Abstract
Quebec platelet disorder (QPD) is an inherited bleeding disorder associated with increased urokinase plasminogen activator (uPA) in platelets but not in plasma, intraplatelet plasmin generation, and α-granule protein degradation. These abnormalities led us to investigate uPA expression by QPD CD34+ progenitors, cultured megakaryocytes, and platelets, and whether uPA was stored in QPD α-granules. Although QPD CD34+ progenitors expressed normal amounts of uPA, their differentiation into megakaryocytes abnormally increased expression of the uPA gene but not the flanking genes for vinculin or calcium/calmodulin-dependent protein kinase IIγ on chromosome 10. The increased uPA production by cultured QPD megakaryocytes mirrored their production of α-granule proteins, which was normal. uPA was localized to QPD α-granules and it showed extensive colocalization with α-granule proteins in both cultured QPD megakaryocytes and platelets, and with plasminogen in QPD platelets. In QPD megakaryocytes, cultured without or with plasma as a source of plasminogen, α-granule proteins were stored undegraded and this was associated with much less uPA-plasminogen colocalization than in QPD platelets. Our studies indicate that the overexpression of uPA in QPD emerges with megakaryocyte differentiation, without altering the expression of flanking genes, and that uPA is costored with α-granule proteins prior to their proteolysis in QPD.
Introduction
Quebec platelet disorder (QPD) is an unusual inherited bleeding disorder, associated with increased expression and storage of the fibrinolytic enzyme urokinase plasminogen activator (uPA) in platelets and delayed-onset bleeding following trauma or surgery that responds only to fibrinolytic inhibitor therapy.1-3 The genetic cause of QPD has recently been linked to inheritance of a region on chromosome 10 that contains the uPA gene (PLAU).4 The normal uPA in QPD urine5 and plasma (prepared with platelet activation inhibitors),6 and apparent increases in uPA message in platelets (based on Northern blot analysis),2 suggest that the increased uPA in QPD platelets results from increased uPA expression by megakaryocytes.1 However, the expression of uPA by CD34+ progenitors, and by normal and QPD megakaryocytes, at different stages of differentiation, has not been characterized or quantified. Furthermore, it has not been determined whether QPD selectively increases uPA mRNA in platelets or whether it also increases mRNA from VCL and CAMK2G, the flanking genes on chromosome 10 that encode vinculin (a protein normally expressed in platelets)7 and calcium/calmodulin-dependent protein kinase IIγ (CAMK2G), a protein expressed by T lymphocytes8 that has not been studied in platelets.
Normally, blood contains similar molar amounts of uPA and tissue plasminogen activator (tPA) for converting plasminogen to plasmin and only small amounts of uPA in platelets (up to 1.3 ng uPA/109 platelets; reviewed in Diamandis et al1 ). Unlike normal platelets, QPD platelets contain sufficient uPA (approximately 400-600 ng uPA/109 platelets)2 to trigger extracellular plasmin generation and premature clot lysis when incorporated into forming or preformed clots.6 Within QPD platelets, single-chain (sc) uPA is not evident, as uPA is stored in active forms that include 2-chain uPA (tcuPA) and low-molecular-weight uPA (LMWuPA). In addition, QPD platelets contain uPA complexed with the active forms of platelet plasminogen activator inhibitor 1 (PAI-1), which are consumed in QPD.2 uPA activation within QPD platelets is postulated to result from exposure to plasmin, as QPD platelets, but not plasma, contain elevated levels of plasmin-α2 plasmin inhibitor complexes.9 uPA-induced, intraplatelet generation of plasmin is thought to trigger degradation of diverse stored α-granule proteins—a hallmark feature of QPD that affects proteins synthesized by megakaryocytes, including thrombospondin-1 (TSP-1), P-selectin, osteonectin, and von Willebrand factor (VWF), and proteins endocytosed from plasma, such as fibrinogen and factor V.10-12 The loss of α-granule multimerin 1 (MMRN1) in QPD11 is also thought to result from plasmin-mediated degradation. Heterogeneity in the protein contents of megakaryocyte and platelet α-granules is now recognized to result in some separation of proteins, such as fibrinogen from VWF,13 and antiangiogenic proteins, such as TSP-1 and endostatin, from proangiogenic proteins, such as vascular endothelial growth factor (VEGF) and basic fibroblast growth factor.14 However, uPA has never been demonstrated within QPD α-granules, and the extent of its colocalization with QPD platelet plasminogen and degraded α-granule proteins has not been evaluated.
To characterize uPA expression during normal and QPD megakaryopoiesis, and investigate uPA storage in QPD α-granules, we studied CD34+ progenitors, cultured megakaryocytes, and platelets. We report that the increased expression of uPA in QPD is not evident in circulating hematopoietic stem cells and that it emerges as QPD megakaryocytes differentiate, resulting in production of platelets that contain increased uPA, but not increased vinculin or CAMK2G mRNA. We also report that uPA is contained within the α-granules of circulating QPD platelets, where it colocalizes with plasminogen and α-granule proteins known to be degraded in QPD, consistent with the proposed mechanism of QPD α-granule protein degradation.
Methods
All studies were conducted with approval of the institutional ethics review boards of all participating institutions and in accordance with the Declaration of Helsinki, as last amended in 2004.
Sample collection
Peripheral blood samples (200 mL/donation) were collected from QPD and healthy control subjects with written informed consent. Samples were collected into sterile acid citrate dextrose anticoagulant (vol/vol = 1:6) containing 1 mM theophylline (Sigma-Aldrich, Oakville, ON), 3 μM prostaglandin E1 (Sigma-Aldrich), and 3 μM aprotinin (Roche Diagnostics, Laval, QC).
Isolation of cells from peripheral blood
Platelets were harvested from peripheral blood, as previously described (upper two-thirds of platelet-rich plasma; minimal leukocyte contamination was verified by cell counting of selected samples).11 CD34+ hematopoietic stem cells with minimal platelet contamination (< 0.5 platelets per nucleated cell) were isolated from peripheral blood by a modification of the method described for isolating CD34+ cells from cord blood.15 Briefly, after removal of platelet-rich plasma, and density gradient separation using Ficoll,15 mononuclear cells were further purified by an OptiPrep (Axis-Shield, Oslo, Norway; 200g, 12-minute centrifugation) density gradient separation before immunomagnetic isolation of CD34+ cells.15 Mononuclear cells, from Ficoll gradient separations, were also used to obtain control T lymphocytes by immunomagnetic isolation (Human CD3+ selection kit; StemCell Technologies, Vancouver, BC) to quantify CAMK2G mRNA. The final CD34+ cell and T-lymphocyte purities, assessed by flow cytometry,15 were consistently more than 90%.
