Abstract
Kaposi sarcoma–associated herpesvirus (KSHV), also known as human herpesvirus 8, is the etiologic agent of Kaposi sarcoma (KS), an angioproliferative lesion characterized by dramatic angiogenesis and inflammatory infiltration. In this study, we report that expression of chemokine CCL20, a potent chemoattractant of dendritic cells and lymphocytes, is strongly induced in cultured cells either by KSHV infection or on ectopic expression of viral FLICE inhibitory protein K13. This induction is caused by transcriptional activation of CCL20 gene, which is mediated by binding of the p65, p50, and c-Rel subunits of the transcription factor nuclear factor–κB (NF-κB) to an atypical NF-κB–binding site present in the CCL20 gene promoter. The CCL20 gene induction is defective in K13 mutants that lack NF-κB activity, and can be blocked by specific genetic and pharmacologic inhibitors of the NF-κB pathway. CCR6, the specific receptor for CCL20, is also induced in cultured cells either by KSHV infection or on K13 expression. Finally, expression of CCL20 and CCR6 is increased in clinical samples of KS. These results suggest that KSHV and K13-mediated induction of CCL20 and CCR6 may contribute to the recruitment of dendritic cells and lymphocytes into the KS lesions, and to tumor growth and metastases.
Introduction
Kaposi sarcoma (KS) is a highly vascular tumor that frequently occurs in the dermis of skin and mucus membranes of immunocompromised patients.1 It is a multifocal angioproliferative lesion that is histologically characterized by the presence of distinctive proliferating spindle cells of endothelial origin, marked neoangiogenesis with edema and extravasation of red blood cells, and infiltration of lymphomononuclear inflammatory cells.1–3 The inflammatory cells are thought to play a central role in the pathogenesis of KS lesions; indeed, it has proposed that early-stage KS is not a true sarcoma but an angiohyperplastic-inflammatory lesion whose growth is driven, in part, by exuberant production of angiogenic and inflammatory cytokines by lymphocytes and macrophages present in the lesion.1,4 Although infiltration by inflammatory cells, including CD8+ T cells, monocytes, macrophages, and dendritic cells, precedes the appearance of spindle cells in the KS lesions,4,5 the nature and source of chemokines responsible for their recruitment remain to be fully characterized.
Infection with Kaposi sarcoma–associated herpesvirus (KSHV) is thought to play a central role in the histogenesis of KS lesions, including its inflammatory component.4 KSHV infection has been detected in the endothelial cells of early KS lesions and is thought to contribute to their phenotypic transformation into spindle cells.6–8 This hypothesis is supported by in vitro studies showing that microvascular and macrovascular endothelial cells latently infected with KSHV acquire a spindle cell morphology.9–11 More importantly, these KSHV-infected endothelial cells were shown to up-regulate the expression of genes encoding several proinflammatory and angiogenic cytokines and chemokines that have been previously implicated in the pathogenesis of KS lesions, such as interleukin-6 (IL-6), IL-8, IL-1, GRO-1, monocyte chemotactic protein-1 (MCP-1), NAP-2, Rantes, and CXCL16.1,12–21
The KSHV genome contains an open reading frame K13, which is one of the few genes to be expressed in latently infected KS spindle cells.22 The K13 gene encodes for a protein with homology to the prodomain of caspase 8/FLICE.23 The K13 protein was originally thought to protect KSHV-infected cells from apoptosis by preventing the activation of caspase 8/FLICE and, as such, was classified as a viral FLICE inhibitory protein (vFLIP).23 However, it was subsequently demonstrated that K13 directly binds to and activates an approximately 700-kDa IκB kinase (IKK) signalosome complex to activate the nuclear factor-κB (NF-κB) pathway.24–26 K13 uses the NF-κB pathway to promote cellular survival, proliferation, transformation, cytokine secretion, and KSHV latency.15,16,27–32 Recent work from our laboratory and others has shown that ectopic expression of K13 in human umbilical vein endothelial cells (HUVECs) induces them to acquire a spindle cell phenotype, which is accompanied by exuberant production of proinflammatory cytokines and chemokines known to be involved in the pathogenesis of KS lesions.31,32
CCL20 is a recently identified chemokine that binds to the CC chemokine receptor 6 (CCR6) and serves as a powerful chemoattractant of a subset of effector/memory T cells, B cells, and immature dendritic cells.33 It has been proposed that CCL20 plays a crucial role in the recruitment of lymphocytes and dendritic cells to the sites of inflammation and in the regulation of inflammatory response, particularly at skin and mucosal surfaces.33 Because the KS lesions show intensive infiltration with inflammatory and dendritic cells and primarily involve the skin and mucosa,4 we have examined the effect of KSHV infection on the induction of CCL20. Our results suggest that latent infection with KSHV strongly induces CCL20 expression, and vFLIP K13 plays a key role in this process. We further demonstrate that mRNA of CCR6 is also strongly induced in KSHV-infected or K13-expressing cells. These studies suggest that KSHV-mediated induction of CCL20 and CCR6 may contribute to the recruitment of dendritic cells and lymphocytes into the KS lesions, and vFLIP K13 may play a key role in this process.
Methods
Cell lines and reagents
HUVECs were purchased from Cambrex (East Rutherford, NJ) and were grown in endothelial cell basal medium-2 (EMB; Lonza, Walkersville, MD) medium containing 10% fetal bovine serum and supplemented with the bullet kit. Cells were used for experiments at passages 2 to 6. HUVECs stably expressing 4-hydroxytamoxifen (4-OHT)–inducible K13-ERTAM have been described previously.31 293T, BC-1, BCBL-1, BJAB, Namalwa, and K562 cells were obtained from ATCC (Manassas, VA). JSC-1 cells were obtained form Dr Richard Ambinder (Johns Hopkins University, Baltimore, MD). Telomerase immortalized HUVECs (iHUVECs) were obtained from Dr Deborah Freedman (Harvard Medical School, Boston, MA). MSCVneo-based retroviral vectors expressing Flag-tagged K13 and K13-ERTAM have been described previously28,31 and were used to generate polyclonal populations of infected cells after selection with G418. Rabbit polyclonal antibodies against p65, p50, p52, RelB, and cRel were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against Flag and tubulin were from Sigma-Aldrich (St Louis, MO), and an antibody against CCL20 was from R&D Systems (Minneapolis, MN). NF-κB inhibitors Bay-11-7082, PS-1145, IKK-inhibitor VI (5-phenyl-2-ureido)thiophene-3-carboxamide, and IKK-inhibitor VIII (ACHP, 2-amino-6-(2-(cyclopropylmethoxy)-6-hydroxyphenyl)-4-(4-piperidinyl)-3-pyridinecarbonitrile) were purchased from Calbiochem (San Diego, CA). Arsenic trioxide (As2O3) was from Sigma-Aldrich.
