Abstract
Quebec platelet disorder (QPD) is an autosomal dominant disorder with high penetrance that is associated with increased risks for bleeding. The hallmark of QPD is a gain-of-function defect in fibrinolysis due to increased platelet content of urokinase plasminogen activator (uPA) without systemic fibrinolysis. We hypothesized that increased expression of uPA by differentiating QPD megakaryocytes is linked to PLAU. Genetic marker analyses indicated that QPD was significantly linked to a 2-Mb region on chromosome 10q containing PLAU with a maximum multipoint logarithm of the odds (LOD) score of +11 between markers D10S1432 and D10S1136. Analysis of PLAU by sequencing and Southern blotting excluded mutations within PLAU and its known regulatory elements as the cause of QPD. Analyses of uPA mRNA indicated that QPD distinctly increased transcript levels of the linked PLAU allele with megakaryocyte differentiation. These findings implicate a mutation in an uncharacterized cis element near PLAU as the cause of QPD.
Introduction
Most inherited bleeding disorders result from genetic defects that reduce hemostatic protein expression, secretion, or function.1 Inherited conditions that cause bleeding by increasing gene expression are uncommon and include Quebec platelet disorder (QPD),2,3 an autosomal dominant disorder with high and possibly complete penetrance4 and a prevalence in Quebec of 1:300 000.4 QPD is associated with a unique gain-of-function abnormality in fibrinolysis due to increased platelet stores of urokinase plasminogen activator (uPA) without systemic fibrinolysis or increased uPA in plasma,2,5 urine,6 or CD34+ hematopoietic progenitors.7 QPD increases risks for a number of bleeding symptoms, including delayed-onset bleeding after hemostatic challenges that responds only to fibrinolytic inhibitor therapy.4 Diagnostic tests for QPD include assays for increased platelet uPA and α-granule protein degradation from intraplatelet plasmin generation.2,4
We considered PLAU, the uPA gene on chromosome 10q24, as a candidate gene for QPD as QPD selectively increases (about 100-fold) PLAU expression during megakaryocyte differentiation.7 In addition, increased PLAU expression in mouse megakaryocytes results in a QPD-like disorder characterized by abnormal bleeding that responds to fibrinolytic inhibitor therapy, stored platelet protein degradation, and accelerated lysis of thrombi.8 PLAU contains 11 exons and 10 introns, and its conserved regulatory elements include a 3′ untranslated region (UTR) that affects mRNA stability (reviewed in Diamandis et al2 ; Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) and a 2.5-kb upstream region that is known to bind transcription factors produced during megakaryopoiesis (reviewed in Kaushansky9 ). These observations led us to investigate PLAU as a candidate gene for QPD.
Methods
This study was carried out in accordance with the Helsinki Protocol for research on human subjects and institutional ethics review board approval from Hamilton Health Sciences, McMaster University, and Centre Hôspitalier Universitaire Sainte Justine. Participants were 28 affected and 110 unaffected individuals who were descendants of a common QPD ancestor,4 and 110 healthy unrelated controls. Subjects provided written informed consent; if under 18 years of age, parental consent was obtained. DNA was collected from all subjects for linkage analysis, DNA sequencing, and Southern blotting. PLAU allelic expression was assessed using RNA harvested from platelets, peripheral blood CD34+ cells, and saliva cells of selected individuals (4 with QPD and 5 controls).
A detailed description of materials and methods is provided in Document S1.
Results and discussion
The 41 individuals selected for initial genotyping of chromosome 10 microsatellites were predicted to be informative, with estimated average (maximum) logarithm of the odds (LOD) score of +8.4 (+10.9) compared with 10.0 (14.1) for all family members. Genotyping of the 41 subjects (Table 1) indicated significant linkage of GATA121A08 and D10S1432 to QPD (respective LOD scores, +3.6 at θ = 0.05 and +3.3 at θ = 0.1; data not shown). Genotyping of all QPD family members, for these and additional markers (Table 1), confirmed that the region containing PLAU was strongly linked to QPD (LOD scores of +3.0 to +8.4), with a common haplotype in individuals with QPD.