Megakaryocyte cultures
CD34+ cells were cultured with thrombopoietin (TPO), as described,15 except 50 ng/mL recombinant human TPO (PeproTech, Rocky Hill, NJ) was used. Cell expansion was quantified at different stages of culture, as described,15 with viability determined by trypan blue exclusion. Cells were evaluated by flow cytometry, as described, to quantify the percentage that expressed the mature megakaryocyte marker CD41a (glycoprotein αIIb).15 Cells were cultured until day 13 when megakaryocyte maturation was evident and cell viability was still acceptable (> 70%). For some analyses, day-7 cultures were supplemented with 10% (vol/vol) sterile, heparinized, platelet-poor plasma (from blood group AB donors)16 as a source of exogenous plasminogen, as the addition of purified plasminogen led to a complete loss of viable cells in control and QPD cultures, and the addition of more plasma reduced megakaryocyte differentiation.
Analyses of mRNA by quantitative reverse-transcription polymerase chain reaction
Total cellular RNA was extracted from different cells of the same subjects, including CD34+ cells, cultured megakaryocytes, and platelets, using RNeasy Mini Kits (QIAGEN, Mississauga, ON) as recommended. RNase-Free DNase (QIAGEN) was added to digest any contaminating DNA. Quality and quantity of extracted RNA were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Mississauga, ON). Isolated RNA was stored at −80°C until analyzed.
For reverse transcription (RT), 72 ng total RNA was incubated at 37°C for 60 minutes in a 20-μL reaction mixture containing recommended Omniscript RT Kit components (QIAGEN), 2.5 μM oligo (deoxythymidine)20 primer (Invitrogen, Burlington, ON), and 20 units RNAse inhibitor RNaseOUT (Invitrogen). Reactions were stopped by a 5-minute incubation at 95°C, followed by rapid cooling on ice.17
RNA from CD34+ cells was used only for quantitative real-time polymerase chain reaction (qPCR) analysis of PLAU transcription, as the quantities harvested precluded other analyses. RNA from day-7 and -13 megakaryocytes and platelets was used for qPCR analysis of PLAU, VWF (control for increased mRNA during megakaryocyte differentiation18 ), and VCL7 transcription. RNA from platelets was also used to evaluate CAMK2G transcription in QPD. Transcription of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was evaluated simultaneously in all samples as an endogenous control to correct for potential variations in template RNA, RT, and qPCR efficiencies. Supplies and instructions for qPCR were from Applied Biosystems (Foster City, CA). The gene-specific sets of oligonucleotide primers and fluorescent probes for qPCR (from predeveloped TaqMan Gene Expression Assays) were as follows: uPA: Hs00170182_m1; VWF: Hs00169795_m1; vinculin: Hs00243320_m1; CAMK2G: Hs00538454_m1; and GAPDH: 4333764T. All selected amplicons spanned intron-exon boundaries. qPCR singleplex reaction mixtures included: 5 μL cDNA, 12.5 μL TaqMan Gene Expression Master Mix (containing DNA polymerase), 1.25 μL 20× TaqMan Gene Expression Assay reagents, and 6.25 μL RNAse-free water. Assays were done in 96-well TaqMan optical reaction plates in triplicate (or duplicate if there were limiting quantities of template RNA and cDNA) using an ABI PRISM 7900HT real-time thermal cycler (Applied Biosystems), with the following thermal profile: 2 minutes at 50°C, 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. No template controls served to exclude contamination.
For each qPCR run, standard curves were obtained by amplifying serially diluted (1:5-1:3125) cDNA that was reverse-transcribed from pooled samples. For uPA, vinculin, and VWF, a pool of total platelet RNA from 5 QPD subjects was used. For CAMK2G, a pool of total T-lymphocyte RNA from 5 control subjects was used. Relative mRNA levels were obtained using the average value for the target gene threshold cycle (Ct), normalized to GAPDH Ct, as described.19
Protein analyses
Cell lysates for protein analysis were prepared from CD34+ cells, cultured megakaryocytes, and platelets, by methods described.2,15 Cell-free supernatants were harvested from megakaryocyte cultures on day 7 and day 13, and for some studies daily from days 7 to 13, as described.16 Samples were stored at −80°C until analyzed.
Enzyme-linked immunosorbent assays (ELISAs) were used to quantify the following: uPA (modified to include a lower concentration standard of 12.5 pg/mL) and PAI-1 (Oncogene Science, Cambridge, MA); uPA-PAI-1 complexes and platelet factor 4 (PF-4; Hyphen Biomed, Neuville-sur-Oise, France); tPA (lower standards used as described)2 ; uPA receptor (uPAR; American Diagnostica, Montreal, QC); TSP-111 ; MMRN111 ; and VWF.15 Cell lysate and culture supernatant results were expressed in quantities per 106 cells, or per milligram of total cellular protein (comparisons of megakaryocyte and platelet lysates).15 Due to the low numbers of CD34+ cells in peripheral blood, a pooled QPD CD34+ cell lysate from 3 donors was compared with 3 pooled control CD34+ cell lysates (each from 3 donors), to determine whether the protein quantities for QPD samples were within the range for control samples.
Western blotting was used to evaluate the mobilities of uPA, TSP-1, VWF, P-selectin, and PAI-1 in platelet lysates (10 μL/lane) and cultured megakaryocyte lysates (45 μL/lane; pooled from 3 individuals to obtain sufficient material) after separation by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), as described.2,9,11,15 Platelet VEGF was also analyzed by Western blotting (primary antibody: 1 μg/mL mouse anti-VEGF [Cedarlane, Burlington, ON]; secondary antibody: 1:25 000 horseradish peroxidase–labeled donkey anti–mouse IgG [Jackson ImmunoResearch Laboratories, West Grove, PA]) after separation (reduced) on a 12% SDS-polyacrylamide gel.
Immunofluorescent microscopy
Immunolabeling.