Plasmids
Plasmids encoding various vFLIPs, phosphorylation-resistant mutants of IκBα and lentiviral vectors encoding control and K13 shRNAs have been described previously.16,26,28 The pGL2-CCL20/MIP-α (CCL20 WT-Luc) and pGL2-CCL20/MIP-3α/κBM (CCL20 ΔNF-κB-Luc) luciferase reporter constructs were kind gifts from Dr Tomoko Kohno (Nagasaki University, Nagasaki, Japan).34 Recombinant lentiviruses were generated and used to infect BCBL-1 cells essentially as described previously.26
Luciferase reporter assay
The 293T cells were transfected in a 24-well plate with various test plasmids along with the wild-type (WT) or mutant CCL20 luciferase reporter constructs (75 ng/well) and a pRSV/LacZ (β-galactosidase) reporter construct (75 ng/well) using calcium phosphate as described previously.24 Cells were lysed 24 to 36 hours later, and extracts were used for the measurement of firefly luciferase and β-galactosidase activities, respectively. Luciferase activity was normalized relative to the β-galactosidase activity to control for the difference in the transfection efficiency. Luciferase reporter assay in MEF cells was conducted essentially as described previously.35
Real-time reverse-transcribed–polymerase chain reaction
RNA was isolated using the RNeasy Mini kit (QIAGEN, Valencia, CA) and quantitative reverse-transcribed–polymerase chain reaction (RT-PCR) performed as described previously.36 Real-time PCRs were performed in triplicate using an ABI Prism 7000 system and SYBR green-Taq polymerase mix to determine the relative change in the expression of CCL20 or CCR6 genes. GNB2L1 (guanine nucleotide binding protein, beta polypeptide 2-like 1) or β-actin was used as a housekeeping control. Quantitative RT-PCR data (Ct values) were analyzed using the 2−ΔΔC method,37 and the data presented as fold change in target gene expression plus or minus SEM. Primers used for real-time PCR are: CCL20, forward: CCAAGAGTTTGCTCCTGGCT; CCL20, reverse: TGCTTGCTGCTTCTGATTCG; CCR6, forward: GGACCGGTACATCTCCATT; CCR6, reverse: TGCTGCGCGGTAGTGTTCT; GNB2L1, forward: GAGTGTGGCCTTCTCCTCTG; GNB2L1, reverse: GCTTGCAGTTAGCCAGGTTC; Actin, forward: TCACCCACACTGTGCCATCTACGA; Actin, reverse: CAGCGGAACCGCTCATTGCCAATGG. Primers for K13 and latency-associated nuclear antigen (LANA) have been described previously.36
NF-κB DNA-binding assay
The DNA-binding activity of the p65, p50, p52, RelB, and cRel subunits was measured in triplicate in the nuclear extracts using an enzyme-linked immunosorbent assay (ELISA)–based NF-κB DNA binding assay, as described previously.36
ELISA
Human CCL20 was measured in the cell-culture supernatant using a CCL20 ELISA kit from R&D Systems and following the recommendations of the manufacturer.
Immunohistochemistry
Biopsy specimens were fixed in 10% buffered formalin and embedded in paraffin. Sections (5 μm) were deparaffinized by sequential immersion in xylene and ethanol and then used for high-temperature antigen retrieval in citrate buffer (pH 6.0) for 20 minutes. All sections were subsequently incubated with 3% hydrogen peroxide to block the endogenous peroxidase activity and incubated with an antibody against LANA (raised in mouse) or an antibody against CCL20 (raised in goat). The reactions were developed using the MACH2 detection system (Biocare Medical, Concord, CA) as recommended by the manufacturer using 3,3-diaminobenzidine (for LANA) and Vulcan fast red (for CCL20) as chromogens. The sections were counterstained lightly with hematoxylin. KS and tonsil samples were used as positive controls for LANA and CCL20 staining, respectively. To examine nonspecific reactivity, samples were analyzed by either omitting the primary antibody or by replacing the primary antibody with an isotype-matched control antibody.
Statistical analysis
Differences in the expression level of CCL20, CCR6, K13, and LANA among control and KSHV-infected samples were studied using nonparametric Mann-Whitney U test using Analyse-it statistical software (Analyse-it, Leeds, United Kingdom). All tests were 2-tailed, and P values less than .05 were considered significant.
Image acquisition
Images in Figure 7 were viewed with an Olympus CKX41 microscope with 40×/0.60 numeric aperature objective Luc Plan PLN lenses and the following stains: hematoxylin and eosin (panel B), DAB-3 (3-diaminobenzidine; panel C), and DAB and Vulcan fast Red (panel D). Images were acquired using a SPOT Insight 4 Firewire camera and Spot Advanced version 4.6 acquisition software (Diagnostic Instruments, Sterling Heights, MI), and processed with Adobe Photoshop CS2 software (Adobe Systems, San Jose, CA).
Results
Induction of CCL20 expression by KSHV
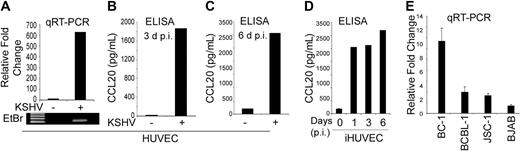
Infection of HUVECs with KSHV is known to induce the expression of a large number of chemokines and cytokines, such as IL-6, IL-8, and CCL5/RANTES, and CXCL16.1,12–21 To determine whether KSHV infection can also induce CCL20 gene expression, we infected HUVECs with KSHV and, approximately 48 hours after infection examined the induction of CCL20 mRNA by quantitative RT-PCR analysis. Significant induction of CCL20 mRNA expression was observed in KSHV-infected HUVECs compared with the mock-infected cells (Figure 1A). CCL20 protein level, as measured by ELISA, was also significantly higher in the supernatant of KSHV-infected HUVECs 3 and 6 days after infection (Figure 1B,C). A sustained increase in CCL20 secretion, which was maintained for up to 6 days after infection, was also seen in telomerase-iHUVECs on KSHV infection (Figure 1D).
KSHV up-regulates CCL20 expression in HUVEC and PEL cell lines. (A) HUVECs were infected with KSHV for 48 hours, and expression of CCL20 was measured by quantitative RT-PCR analysis and normalized to GNB2L1 (housekeeping control). PCRs were performed in triplicate and the data presented as fold change in target gene expression (mean ± SE). The results of the quantitative RT-PCR analysis were confirmed by separating the products (5.0 μL) on a 1.5% agarose gel followed by staining with ethidium bromide (bottom panel). (B,C) Cellular supernatants from KSHV-infected HUVECs were collected 3 days (B) and 6 days (C) after infection and used to measure the secretion of CCL20 by ELISA. The values shown are averages (mean ± SE) of 1 representative experiment of 3 in which the level of CCL20 secretion was measured in duplicate. (D) Kinetics of CCL20 up-regulation in KSHV-infected iHUVEC as measured by ELISA. (E) Level of CCL20 mRNA expression, as measured by quantitative RT-PCR, in PEL cell lines naturally infected with KSHV (BC-1, BCBL-1, and JSC-1). The KSHV-negative BJAB cell line was used as negative control.
