria or released upon bacterial lysis. In the current study, we were able to demonstrate for the first time that mLCs express functional NOD1 and NOD2. Although their ligands induced significant maturation of mLCs, effects on HIV infection were quite limited compared with PGN. Furthermore, TLR2 mAb significantly blocked the capacity of PGN to enhance HIV infection, suggesting that PGN is modulating HIV infection levels in LCs via TLR2-MyD88 rather than through NOD pathways.
In most healthy women, the vaginal microflora comprises large quantities of a limited number of lactobacilli species, which appears to confer some protection against HIV acquisition.1 BV is characterized by a reduction in the number of lactobacilli and the growth of several species of anaerobic bacteria. Interestingly, in this regard, we found that Listeria spp as well as anaerobic bacteria did not significantly affect HIV infection levels in langerin+ mLCs. The failure of anaerobic bacteria to enhance HIV susceptibility in LCs might be because anaerobic bacteria, including P bivia and B fragilis, are Gram− bacteria recognized mainly by TLR4.50 In addition, it has been recently reported that Listeria spp did not induce TNF-α secretion and HIV LTR activation in THP-1 cells that express TLR2.37 Taken together, in addition to bacterial components, we also demonstrated that whole Gram+ bacteria, including L monocytogenes, S aureus, and group B streptococcus, enhanced HIV susceptibility in langerin+ mLCs. Nonulcerative STDs, such as Neisseria gonorrhoeae, Chlamydia trichomatis, and Candida spp, have been shown to stimulate cells via TLR2,28,48,49 suggesting that these organisms may also enhance HIV infections in LCs like in other immune cells.27-31
We demonstrated that Gram+ bacterial components directly augmented HIV infection in LCs by activating TLR2. It is possible that TLR2 stimulation of LC infection underlies, at least in part, enhanced sexual transmission of HIV in settings where there is concomitant STD infection. Based on this theory, blockade of TLR2 signaling might be considered as an alternate approach to decrease sexual transmission of HIV.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Naohiro Inohara, Naotaka Shibagaki, and Atsuya Yamashita for their helpful discussions, and Yoshie Hayakawa and Takashi Uchida for their technical assistance.
Authorship
Contribution: Y.O. performed experiments and analyzed data; T. Kawamura directed and performed experiments, analyzed data, and wrote the manuscript; T. Kimura performed experiments and analyzed data; M.I. contributed analytical tools; A.B. codirected experiments and wrote the manuscript; and S.S. codirected experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Tatsuyoshi Kawamura, 1110 Shimokato, Chuo, Yamanashi 409-3898, Japan; e-mail: tkawa@yamanashi.ac.jp.