Cultured megakaryocytes and platelets were double labeled as described,16 except resting platelets (2 × 108/mL, prepared as described2 ) were fixed in suspension (1% paraformaldehyde, room temperature, 10 minutes) and dried on aptex-coated slides overnight, before permeabilization, quenching, and blocking as described.16 Primary antibodies included the following: mouse antibodies to uPA (10 μg/mL; Monosan, Uden, The Netherlands), TSP-1 (CH-1, 3.5 μg/mL),20 MMRN1 (JS-1, 10 μg/mL),20 αIIbβ3 (10 μg/mL; BD Biosciences, Mississauga, ON), VEGF (2 μg/mL; Cedarlane), and osteonectin (5 μg/mL; Haematologic Technologies, Essex Junction, VT); rabbit antibodies to uPA (10 μg/mL; Monosan), uPAR (6.4 μg/mL; Abcam, Cambridge, MA), plasminogen (1:100; Biogenesis, Poole, Great Britain), fibrinogen (1:5000; Behring, Marburg, Germany), and VWF (1:300; Dako, Mississauga, ON); and sheep antibody to factor V (10 μg/mL; Affinity Biologicals, Ancaster, ON). Secondary antibodies (Alexa Fluor 594– or Alexa Fluor 488–conjugated) were used as described.16 Controls included single labeled cells, cells labeled without primary or secondary antibodies, or with normal mouse, rabbit, or sheep IgG. Additional controls included cells single labeled using the opposite secondary antibodies to ensure no antibody cross-reactivity. Labeled cells were mounted with Permafluor mounting media (Beckman Coulter, Marseille, France).
Image acquisition and processing.
Immunolabeled samples were viewed on a DMI6000B wide fluorescent microscope (Leica Microsystems, Richmond Hill, ON) with an Orca ER-AG camera (Hamamatsu, Bridgewater, NJ) and Volocity 4 acquisition software (Improvision, Waltham, MA). Fluorescent crossover between channels was verified to be negligible. Z-series were acquired at 22°C with 100×/1.4NA DIC oil-plan apo objective, using identical microscope settings for samples and negative controls. Images were restored in Volocity 4 using the iterative deconvolution function,21 before importing into MBF-ImageJ22 (McMaster University Biophotonics Facility, http://www.macbiophotonics.ca/imagej/) for linear, uniform brightness and contrast adjustments, background subtraction, and scale bar labeling.
Quantitative colocalization analysis.
The degree of labeled protein colocalization was assessed using the MBF-ImageJ Intensity Correlation Analysis plugin to determine the Pearson correlation coefficient (rp, range: −1 to 1, with 1 equivalent to complete colocalization)23 and the Manders overlap coefficient (r, range: 0 to 1, with 1 equivalent to complete colocalization).24 A minimum of 10 day-13 cultured megakaryocytes and 30 platelets were evaluated for each protein comparison and data were confirmed using cells from 2 additional subjects.
Immunoelectron microscopy
To determine the subcellular distribution of uPA, frozen thin sections of platelets were prepared and immunolabeled, as described15 with rabbit anti-uPA (Monosan) and a gold-coupled secondary antibody (British Biocell, Cardiff, United Kingdom). After counterstaining with uranyl acetate, platelets were examined with a Philips CM10 electron microscope (Philips Healthcare, Surenes Cedex, France).
Statistical analysis
Most quantitative data were expressed as mean plus or minus SEM (range) and compared by 2-tailed unpaired t test. Immunolocalization data (rp and r; expressed as mean ± SD, range) were analyzed by Welch ANOVA with Satterthwaite t post-hoc analysis. Significance was established at P less than .05.
Results
Characteristics of control and QPD-cultured megakaryocytes
QPD and control megakaryocytes showed the typical phenotype and expansion of megakaryocytes grown in culture with TPO from peripheral blood progenitors25 (fold expansion: 8.3 ± 0.8 [range, 5.3-10.0] for 6 QPD cultures; 7.4 ± 2.6 [range, 1.3-16.5] for 5 control cultures; P = .8). Although QPD cultures contained a lower proportion of cells expressing αIIbβ3 on day 7 (QPD: 5.2% ± 1.3%, [range, 2%-10%]; control: 21% ± 4% [range, 12%-34%], P = .01), by day 13, the difference was not significant (QPD: 57% ± 6% [range, 45%-76%]; control: 69% ± 4% [range, 61%-84%]; P = .13).
Evaluation of gene expression in CD34+ cells, cultured megakaryocytes, and platelets
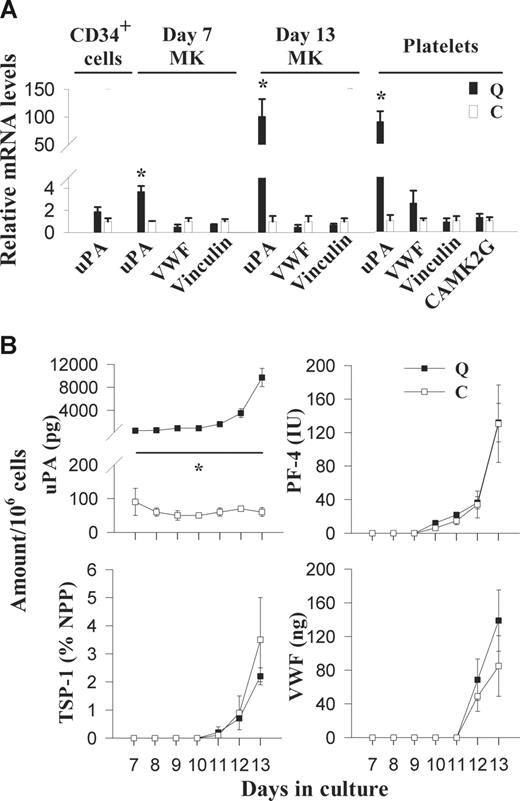
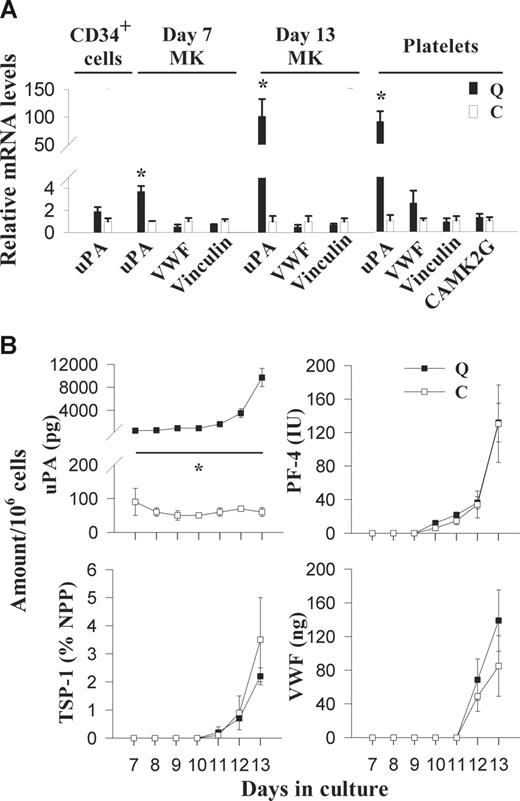
RT-qPCR analyses indicated that uPA mRNA was not increased in QPD CD34+ cells (Figure 1A); however, it was increased in day-7 (3.7 ± 0.5-fold higher than controls) and day-13 (101 ± 31-fold higher than controls) cultured QPD megakaryocytes (Figure 1A), and in QPD platelets (90.1- ± 18.6-fold higher than control platelets; Figure 1A). Unlike uPA mRNA, VWF and vinculin mRNA were not increased in QPD platelets or megakaryocytes (Figure 1A). In addition, CAMK2G mRNA was not increased in QPD platelets (Figure 1A).