KSHV up-regulates CCL20 expression in HUVEC and PEL cell lines. (A) HUVECs were infected with KSHV for 48 hours, and expression of CCL20 was measured by quantitative RT-PCR analysis and normalized to GNB2L1 (housekeeping control). PCRs were performed in triplicate and the data presented as fold change in target gene expression (mean ± SE). The results of the quantitative RT-PCR analysis were confirmed by separating the products (5.0 μL) on a 1.5% agarose gel followed by staining with ethidium bromide (bottom panel). (B,C) Cellular supernatants from KSHV-infected HUVECs were collected 3 days (B) and 6 days (C) after infection and used to measure the secretion of CCL20 by ELISA. The values shown are averages (mean ± SE) of 1 representative experiment of 3 in which the level of CCL20 secretion was measured in duplicate. (D) Kinetics of CCL20 up-regulation in KSHV-infected iHUVEC as measured by ELISA. (E) Level of CCL20 mRNA expression, as measured by quantitative RT-PCR, in PEL cell lines naturally infected with KSHV (BC-1, BCBL-1, and JSC-1). The KSHV-negative BJAB cell line was used as negative control.
To examine whether CCL20 gene expression in KSHV-infected cells was limited to vascular endothelial cells, we examined its expression in 3 primary effusion lymphoma (PEL) cell lines (BC-1, BCBL-1, and JSC-1), which are naturally infected with KSHV. As shown in Figure 1E, strong CCL20 gene expression was observed in the BC-1 cell line, whereas weak expression was seen in the BCBL-1 and JSC-1 cell lines. The level of expression of CCL20 in the 3 PEL cell lines correlated well with their previously reported level of vFLIP K13 expression and NF-κB activity.25,36 In contrast, very little CCL20 expression was seen in the human B-cell lymphoma cell line BJAB (Figure 1E), which is not infected with KSHV and demonstrates weak NF-κB activity.38
Induction of CCL20 expression by KSHV-encoded vFLIP K13
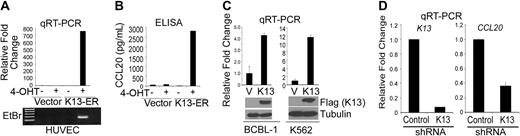
We have previously generated HUVECs stably expressing a K13-ERTAM fusion construct, in which the K13 cDNA is fused in-frame to the ligand-binding domain of a mutated estrogen receptor.31 The mutated estrogen receptor does not bind to the physiologic ligand estrogen but binds with very high affinity to the synthetic ligand 4-OHT and regulates the activity of its fusion partner K13 in a 4-OHT–dependent fashion.31 Induction of K13 activity in these cells on treatment with 4-OHT results in the acquisition of spindle cell morphology and secretion of pro-inflammatory cytokines and chemokines, thus mimicking the effect of KSHV infection.31 We used this cell line model to examine whether induction of K13 activity in HUVEC-K13-ERTAM can also mimic the effect of KSHV infection on CCL20 gene induction. Treatment of HUVEC-K13-ERTAM cells with 4-OHT led to a dramatic induction of CCL20 gene expression as determined by quantitative RT-PCR analysis (Figure 2A). This was accompanied by a parallel increase in CCL20 secretion in the supernatant, as determined by ELISA (Figure 2B). 4-OHT treatment had no effect on CCL20 mRNA or CCL20 protein expression in the control vector-expressing HUVECs, thus demonstrating that the observed effect was specific to the induction of K13 activity (Figure 2A,B).
K13 induces CCL20 expression in HUVEC and PEL cell lines. (A) HUVECs stably expressing a control vector or K13-ERTAM were mock-treated or treated with 4-OHT (50 nM) for 48 hours after which RNA was extracted and used for quantitative RT-PCR. The results of the quantitative RT-PCR analysis were confirmed by agarose gel electrophoresis (bottom panel). (B) Supernatants from cells treated in panel A were used for the measurement of CCL20 protein by ELISA. (C) Quantitative RT-PCR analysis showing increased expression of CCL20 mRNA in BCBL-1 and K562 cells stably transduced with a Flag-K13–expressing retroviral vector (top panel). The expression of Flag-tagged K13 in cell lysates was confirmed by immunoblotting (bottom panel). (D) Quantitative RT-PCR analyses showing down-regulation of K13 and CCL20 mRNAs in BCBL-1 cells infected with a K13 shRNA-expressing lentiviral vector compared with cells infected with a control shRNA vector.
K13 induces CCL20 expression in HUVEC and PEL cell lines. (A) HUVECs stably expressing a control vector or K13-ERTAM were mock-treated or treated with 4-OHT (50 nM) for 48 hours after which RNA was extracted and used for quantitative RT-PCR. The results of the quantitative RT-PCR analysis were confirmed by agarose gel electrophoresis (bottom panel). (B) Supernatants from cells treated in panel A were used for the measurement of CCL20 protein by ELISA. (C) Quantitative RT-PCR analysis showing increased expression of CCL20 mRNA in BCBL-1 and K562 cells stably transduced with a Flag-K13–expressing retroviral vector (top panel). The expression of Flag-tagged K13 in cell lysates was confirmed by immunoblotting (bottom panel). (D) Quantitative RT-PCR analyses showing down-regulation of K13 and CCL20 mRNAs in BCBL-1 cells infected with a K13 shRNA-expressing lentiviral vector compared with cells infected with a control shRNA vector.
Because induction of K13 activity in HUVEC K13-ERTAM cells is accompanied by acquisition of spindle cell morphology,31,32 it was conceivable that K13 does not up-regulate CCL20 expression directly but rather does so indirectly by inducing spindle cell differentiation of HUVECs. In addition, because KSHV infection has been also linked to the pathogenesis of PEL, it was important to examine whether K13 could up-regulate CCL20 expression in PEL cells. To address these questions, we examined the effect of exogenous K13 expression in the PEL-derived BCBL-1 cell line, which expresses very low levels of K13 endogenously36 and shows weak endogenous CCL20 expression (Figure 1C). BCBL-1 cells that had been stably transduced with a Flag-K13–expressing retroviral vector demonstrated approximately 4-fold increase in CCL20 mRNA compared with the vector-expressing cells (Figure 2C). Essentially similar results were obtained on ectopic K13 expression in the human chronic myeloid leukemia cell line K562 (Figure 2C). Collectively, these results demonstrate that K13 stimulates CCL20 gene expression in cells of different lineage, and this effect is not related to the acquisition of spindle cell phenotype by vascular endothelial cells.
shRNA-mediated silencing of K13 down-regulates CCL20 gene expression
We have previously reported successful silencing of K13 expression in PEL cells using lentiviral-mediated shRNA delivery.26 To confirm the involvement of K13 in the constitutive CCL20 expression in PEL cells, we infected BCBL-1 cells with lentiviral vectors encoding a control shRNA or an shRNA against K13. The lentiviral constructs also expressed a green fluorescent protein–blasticidin fusion protein to help with monitoring the infection efficiency. Approximately 80% to 90% of the cells expressed green fluorescent protein after a single round of infection with the lentiviral constructs, as measured by fluorescence microscopy (not shown). A quantitative RT-PCR analysis performed 72 hours after infection revealed an approximately 90% reduction in K13 expression in the cells infected with the lentiviral vector encoding shRNA targeting this protein compared with the cells infected with the control shRNA vector (Figure 2D). More importantly, this reduction in the K13 mRNA was accompanied by an approximately 70% reduction in the expression of CCL20 mRNA (Figure 2D), thereby confirming a major role of K13 in the constitutive CCL20 expression in the PEL cells.
K13 stimulates CCL20 promoter activity via NF-κB activation
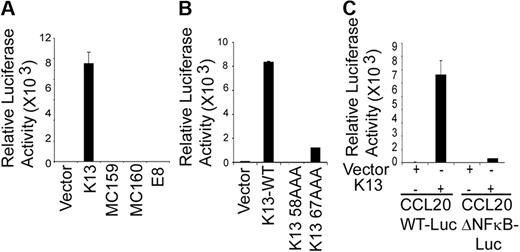
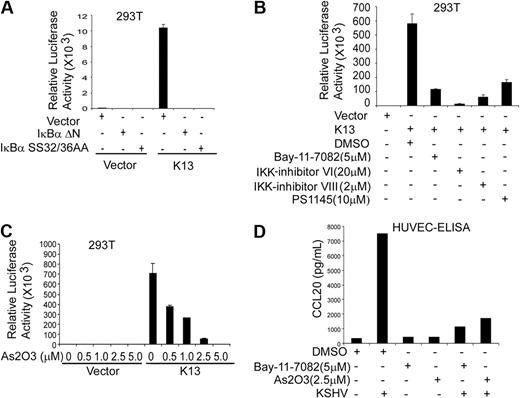
To examine whether the CCL20 promoter responds to K13, we transfected human embryonic kidney 293T cells with a luciferase-based reporter construct containing 874 bp of the CCL20 gene promoter. Coexpression of K13 led to approximately 10 000-fold increase in CCL20 promoter-driven reporter activity, suggesting that K13 activates the CCL20 gene at the transcriptional level (Figure 3A). In contrast, vFLIPs MC159 and MC160 from the molluscum contagiosum virus and the vFLIP E8 from the equine herpesvirus 2, which resemble K13 in structure but lack the ability to activate the NF-κB pathway,24,39 were unable to activate the CCL20 promoter (Figure 3A). The involvement of the NF-κB pathway in K13-induced CCL20 of promoter activation was further examined using 2 previously described point mutants of K13, K13-58AAA and K13-67AAA, which demonstrate total and partial loss of NF-κB activity, respectively.28 Consistent with their loss of NF-κB activating ability, the mutant 58AAA demonstrated a complete loss of CCL20 reporter activation, whereas the mutant 67AAA demonstrated partial activity (Figure 3B). Collectively, these results suggest that the ability of K13 to transactivate the CCL20 promoter is probably linked to its ability to activate the NF-κB pathway.
K13-induced NF-κB activity is critical for the activation of CCL20 promoter. (A) The 293T cells were transfected with an empty vector or the indicated vFLIPs (250 ng/well) along with a WT CCL20 promoter luciferase construct (75 ng/well) and a pRSV/LacZ (β-galactosidase) reporter construct (75 ng/well), and the reporter assay performed as described in “Luciferase reporter assay.” The values shown are averages (mean ± SE) of 1 representative experiment of 3 in which each transfection was performed in duplicate. (B) Ectopic expression of WT K13 but not its NF-κB–defective mutants (K13 58AAA and K13 67AAA) induces CCL20 promoter activity. The experiment was performed essentially as described in panel A. (C) The NF-κB site in the CCL20 promoter is critical for activation by K13. The 293T cells were transfected with a control vector or a vector encoding K13 along with either CCL20-WT-Luc or CCL20-ΔNF-κB-Luc reporter constructs, and the luciferase reporter assay performed as described in panel A. The values shown are averages (mean ± SE) of 1 representative experiment of 3 in which each transfection was performed in duplicate.
K13-induced NF-κB activity is critical for the activation of CCL20 promoter. (A) The 293T cells were transfected with an empty vector or the indicated vFLIPs (250 ng/well) along with a WT CCL20 promoter luciferase construct (75 ng/well) and a pRSV/LacZ (β-galactosidase) reporter construct (75 ng/well), and the reporter assay performed as described in “Luciferase reporter assay.” The values shown are averages (mean ± SE) of 1 representative experiment of 3 in which each transfection was performed in duplicate. (B) Ectopic expression of WT K13 but not its NF-κB–defective mutants (K13 58AAA and K13 67AAA) induces CCL20 promoter activity. The experiment was performed essentially as described in panel A. (C) The NF-κB site in the CCL20 promoter is critical for activation by K13. The 293T cells were transfected with a control vector or a vector encoding K13 along with either CCL20-WT-Luc or CCL20-ΔNF-κB-Luc reporter constructs, and the luciferase reporter assay performed as described in panel A. The values shown are averages (mean ± SE) of 1 representative experiment of 3 in which each transfection was performed in duplicate.
The NF-κB site at −82 in the CCL20 promoter is essential for K13-mediated activation
The CCL20 promoter region contains an atypical NF-κB binding-element (GGGAAAACCC) beginning at position −82 bp from the putative transcription start site, designated −82 κB, which has been shown to play a key role in response to TNF-α stimulation.40,41 To examine the involvement of the −82 κB site in K13-induced CCL20 transcription, we transfected 293T cells with either a luciferase reporter construct containing the WT CCL20 promoter (CCL20-WT-Luc) or a reporter construct (CCL20-Δ-NF-κB-Luc) bearing mutations (GGGAAAAAAAC) in this site. As shown in Figure 3C, K13-induced CCL20 promoter activity was nearly abolished in the CCL20-Δ NF-κB-Luc construct, confirming the importance of the −82 κB site in K13-mediated CCL20 gene expression.