Expression of uPA, α-granule proteins, vinculin, and CAMK2G in QPD (Q) and control (C) CD34+ cells, cultured megakaryocytes, and platelets. (A) RT-qPCR analysis of uPA, VWF, vinculin, and CAMK2G mRNA levels in platelets and/or CD34+ cells, and day-7 and -13 megakaryocytes, relative to controls (arbitrarily set to 1) to identify altered patterns of gene expression in QPD. (B) Comparison of uPA, PF-4, TSP-1, and VWF antigen in megakaryocyte culture supernatants, evaluated by ELISA at different stages of culture. Undetectable levels are presented as zero. Data represent mean values; error bars indicate SEM for data from 3 to 5 subjects. *Significant increases (P < .05) in QPD, compared with control.
Expression of uPA, α-granule proteins, vinculin, and CAMK2G in QPD (Q) and control (C) CD34+ cells, cultured megakaryocytes, and platelets. (A) RT-qPCR analysis of uPA, VWF, vinculin, and CAMK2G mRNA levels in platelets and/or CD34+ cells, and day-7 and -13 megakaryocytes, relative to controls (arbitrarily set to 1) to identify altered patterns of gene expression in QPD. (B) Comparison of uPA, PF-4, TSP-1, and VWF antigen in megakaryocyte culture supernatants, evaluated by ELISA at different stages of culture. Undetectable levels are presented as zero. Data represent mean values; error bars indicate SEM for data from 3 to 5 subjects. *Significant increases (P < .05) in QPD, compared with control.
Production of uPA and other proteins during QPD megakaryopoiesis
Pooled QPD CD34+ cells contained approximately 60 pg uPA/106 cells, which was within the range observed for pooled control samples (n = 3; 40 ± 20 pg/106 [range, 10-70 pg/106]). Although day-7 and day-13 QPD megakaryocyte cultures contained normal amounts of PAI-1, PF-4, TSP-1, VWF, and MMRN1, they contained increased amounts of uPA (Table 1). Some day-13 QPD megakaryocyte cultures also contained small amounts of uPA–PAI-1 complexes (Table 1). The increased uPA production by QPD megakaryocytes coincided with the increased production of PF-4, TSP-1, and VWF in QPD and control cultures (Figure 1B). By day 13, QPD megakaryocytes contained 19% to 27% of the QPD platelet uPA/mg cellular protein (paired analyses of samples from 4 QPD individuals). tPA was undetectable (< 1.6 ng/mL) in all CD34+ cells (n = 3 QPD and 3 control lysates) and megakaryocyte cultures (n = 4 QPD and 6 control day-7 and day-13 lysates and supernatants), indicating that uPA was the only plasminogen activator produced during megakaryopoiesis.
Plasminogen was undetectable in all megakaryocyte cultures (< 0.8 ng/mL in n = 3 QPD and 3 control day-13 lysates and supernatants) unless the cells were cultured in media containing plasma (ng/106 cells, in lysate from cultures with plasma, n = 3: QPD: 73 ± 13 [range, 46-90], control: 27 ± 8 [range, 11-36]; P = .06).
The amounts of uPAR in cultured megakaryocytes were not quantified as there were similar, low amounts of uPAR antigen in control and QPD platelets (ng/109 platelets, n = 4: QPD: 0.7 ± 0.1 [range, 0.6-0.9]; control: 0.4 ± 0.1 [range, 0.2-0.9]; P = .1), which were undetectable by immunostaining of platelets and megakaryocytes (not shown).