K13-induced binding of NF-κB subunits to the CCL20 promoter
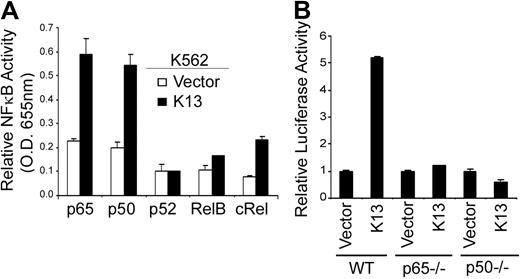
Because the mutation analysis of the CCL20 promoter suggested that K13 probably activated transcription through the −82 κB site, we next examined the nuclear proteins that bind to this site. For this purpose, we took advantage of an ELISA-based NF-κB DNA-binding assay and examined the recruitment of different NF-κB subunits to a synthetic oligonucleotide duplex containing the −82 κB site. Nuclear extracts from K13-expressing K562 cells demonstrated significant recruitment of p65 and p50 subunits and modest recruitment of c-Rel subunit to the −82 κB site compared with the nuclear extracts from the control vector cells (Figure 4A). In contrast, no significant recruitment of p52 and RelB subunits to the −82 κB site was observed (Figure 4A). To confirm the functional significance of the p65 and p50 subunits in K13-induced CCL20 gene expression, we use mouse embryonic fibroblast (MEF) cells lacking the expression of these subunits. As shown in Figure 4B, K13 failed to activate the CCL20 luciferase reporter construct in p65−/− and p50−/− MEFs, compared with the WT MEFs. Taken collectively, these results support the hypothesis that K13-induced NF-κB activity is directly involved in CCL20 promoter activation and this process is not dependent on the presence of an intermediate protein.
K13 triggers the recruitment of p65, p50, and c-Rel to the CCL20 promoter. (A) An ELISA-based NF-κB binding assay showing increased binding of p65, p50, and c-Rel subunits in nuclear extracts derived from K13-expressing cells to the NF-κB site present in the CCL20 promoter. (B) K13 fails to activate CCL20 promoter in p65 and p50 knockout cells. WT and p65−/− and p50−/− MEFs were transfected with an empty vector or K13 along with a CCL20-WT-Luc (75 ng/well) and a Renilla reporter construct using Lipofectamine. The luciferase assay was performed 48 hours after transfection as described previously.35
K13 triggers the recruitment of p65, p50, and c-Rel to the CCL20 promoter. (A) An ELISA-based NF-κB binding assay showing increased binding of p65, p50, and c-Rel subunits in nuclear extracts derived from K13-expressing cells to the NF-κB site present in the CCL20 promoter. (B) K13 fails to activate CCL20 promoter in p65 and p50 knockout cells. WT and p65−/− and p50−/− MEFs were transfected with an empty vector or K13 along with a CCL20-WT-Luc (75 ng/well) and a Renilla reporter construct using Lipofectamine. The luciferase assay was performed 48 hours after transfection as described previously.35
Abrogation of K13-induced CCL20 promoter activation by genetic and pharmacologic inhibitors of the NF-κB pathway
K13 is known to activate the NF-κB pathway by inducing phosphorylation of IκBα at residues Ser32 and Ser36, which leads to its ubiquitination and subsequent proteasome-mediated degradation.24,25 Consistent with this mechanism, K13-induced NF-κB activity can be blocked by phosphorylation-resistant mutants of IκBα.24 Therefore, we examined whether phosphorylation-resistant mutants of IκBα can also block K13-induced CCL20 promoter activity. As shown in Figure 5A, K13-induced CCL20 promoter activity was completely blocked by either a phosphorylation-resistant mutant of IκBα in which the 2 critical serine residues have been mutated to alanine (IκBα SS32/36AA), or a deletion mutant of IκBα, lacking the N-terminal 36 amino acids (IκBαΔN).
Role of NF-κB pathway in K13-induced CCL20 promoter activation. (A) Dominant-negative mutants of IκBα (IκBαΔN and IκBαSS32/36AA) block K13-induced CCL20 promoter activity. The 293T cells were transfected either with an empty vector or K13, along with a CCL20 luciferase reporter construct and a β-galactosidase reporter construct, as described in Figure 3A. The amount of IκBα mutant plasmids (500 ng/well) was 5 times the amount of vector or K13 (100 ng/well) plasmid, and the total amount of transfected DNA was kept constant by adding empty vector. The values shown are averages (mean ± SE) of 1 representative experiment of 3 in which each transfection was performed in duplicate. (B,C) Pharmacologic inhibitors of NF-κB block K13-induced CCL20 promoter activation. The 293T cells were transfected with an empty vector or a vector encoding K13, and 30 minutes after transfection treated with dimethyl sulfoxide (vehicle) or the indicated compounds for 16 hours before cell lysis. Reporter assay was performed as described for Figure 3A. (D) HUVECs were pretreated with Bay-11-7082 (5 μM) or As2O3 (2.5 μM) for 2 hours and subsequently infected with KSHV as described previously.31 Four hours after infection, the medium was changed with fresh medium containing Bay-11-7082 or As2O3. After overnight incubation, supernatant was collected and CCL20 secretion was measured as described in Figure 1B.
Role of NF-κB pathway in K13-induced CCL20 promoter activation. (A) Dominant-negative mutants of IκBα (IκBαΔN and IκBαSS32/36AA) block K13-induced CCL20 promoter activity. The 293T cells were transfected either with an empty vector or K13, along with a CCL20 luciferase reporter construct and a β-galactosidase reporter construct, as described in Figure 3A. The amount of IκBα mutant plasmids (500 ng/well) was 5 times the amount of vector or K13 (100 ng/well) plasmid, and the total amount of transfected DNA was kept constant by adding empty vector. The values shown are averages (mean ± SE) of 1 representative experiment of 3 in which each transfection was performed in duplicate. (B,C) Pharmacologic inhibitors of NF-κB block K13-induced CCL20 promoter activation. The 293T cells were transfected with an empty vector or a vector encoding K13, and 30 minutes after transfection treated with dimethyl sulfoxide (vehicle) or the indicated compounds for 16 hours before cell lysis. Reporter assay was performed as described for Figure 3A. (D) HUVECs were pretreated with Bay-11-7082 (5 μM) or As2O3 (2.5 μM) for 2 hours and subsequently infected with KSHV as described previously.31 Four hours after infection, the medium was changed with fresh medium containing Bay-11-7082 or As2O3. After overnight incubation, supernatant was collected and CCL20 secretion was measured as described in Figure 1B.
We also examined whether K13-induced CCL20 promoter activity could be antagonized by pharmacologic inhibitors of the NF-κB pathway. We have previously reported that Bay-11-7082, a specific inhibitor of the NF-κB, can block K13-induced NF-κB activity and spindle cell transformation of HUVECs.31 Consistent with these results, treatment with Bay-11-7082 also blocked K13-induced CCL20 promoter activity in 293T cells (Figure 5B). Because K13 activates the NF-κB pathway by activating the IKK complex,24,25 we examined the ability of 3 recently described specific inhibitors of this complex, PS1145,42 IKK inhibitor VI,43 and IKK inhibitor VIII,44 to block K13-induced CCL20 promoter activity. As shown in Figure 5B, all these inhibitors effectively blocked K13-transactivated CCL20 promoter activity. Arsenic trioxide, a known inhibitor of K13-induced NF-κB activation, also blocked K13-induced CCL20 promoter activity in a dose-dependent fashion (Figure 5C). We also examined the effect of NF-κB inhibitors on KSHV-induced CCL20 up-regulation in HUVECs. As shown in Figure 5D, treatment with Bay-11-7082 and arsenic trioxide led to near-complete inhibition of CCL20 protein secretion in the supernatant of KSHV-infected HUVECs, as determined by ELISA. Taken collectively, these results confirm the importance of the IKK complex and the NF-κB pathway in KSHV- and K13-induced CCL20 expression.
KSHV- and K13-mediated induction of CCR6
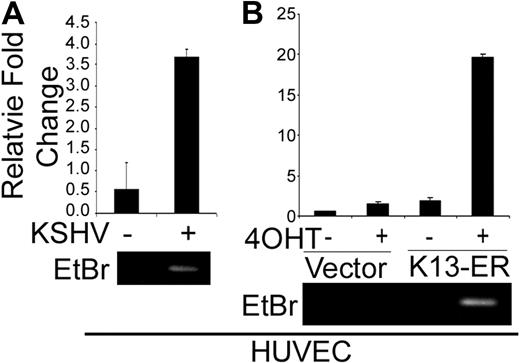
CCL20 is known to modulate its cellular effects through its receptor CCR6.45 Therefore, we studied the effect of KSHV infection on CCR6 gene expression. CCR6 mRNA expression, as assayed by quantitative RT-PCR, was highly induced in HUVECs that were infected with KSHV (Figure 6A). CCR6 mRNA expression was also induced by treatment with 4-OHT in the HUVEC-K13-ERTAM cells, whereas 4-OHT had no effect in the control vector-expressing HUVECs (Figure 6B).
KSHV infection and K13 expression induce the expression of CCL20 receptor CCR6 in HUVECs. Quantitative RT-PCR analysis showing induction of CCR6 mRNA in HUVECs after infection with KSHV (A) and in HUVEC-K13-ERTAM cells on treatment with 4-OHT (B). The experiments were performed essentially as described in Figures 1A and 2A, respectively.
KSHV infection and K13 expression induce the expression of CCL20 receptor CCR6 in HUVECs. Quantitative RT-PCR analysis showing induction of CCR6 mRNA in HUVECs after infection with KSHV (A) and in HUVEC-K13-ERTAM cells on treatment with 4-OHT (B). The experiments were performed essentially as described in Figures 1A and 2A, respectively.
Increased CCL20 and CCR6 expression in KS clinical samples
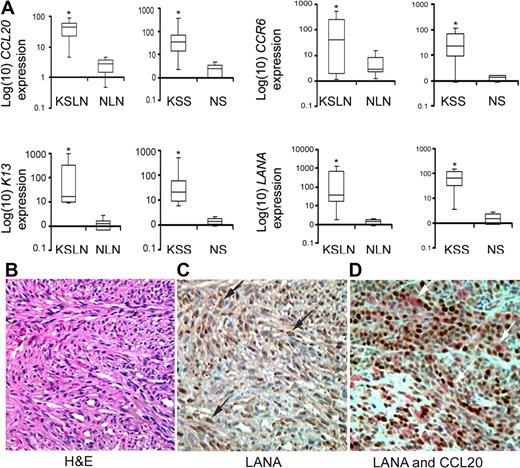
To determine the clinical significance of our results, we examined the expression of CCL20 and CCR6 in KS lymph nodes (n = 5) and KS skin samples (n = 10) by quantitative RT-PCR analysis. We included benign lymph nodes (n = 6) and normal skin (n = 10) samples as controls. As shown in Figure 7A, the expression levels of CCL20 and CCR6 were significantly higher in the KS lymph nodes and KS skin samples compared with the corresponding healthy skin or lymph node samples (P < .05, Mann-Whitney U test). The expression levels of LANA and K13, 2 KSHV latent genes, were also significantly higher in the KS samples compared with the corresponding controls (P < .05, Mann-Whitney U test), thereby confirming the presence of KSHV in the KS lesions (Figure 7A).
Up-regulation of CCL20 and its receptor CCR6 in KS samples. (A) Box-whisker plots with 95% confidence intervals for the RNA expression of CCL20, K13, LANA, and CCR6 in KSHV-infected patients' lymph nodes (KS lymph nodes, n = 5) and skin samples (KS skin samples, n = 10) along with corresponding benign lymph nodes (n = 6) and normal skin (n = 10) samples. Box-whisker plot indicates the median expression of respective genes relative to β-actin (line in the box) and their interquartile range (box). The whiskers present an entire range of expression level of indicated genes in a group, the upper value being the largest observation ≥ 75th percentile + 1.5 × (interquartile range), with the lowest value being the smallest observation ≤ 25th percentile − 1.5 × (interquartile range). The RNA was isolated from flash frozen tissues, and the expression levels of the indicated genes were determined by quantitative RT-PCR. The expression levels of CCL20 and its receptor CCR6 were significantly higher in the KS samples compared with corresponding uninfected controls (*P < .05, Mann-Whitney U test). Similarly, the expression level of LANA and K13, 2 key genes of KSHV infectivity, were significantly higher in KS samples than the corresponding healthy skin or lymph node samples (*P < .05, Mann-Whitney U test). (B) Hematoxylin and eosin staining shows the presence of characteristic spindle cells in a KS biopsy specimen. (C) Immunohistochemistry shows the presence of LANA-positive cells with typical brown punctate nuclear staining (black arrows) in the KS lesion. (D) Double immunostaining with LANA and CCL20 antibodies shows that a majority of CCL20-positive cells (red; white arrows) are positive for LANA (brown). (B-D) original magnification ×400.