Distribution of uPA in cultured QPD megakaryocytes and platelets
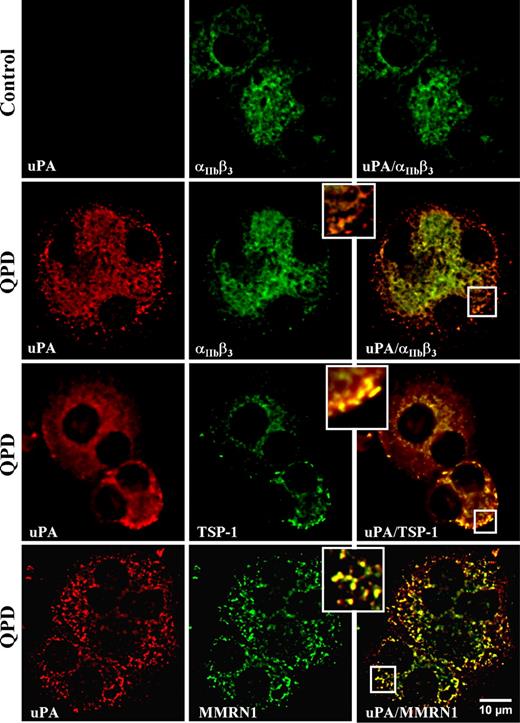
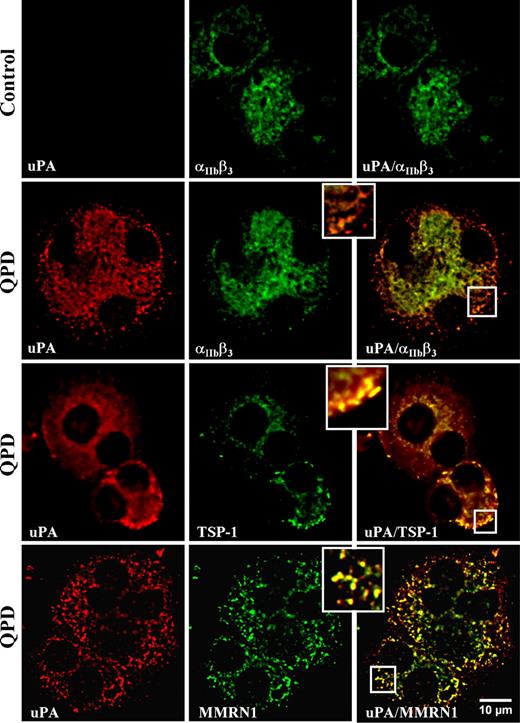
Immunofluorescent microscopy (Figure 2) indicated that there was strong uPA labeling of all differentiated QPD cells that showed strong labeling for αIIbβ3 or TSP-1 (representing 45%-52% of all cells in culture). In contrast, control megakaryocytes showed only faint uPA immunolabeling (Figure 2), which was indistinguishable from background (not shown). Within QPD-cultured megakaryocytes, uPA colocalized with αIIbβ3,TSP-1, and MMRN1 in perinuclear and granular structures (Figure 2), which was confirmed by quantitative analysis (rp: uPA and αIIbβ3: 0.93 ± 0.03 [range, 0.88-0.96]; uPA and TSP-1: 0.92 ± 0.04 [range, 0.80-0.97]; uPA and MMRN1: 0.85 ± 0.04 [range, 0.79-0.90]) (r: uPA and αIIbβ3: 0.97 ± 0.01 [range, 0.96-0.99]; uPA and TSP-1: 0.96 ± 0.02 [range, 0.88-0.98]; uPA and MMRN1: 0.90 ± 0.02 [range, 0.84-0.92]).
Intracellular distribution of uPA compared with αIIbβ3, and α-granule proteins TSP-1 and MMRN1 in day-13 QPD and control cultured megakaryocytes. Panels show deconvolved immunofluorescent images of megakaryocytes, double immunolabeled with rabbit anti-uPA (red, left panels) and mouse monoclonal antibodies to αIIbβ3, TSP-1, or MMRN1 (green, middle panels) (merged images on right). Insets show magnified images of peripheral cytoplasm. Experiments were performed as outlined in “Immunofluorescent microscopy.”
Intracellular distribution of uPA compared with αIIbβ3, and α-granule proteins TSP-1 and MMRN1 in day-13 QPD and control cultured megakaryocytes. Panels show deconvolved immunofluorescent images of megakaryocytes, double immunolabeled with rabbit anti-uPA (red, left panels) and mouse monoclonal antibodies to αIIbβ3, TSP-1, or MMRN1 (green, middle panels) (merged images on right). Insets show magnified images of peripheral cytoplasm. Experiments were performed as outlined in “Immunofluorescent microscopy.”
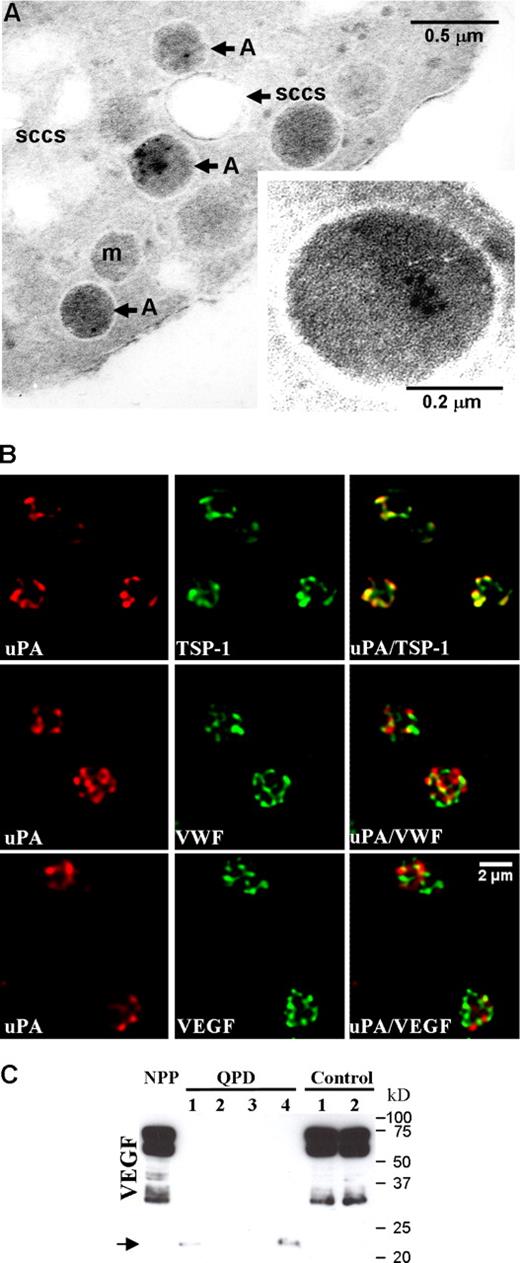
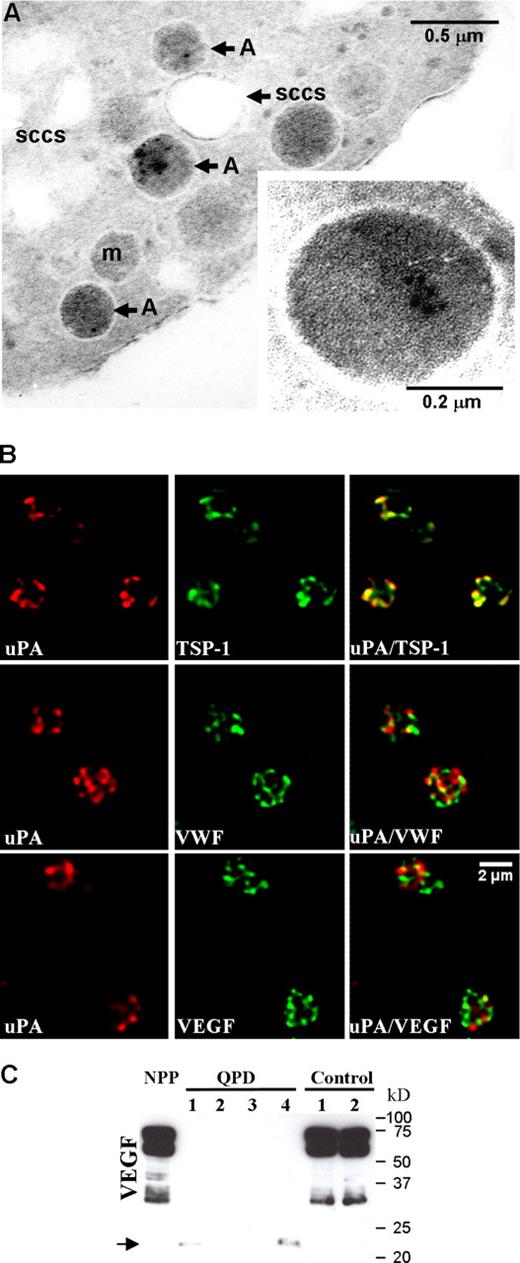
Immunoelectron microscopy of platelets confirmed that uPA was stored in QPD α-granules, without significant labeling of other platelet structures (Figure 3A) or control platelets (not shown). Further analyses by immunofluorescent microscopy indicated that there was extensive colocalization of QPD platelet uPA with TSP-1 (Figure 3B; Table 2) and plasminogen (Figure 4Q plt; Table 2), and extensive but less complete colocalization of QPD platelet uPA with VWF, VEGF (Figure 3B; Table 2), osteonectin, fibrinogen, and factor V (Table 2). Colocalization of QPD platelet uPA with MMRN1 could not be quantified, as unlike QPD megakaryocytes (Figure 2), QPD platelets showed only very faint immunolabeling for MMRN1 (not shown). Western blot analyses indicated that like other α-granule proteins, QPD platelet VEGF was abnormally degraded as QPD platelets contained decreased amounts of VEGF (Figure 3C) that also had an abnormally lower mobility (bands indicated by arrow in Figure 3C that were evident in all QPD samples on longer exposures not shown).