Up-regulation of CCL20 and its receptor CCR6 in KS samples. (A) Box-whisker plots with 95% confidence intervals for the RNA expression of CCL20, K13, LANA, and CCR6 in KSHV-infected patients' lymph nodes (KS lymph nodes, n = 5) and skin samples (KS skin samples, n = 10) along with corresponding benign lymph nodes (n = 6) and normal skin (n = 10) samples. Box-whisker plot indicates the median expression of respective genes relative to β-actin (line in the box) and their interquartile range (box). The whiskers present an entire range of expression level of indicated genes in a group, the upper value being the largest observation ≥ 75th percentile + 1.5 × (interquartile range), with the lowest value being the smallest observation ≤ 25th percentile − 1.5 × (interquartile range). The RNA was isolated from flash frozen tissues, and the expression levels of the indicated genes were determined by quantitative RT-PCR. The expression levels of CCL20 and its receptor CCR6 were significantly higher in the KS samples compared with corresponding uninfected controls (*P < .05, Mann-Whitney U test). Similarly, the expression level of LANA and K13, 2 key genes of KSHV infectivity, were significantly higher in KS samples than the corresponding healthy skin or lymph node samples (*P < .05, Mann-Whitney U test). (B) Hematoxylin and eosin staining shows the presence of characteristic spindle cells in a KS biopsy specimen. (C) Immunohistochemistry shows the presence of LANA-positive cells with typical brown punctate nuclear staining (black arrows) in the KS lesion. (D) Double immunostaining with LANA and CCL20 antibodies shows that a majority of CCL20-positive cells (red; white arrows) are positive for LANA (brown). (B-D) original magnification ×400.
To confirm the results of quantitative RT-PCR analysis and to investigate the cells of origin of CCL20, we performed immunohistochemistry on KS biopsy specimens. The diagnosis of KS in the biopsy specimens was confirmed by the presence of characteristic spindle-shaped cells, as determined by hematoxylin and eosin staining (Figure 7B) and by characteristic punctate nuclear staining for LANA, as determined by immunohistochemistry (Figure 7C). Importantly, double immunohistochemical staining for LANA and CCL20 revealed that a majority of cells that were positive for CCL20 were also positive for LANA (Figure 7D). Taken collectively, these results suggest that KSHV-mediated stimulation of CCL20 and CCR6 expression that we demonstrated in vitro also occurs in vivo.
Discussion
In addition to the presence of distinctive spindle cells, KS lesions are characterized by infiltration with inflammatory cells, including dendritic cells, macrophages, plasma cells, and lymphocytes.4,46 Although the inflammatory cell infiltrate is the first histologic change in the KS lesions that precedes the appearance of spindle cells,4,5 the factors responsible for the accumulation of inflammatory cells in KS lesions have not been completely delineated. It has been proposed that the inflammatory cytokines produced in the KS lesions induce the expression of chemokines, such as MCP-1, MIP-1α, MIP-1β, IL-8, and Rantes, which promote recruitment of inflammatory cells into the lesions.47 An additional mechanism for chemokine production in the KS lesions is infection with KSHV. Whereas lytic replication of KSHV is associated with the expression of viral proteins with known proinflammatory and chemotactic activities, such as viral G protein–coupled receptor, viral interleukin-6 (vIL-6), and several chemokine homologs,48 latent infection with KSHV up-regulates the expression of several cellular chemokines, such as IL-8, IL-1, GRO-1, MCP-1, NAP-2, Rantes, and CXCL16.12–19,32,49 Here we demonstrate that infection with KSHV also induces the expression of CCL20, a chemokine that is not only a chemoattractant for lymphocytes but is also the most potent chemoattractant for immature dendritic cells.
The CCL20 gene was discovered through bioinformatics-based searches of DNA databases.50–52 Under physiologic conditions, CCL20 is expressed by a variety of epithelial cells, including keratinocytes, pulmonary epithelial cells, and intestinal epithelial cells, where it promotes the assembly and maintenance of organized lymphoid structures located beneath epithelial surfaces.33 Although CCL20 is usually expressed at a low basal level, its expression is highly induced by proinflammatory cytokines (eg, TNF-α, IL-1, and interferon-γ), and Toll-like receptor agonists (eg, lipopolysaccharide).33 The increased level of CCL20 observed during inflammatory stimuli is thought to help recruit CCR6-expressing cells and serve an antimicrobial function.33
The mechanism of CCL20 induction after inflammatory stimuli has been clarified by the analysis of its promoter region. Although the CCL20 promoter contains putative binding sites for several transcription factors, the NF-κB transcription factors are thought to be primarily responsible for its gene induction after inflammatory insults.40,41 Consistent with these prior studies, we demonstrate that the NF-κB pathway is also primarily responsible for KSHV-induced CCL20 induction. This claim is supported by the following lines of evidence. First, in vitro infection of vascular endothelial cells with KSHV results in NF-κB activation,31,32,53 which supports the involvement of this pathway in CCL20 gene induction after acute KSHV infection of HUVECs observed in the current study. Second, the NF-κB pathway is also constitutively active in PEL cell lines that are latently infected with KSHV, although the level of NF-κB activity varies between the different cell lines.25,54 Interestingly, the level of CCL20 mRNA in the PEL cell lines observed in the current study correlates closely with their previously reported level of NF-κB activity,25,36,54 suggesting a causal association. Thus, the BC-1 cell line, which possesses high constitutive NF-κB activity, showed high CCL20 mRNA expression, whereas BCBL-1 and JSC-1 cell lines, which have low NF-κB activity, showed weak CCL20 expression. Finally, KSHV-induced CCL20 induction in HUVECs could be effectively blocked by Bay-11-7082, a specific inhibitor of the NF-κB (Figure 5D).
It was recently reported that overexpression of KSHV-encoded K15 protein, a known activator of the NF-κB pathway, induces CCL20 gene expression in HeLa cells.55 However, K15 is primarily expressed during the lytic phase of the KSHV life cycle.55–57 Therefore, we focused our attention on vFLIP K13 because it is not only a powerful activator of the NF-κB pathway but has been also shown to be primarily responsible for NF-κB activity in latently infected cells.25,54 We and others have previously reported that ectopic expression of K13 in vascular endothelial cells is sufficient to mimic the effect of KSHV on the acquisition of spindle cell phenotype.31,32 In this report, we demonstrate that ectopic expression of K13 in HUVECs is also sufficient to induce CCL20 gene and protein expression, thereby suggesting that K13 plays a key role in KSHV-induced CCL20 gene induction. Furthermore, we confirm the involvement of K13 in constitutive CCL20 gene expression in KSHV-infected PEL cells by its shRNA-mediated gene-silencing.