The uPA storage site in QPD platelets. (A) Immunogold labeling of uPA in QPD platelets, showing occasional uPA labeling in α-granules, with no significant labeling of the surface-connected canalicular system (SCCS), mitochondria (m), or plasma membrane. Inset shows a QPD platelet α-granule under higher magnification. Experiments were performed as outlined in “Immunoelectron microscopy.” (B) Deconvolved immunofluorescent images of QPD platelets, double immunolabeled with an antibody to uPA (red, left panels) and antibodies to TSP-1, VWF, or VEGF (green, middle panels) (merged images on right). Experiments were performed as outlined in “Immunofluorescent microscopy.” (C) Western blot of VEGF in pooled platelets from 20 healthy individuals (NPP), and in platelets from 4 QPD and 2 control individuals, evaluated after SDS-PAGE. Arrow indicates VEGF forms with lower mobility in QPD platelets.
The uPA storage site in QPD platelets. (A) Immunogold labeling of uPA in QPD platelets, showing occasional uPA labeling in α-granules, with no significant labeling of the surface-connected canalicular system (SCCS), mitochondria (m), or plasma membrane. Inset shows a QPD platelet α-granule under higher magnification. Experiments were performed as outlined in “Immunoelectron microscopy.” (B) Deconvolved immunofluorescent images of QPD platelets, double immunolabeled with an antibody to uPA (red, left panels) and antibodies to TSP-1, VWF, or VEGF (green, middle panels) (merged images on right). Experiments were performed as outlined in “Immunofluorescent microscopy.” (C) Western blot of VEGF in pooled platelets from 20 healthy individuals (NPP), and in platelets from 4 QPD and 2 control individuals, evaluated after SDS-PAGE. Arrow indicates VEGF forms with lower mobility in QPD platelets.
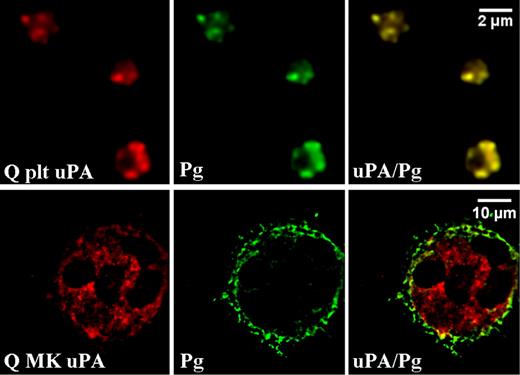
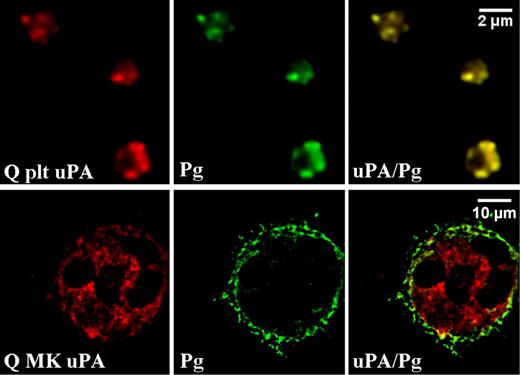
Distributions of plasminogen and uPA in day-13 QPD cultured megakaryocytes compared with platelets. Deconvolved immunofluorescent images of QPD platelets (Q plt) and QPD megakaryocytes (Q MK) grown with plasma. Cells were double immunolabeled with antibodies to uPA (red) and plasminogen (Pg, green; merged images at right). Experiments were performed as outlined in “Immunofluorescent microscopy.”
Distributions of plasminogen and uPA in day-13 QPD cultured megakaryocytes compared with platelets. Deconvolved immunofluorescent images of QPD platelets (Q plt) and QPD megakaryocytes (Q MK) grown with plasma. Cells were double immunolabeled with antibodies to uPA (red) and plasminogen (Pg, green; merged images at right). Experiments were performed as outlined in “Immunofluorescent microscopy.”
In day-13 QPD megakaryocytes cultured with plasma, plasminogen was found predominantly in peripheral structures and not in uPA-containing granules (Figure 4Q MK). Quantitative analyses confirmed that there was less colocalization of uPA and plasminogen in day-13 QPD megakaryocytes (rp: 0.39 ± 0.08 [range, 0.30-0.53]; r: 0.67 ± 0.07 [range, 0.55-0.74]) compared with QPD platelets (Table 2; P < .001). These data indicated that there was incomplete trafficking of plasminogen into uPA-containing structures in cultured QPD megakaryocytes compared with QPD platelets.
Forms of uPA and α-granule proteins in QPD cultured megakaryocytes
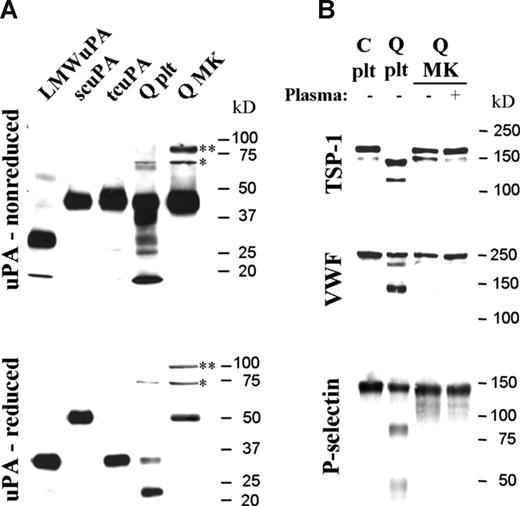
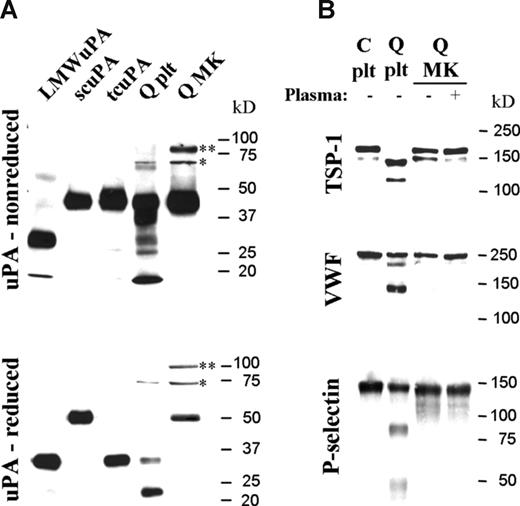
In contrast to QPD platelets, which contained mostly tcuPA, day-13 QPD cultured megakaryocytes contained mainly scuPA (Figure 5A), even when cultured with plasma (not shown). Like QPD platelets, QPD megakaryocytes contained some high-molecular-weight uPA complexes (Figure 5A, bands indicated by * and **) that included forms (*) recognized by PAI-1 antibodies (data not shown), as suggested by uPA–PAI-1 complex ELISA (Table 1). Control megakaryocytes contained undetectable uPA by Western blotting (not shown).
Forms of uPA and α-granule proteins stored in day-13 QPD cultured megakaryocytes compared with platelets. Panels show proteins evaluated by Western blotting after SDS-PAGE. (A) Nonreduced (top panel) and reduced (bottom panel) uPA in QPD megakaryocytes (Q MK) compared with platelets (Q plt). Recombinant low-molecular-weight (LMW), single-chain (sc), and 2-chain (tc) uPA are shown for reference. High-molecular-weight complexes (* and **) and forms recognized by PAI-1 antibodies (*) are indicated. (B) TSP-1, VWF, and P-selectin in QPD megakaryocytes (Q MK), grown with (+) or without (−) plasma, compared with control (C plt) and QPD (Q plt) platelets.
Forms of uPA and α-granule proteins stored in day-13 QPD cultured megakaryocytes compared with platelets. Panels show proteins evaluated by Western blotting after SDS-PAGE. (A) Nonreduced (top panel) and reduced (bottom panel) uPA in QPD megakaryocytes (Q MK) compared with platelets (Q plt). Recombinant low-molecular-weight (LMW), single-chain (sc), and 2-chain (tc) uPA are shown for reference. High-molecular-weight complexes (* and **) and forms recognized by PAI-1 antibodies (*) are indicated. (B) TSP-1, VWF, and P-selectin in QPD megakaryocytes (Q MK), grown with (+) or without (−) plasma, compared with control (C plt) and QPD (Q plt) platelets.
Unlike the degraded α-granule proteins in QPD platelets, the TSP-1, VWF, and P-selectin in day-13 QPD cultured megakaryocytes, grown with or without added plasma, had the mobility of the normal forms in control platelets (Figure 5B) and megakaryocytes (not shown). QPD megakaryocytes also contained normal quantities of MMRN1, even when cultured in media with plasma (Table 1 and data not shown). Analyses of cultures with plasma, by Western blotting, indicated that there was no detectable conversion of plasminogen to plasmin in QPD megakaryocytes (not shown), unlike QPD platelets.
Discussion
The magnitude and timing of increased uPA transcription and storage during QPD megakaryopoiesis has been uncertain. Our study indicates the differentiation of QPD CD34+ progenitors into megakaryocytes results in differentiation-dependent, log-fold increases in uPA message and protein, without increasing VWF, vinculin, or CAMK2G expression (Table 1; Figure 1). Interestingly, the temporal increased production of uPA, during QPD megakaryopoiesis, mirrored the increased production of α-granule proteins, which were made in normal quantities (Table 1; Figure 1B) and costored with uPA within QPD α-granules (Figures 3,4; Table 2). We found that unlike QPD platelets, QPD megakaryocytes contained undegraded α-granule proteins and scuPA (Figure 5), which has low catalytic activity.26 These data, and the different distributions of plasminogen in cultured QPD megakaryocytes compared with platelets (Figure 4), suggest that the activation of uPA and proteolysis of α-granule proteins occur late, after plasminogen traffics into QPD α-granules and is converted to plasmin. Importantly, our study provides new evidence that profibrinolytic abnormalities of QPD platelets reflect increased expression of the uPA gene as hematopoietic progenitors differentiate into megakaryocytes, without altering expression of the flanking genes on chromosome 10 that encode vinculin and CAMK2G.