The CCL20 promoter region contains binding sites for different transcription factors, such as activator protein 1 and activator protein 2, CAAT/enhancer-binding protein, stimulating protein 1, and the epithelium-specific Ets nuclear factor ESE-1.40,41 However, a nonstandard NF-κB binding site (5′-GGGAAAACCC-3′), designated −82 κB site, is thought to be primarily responsible for induction of its gene expression by proinflammatory cytokines.40,41 The involvement of the NF-κB pathway and the −82 κB site in K13-induced CCL20 gene induction is supported by the following lines of evidence. First, we observed a strong correlation between the ability of vFLIPs to activate NF-κB and their ability to activate CCL20 promoter. Thus, although WT K13 strongly activated CCL20 promoter, its NF-κB–defective mutants failed to do so. Similarly, no CCL20 promoter activation was observed on expression of vFLIPs MC159, MC160, and E8, which lack NF-κB activity. Second, mutations in the −82 κB site nearly abolished K13-induced CCL20 promoter activity. Third, K13 failed to induce CCL20 promoter activity in cells lacking the p65 and p50 subunits of NF-κB. Finally, K13-induced CCL20 promoter activity was blocked by genetic (ie, IκBα SS32/36AA and ΔNIκBα) and pharmacologic (ie, Bay-11-7082, PS1145, IKK inhibitors VI and VIII, and arsenic trioxide) inhibitors of the NF-κB pathway.
Because KSHV infection and ectopic K13 expression in primary endothelial cells are accompanied by their spindle cell differentiation, it was conceivable that the induction of CCL20 is a secondary consequence of spindle cell differentiation. Support for such as mechanism is provided by a recent study in which increased expression of chemokine CXCL16 was observed in HUVECs after latent infection with KSHV or ectopic expression of K13.15 However, interestingly, this effect was limited to primary endothelial cells, such as HUVECs and primary microvascular endothelial cells of blood or lymphatic lineage, which are known to undergo spindle cell differentiation on KSHV infection or K13 expression, and neither KSHV infection nor ectopic K13 expression was able to induce CXCL16 expression in cells that are generally resistant to this effect.15 As most cells expressing K13 display NF-κB induction, these results led the authors of this study to the hypothesis that the ability to up-regulate CXCL16 mRNA is probably controlled by post–NF-κB regulatory steps/factors that are present in HUVECs but are absent in the nonresponsive cells.15 To rule out the possibility that K13-induced CCL20 gene induction is similarly mediated by another protein controlled by NF-κB, we assayed binding of the different NF-κB subunits to the −82 κB site present in the CCL20 promoter region and found that p65, p50, and c-Rel bind to this site. Furthermore, in contrast to the results with CXCL16, we found that KSHV- or K13-induced CCL20 expression is not limited to HUVECs but is observed in a variety of established cell lines of different lineage, such as K562, BCBL-1, and 293T, none of which undergoes spindle cell differentiation on K13 expression. Taken collectively, these results demonstrate that K13-induced NF-κB activity itself is the direct mediator of CCL20 gene induction, and no intermediate protein or mechanism, such as spindle cell differentiation, is involved in this process.
An unexpected result of our study was that infection with KSHV or induction of K13 activity in HUVECs not only induced the expression of CCL20 but also up-regulated the expression of CCR6, a G protein–coupled receptor that acts as the specific receptor for CCL20. Unlike CCL20, the transcriptional regulation of CCR6 gene has not been clarified yet. However, because K13 primarily activates the NF-κB pathway,38 it is possible that this pathway is also involved in CCR6 gene induction, either directly or indirectly. Studies are currently in progress to address this question by analysis of the CCR6 promoter region.
We observed increased expression of CCL20 and CCR6 mRNAs in clinical samples of KS. Concomitant induction of CCR6 and CCL20 on the same cell types has been previously documented in pancreatic cancer and human T-cell leukemia-1–infected cells.34,58 Importantly, exogenous CCL20 was shown to stimulate the growth of one pancreatic cell line and enhance the migration of another cell line showing CCR6 expression.58 Therefore, it is conceivable that KSHV-infected endothelial cells (and possibly lymphocytes), with up-regulated expression of CCL20 and CCR6, could similarly proliferate or migrate in an autocrine/paracrine-dependent manner, thereby contributing to the pathogenesis of KSHV-associated malignancies. It is important to note in this context that circulating endothelial cells in patients with KS have evidence of KSHV infection.59 Recent studies further suggest that new blood vessels in tumors develop not only from existing vessels but also from circulating endothelial progenitor cells originating from the bone marrow,60 which has led to the suggestion that KSHV-infected circulating endothelial precursor cells may home to the permissive sites and propagate to produce KS lesions.61 Therefore, increased expression of CCR6 on KSHV-infected endothelial progenitor and immune cells may promote their migration to sites of injury and inflammation, locations that are rich in CCL20.33 The preferential migration of KSHV-infected cells to sites of inflammation and injury might, in part, provide a mechanistic explanation for the clinical observation that KS lesions sometimes arise precisely at the site of antecedent inflammation or injury, a property known as the Koebner phenomenon.62 Thus, in addition to promoting the recruitment of inflammatory and dendritic cells into KS lesions, KSHV- and K13-induced CCL20 and CCR6 up-regulation may directly contribute to tumor initiation, proliferation, and metastases. Furthermore, because CCL20 is known to inhibit the proliferation of hematopoietic progenitor cells in response to multiple growth factors,52 it may also contribute to the pathogenesis of bone marrow failure syndromes associated with KSHV infection.63,64 It needs to be pointed out, however, that because KSHV infection increases the expression of several chemokines and their receptors,14–16,31,32,49 CCL20/CCR6 signaling probably acts in conjunction with signaling through other chemokine receptors in the pathogenesis of KSHV-associated malignancies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Hiroyasu Nakano and Dr Gutian Xiao for providing MEFs lacking the NF-κB p65 and p50 subunits, respectively; Dr Tomoko Kohno for pGL2-CCL20/MIP-α (CCL20 WT-Luc) and pGL2-CCL20/MIP-3α/κBM (CCL20 ΔNF-κB-Luc) for luciferase reporter constructs; the National Cancer Research Institute–sponsored AIDS and Cancer Specimen Resources at University of California, San Francisco, CA, and George Washington University, Washington, DC, for providing patient samples; Marie Acquafondata (Tissue and Research Pathology Services) for help with immunohistochemistry; and Dr Siddhartha Kar and Aletheia Tamewitz for critical reading of the manuscript.
This work was supported by the National Institutes of Health (Bethesda, MD; grants CA85177, CA124621, and HL085189), the Leukemia & Lymphoma Society (White Plains, NY), and the Mario Lemieux Foundation (Pittsburgh, PA).
National Institutes of Health
Authorship
Contribution: V.P., H.M., S.S., and T.Y. performed research; V.P. and H.M. prepared figures; Y.C. provided the LANA antibody and KS specimens and helped with immunohistochemistry; and V.P., H.M., and P.M.C. designed research, analyzed the data, and wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Preet M. Chaudhary, Hillman Cancer Center, 5117 Centre Ave, Suite 1.19A, Pittsburgh, PA 15213-1863; e-mail: chaudharypm@upmc.edu.
References
Author notes
*V.P. and H.M. contributed equally to this work.