The expression of genes during hematopoietic stem cell differentiation into megakaryocytes is highly regulated and coordinated by transcription factors synthesized during megakaryopoiesis (for reviews, see Battinelli et al,27 Pang et al,28 and Chang et al29 ). This likely contributes to the sustained low levels of uPA expression, and increased expression of PAI-1 and other α-granule proteins during normal megakaryopoiesis,30 that accompany production of normal, antifibrinolytic platelets.6 PLAU is normally expressed by many different cell types,31 and we found that its transcription remains fairly stable during normal megakaryopoiesis. Further studies are required to determine whether QPD increases uPA expression when hematopoietic stem cells differentiate along other lineages. The mechanism that leads to increased PLAU expression during QPD megakaryopoiesis, without changing α-granule protein production, or the expression of VCL or CAMK2G (the genes that flank PLAU), is presently unknown. This selective increase suggests QPD results from a gain-of-function mutation in a binding site(s) for regulatory factors that increase the expression of genes encoding α-granule proteins and/or other platelet-restricted genes or that normally repress PLAU transcription during megakaryopoiesis. The transcription factors that increase gene expression during megakaryopoiesis include GATA-1; FOG-1; GATA-2; ETS family members ETS-1, FLI-1, TEL, and GABPα; RUNX-1; NF-E2; SCL; GFI-1B; and ZBP-89.27-29,32 In addition, c-MYB, ETS family member PU.1, and EKLF are negative regulators of megakaryocyte differentiation,29,33 and ETO-2 has recently been identified as a corepressor that binds the GATA-1–containing pentameric complex to prevent early expression of some genes expressed late during megakaryopoiesis, such as PF-4.34 The 2.5-kb upstream region of PLAU contains conserved elements that bind some of these transcription factors (eg, GATA, ETS, RUNX-1), and several other silencers and enhancers.35 Recently, QPD was linked to a cis regulatory defect in a 2-megabase region of chromosome 10 that includes PLAU, however, DNA sequencing excluded mutations within PLAU and its characterized regulatory elements, including the binding sites for GATA proteins, ETS family members, and RUNX-1.4 This suggests that QPD results from a mutation in more distal, uncharacterized regulatory element(s) of PLAU that binds one or more transcription factors to increase PLAU transcription during megakaryopoiesis, leading to the production of platelets enriched in stored uPA.
Heterogeneity has been noted in the contents of normal platelet α-granules,13,14 and we documented this in QPD platelets by rigorous, quantitative estimates of protein colocalization (Table 2), previously used to analyze other cell types.36,37 We identified that the increased production of uPA in QPD megakaryocytes results in uPA trafficking to the majority of α-granules, including the subpopulation of these granules that store proangiogenic (VEGF) and antiangiogenic (TSP-1) proteins,14 and the proteins fibrinogen and VWF.13 Immunoelectron microscopy confirmed uPA was stored in QPD α-granules (Figure 3), but we did not attempt to estimate its colocalization with other α-granule proteins by immunoelectron microscopy as the amounts of uPA contained in QPD platelets (< 400 ng/mg cellular protein) were challenging to detect. These findings, and the extensive costorage of uPA and plasminogen in QPD platelet α-granules, offer an explanation for the diversity of the secretory proteins degraded within QPD platelets. The degree to which individual α-granule proteins colocalize with uPA and plasminogen likely influences their degradation in QPD, although this is difficult to formally evaluate as the proteolysis of some QPD α-granule proteins (eg, MMRN1 and factor V)10-12 limits their detection in platelets. At present, the sorting mechanisms that lead to heterogeneous platelet α-granule protein contents are uncertain but could be influenced by the timing of protein production, endocytosis, and also homotypic and heterotypic binding interactions.13,14,38
The exposure of plasminogen to scuPA is known to generate plasmin.26 Our current study provides indirect evidence that the uptake of plasminogen for costorage with uPA is required to generate sufficient plasmin to trigger α-granule protein degradation in QPD, as the trafficking of plasminogen to forming α-granules was not recapitulated in cultures with or without added plasma. The mechanism of plasminogen uptake into α-granules is unknown, although bulk transport or receptor-mediated endocytosis (as demonstrated for other proteins)39-43 seems plausible given that plasminogen production by cultured megakaryocytes was undetectable. In vivo, the uptake of plasma proteins (eg, fibrinogen) into α-granules occurs late during megakaryopoiesis.42-44 It is possible that plasminogen and uPA costorage, and intra–α-granular plasmin generation, occurs in the bone marrow environment in QPD, where maturing megakaryocytes are exposed to plasma proteins throughout megakaryopoiesis. More definitive proof for a key role of platelet plasminogen in triggering QPD α-granule protein degradation might come from crossing plasminogen-deficient mice45 with mice that have QPD-like platelet α-granule protein degradation from overexpression of uPA in megakaryocytes.46
Our current study establishes that overexpression of uPA in QPD emerges as QPD hematopoietic progenitors differentiate into megakaryocytes without increasing expression of the flanking genes or the production of other α-granule proteins. An important next step will be to identify the QPD mutation that switches the pattern of uPA expression during megakaryopoiesis from sustained low levels to the dramatic (log fold) increased expression typical of an α-granule protein.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ms Francine Derome for sample collections, Mr Jean-Marc Massé for electron microscopy, and Dr Kalathil Suresh for help with planning RT-qPCR experiments.
This work was supported by grant T5888 from Heart and Stroke Foundation of Ontario, Toronto, ON (C.P.M.H.) and Bayer Canada, Toronto, ON (G.E.R.). D.K.V. is the recipient of a Canadian Institute of Health Research/Heart and Stroke Foundation of Canada Focus on Stroke Doctoral Research Award, Ottawa, ON. M.D. is the recipient of an Ontario Graduate Student Scholarship, Thunder Bay, ON. C.P.M.H. is the recipient of a Career Investigator Award from Heart and Stroke Foundation of Ontario, Toronto, ON, and a Canada Research Chair in Molecular Hemostasis from the Government of Canada, Ottawa, ON.
Authorship
Contribution: D.K.V. recruited subjects, designed and performed experiments, interpreted results, and wrote the paper; G.E.R. recruited subjects and participated in writing of the paper; M.D. and J.B. did experimental work and participated in writing of the paper; E.M.C.-B. designed experiments, interpreted results, and participated in writing of the paper; and C.P.M.H. supervised the project, designed experiments, interpreted results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine P. M. Hayward, McMaster University Health Sciences Centre, Room 2N30, 1200 Main Street West, Hamilton, Ontario, L8N 3Z5 Canada; e-mail: haywrdc@mcmaster.ca.