Two-point linkage analysis of chromosome 10 microsatellite markers and SNPs
| Marker name . | Rutgers position . | Build 36.1 position . | No. of QPD family members genotyped . | LOD score at φ (recombination fraction) . | ||||
|---|---|---|---|---|---|---|---|---|
| 0.00 . | 0.01 . | 0.05 . | 0.1 . | 0.2 . | ||||
| D10S1453* | 6 615 628 | 41 | −∞ | −16.04 | −7.74 | −4.26 | −1.35 | |
| D10S189* | 19.62 | 6 761 879 | 41 | −∞ | −4.69 | −2.98 | −2.11 | −1.05 |
| D10S2325* | 31.35 | 12 832 951 | 41 | −∞ | −2.52 | 0.07 | 0.93 | 1.23 |
| D10S1423* | 44.78 | 19 477 909 | 41 | −∞ | −2.98 | 0.09 | 1.06 | 1.46 |
| D10S1426 | 57.44 | 30 535 790 | 41 | −∞ | −1.99 | 1.05 | 1.94 | 2.12 |
| D10S1208* | 62.04 | 35 297 637 | 41 | −∞ | −5.2 | −0.25 | 1.33 | 1.97 |
| D10S1221* | 57 199 892 | 41 | −∞ | −10.04 | −4.71 | -2.6 | −0.83 | |
| D10S1225 | 80.04 | 64 425 004 | 41 | −∞ | −0.92 | 0.31 | 0.65 | 0.66 |
| GATA121A08† | 85.32 | 70 182 554 | 138 | −∞ | 4.39 | 4.55 | 4.17 | 3.14 |
| rs12356915* | 73 931 323 | 138 | −2.41 | −2.15 | −1.14 | −0.66 | −0.28 | |
| D10S1432† | 93.7 | 74 329 402 | 138 | −∞ | 4.69 | 6.65 | 6.75 | 5.56 |
| D10S188† | 93.7 | 75 075 749 | 138 | −∞ | 2.98 | 3.23 | 2.95 | 2.11 |
| D10S1737† | 93.7 | 75 259 301 | 138 | −∞ | 1.06 | 3.75 | 4.2 | 3.42 |
| QPD-1† | 75 327 173 | 138 | 11.79 | 11.59 | 10.71 | 9.5 | 6.8 | |
| PLAU | 75 340 896 | |||||||
| rs4065 | 93.7 | 75 346 470 | 138 | 0.76 | 0.72 | 0.59 | 0.45 | 0.23 |
| rs1908339† | 75 513 106 | 138 | 2.88 | 3.0 | 3.0 | 2.72 | 1.97 | |
| D10S535† | 93.7 | 76 010 712 | 138 | −∞ | 1.64 | 3.15 | 3.31 | 2.69 |
| rs7899137† | 76 338 468 | 138 | 3.0 | 2.86 | 2.58 | 2.27 | 1.62 | |
| D10S556 | 94.38 | 76 922 909 | 138 | −∞ | 1.38 | 2.33 | 2.37 | 1.81 |
| D10S195* | 94.38 | 77 289 176 | 138 | −∞ | −3.38 | −1.02 | −0.15 | 0.4 |
| D10S1136† | 95.77 | 78 009 813 | 138 | −∞ | 7.83 | 8.42 | 7.94 | 6.17 |
| D10S1730 | 96.85 | 78 600 935 | 41 | 0 | −0.4 | 1.69 | 2.03 | 1.86 |
| D10S2327* | 98.55 | 80 382 014 | 41 | −11.38 | −3.88 | −1.49 | −0.56 | 0.06 |
| D10S1419 | 92 354 576 | 41 | −∞ | −1.3 | −0.06 | 0.32 | 0.43 | |
| D10S677* | 114.71 | 95 954 428 | 41 | −∞ | −8.1 | −2.33 | −0.29 | 0.98 |
| D10S1239* | 121.96 | 103 186 224 | 41 | −∞ | −3.5 | −0.51 | 0.44 | 0.87 |
| D10S1237 | 133.64 | 116 109 968 | 41 | −∞ | 1.04 | 2.09 | 2.22 | 1.83 |
| D10S1230* | 145.08 | 122 732 623 | 41 | −∞ | −13.29 | −6.56 | −3.9 | −1.59 |
| D10S1213 | 150.11 | 125 396 627 | 41 | 0.26 | 0.24 | 0.15 | 0.06 | −0.02 |
| D10S1248* | 166.09 | 130 982 451 | 41 | −∞ | −7.09 | −3.82 | −2.06 | −0.53 |
| D10S212* | 174.19 | 134 299 489 | 41 | −8.49 | −2.61 | −0.27 | 0.51 | 0.85 |
| Marker name . | Rutgers position . | Build 36.1 position . | No. of QPD family members genotyped . | LOD score at φ (recombination fraction) . | ||||
|---|---|---|---|---|---|---|---|---|
| 0.00 . | 0.01 . | 0.05 . | 0.1 . | 0.2 . | ||||
| D10S1453* | 6 615 628 | 41 | −∞ | −16.04 | −7.74 | −4.26 | −1.35 | |
| D10S189* | 19.62 | 6 761 879 | 41 | −∞ | −4.69 | −2.98 | −2.11 | −1.05 |
| D10S2325* | 31.35 | 12 832 951 | 41 | −∞ | −2.52 | 0.07 | 0.93 | 1.23 |
| D10S1423* | 44.78 | 19 477 909 | 41 | −∞ | −2.98 | 0.09 | 1.06 | 1.46 |
| D10S1426 | 57.44 | 30 535 790 | 41 | −∞ | −1.99 | 1.05 | 1.94 | 2.12 |
| D10S1208* | 62.04 | 35 297 637 | 41 | −∞ | −5.2 | −0.25 | 1.33 | 1.97 |
| D10S1221* | 57 199 892 | 41 | −∞ | −10.04 | −4.71 | -2.6 | −0.83 | |
| D10S1225 | 80.04 | 64 425 004 | 41 | −∞ | −0.92 | 0.31 | 0.65 | 0.66 |
| GATA121A08† | 85.32 | 70 182 554 | 138 | −∞ | 4.39 | 4.55 | 4.17 | 3.14 |
| rs12356915* | 73 931 323 | 138 | −2.41 | −2.15 | −1.14 | −0.66 | −0.28 | |
| D10S1432† | 93.7 | 74 329 402 | 138 | −∞ | 4.69 | 6.65 | 6.75 | 5.56 |
| D10S188† | 93.7 | 75 075 749 | 138 | −∞ | 2.98 | 3.23 | 2.95 | 2.11 |
| D10S1737† | 93.7 | 75 259 301 | 138 | −∞ | 1.06 | 3.75 | 4.2 | 3.42 |
| QPD-1† | 75 327 173 | 138 | 11.79 | 11.59 | 10.71 | 9.5 | 6.8 | |
| PLAU | 75 340 896 | |||||||
| rs4065 | 93.7 | 75 346 470 | 138 | 0.76 | 0.72 | 0.59 | 0.45 | 0.23 |
| rs1908339† | 75 513 106 | 138 | 2.88 | 3.0 | 3.0 | 2.72 | 1.97 | |
| D10S535† | 93.7 | 76 010 712 | 138 | −∞ | 1.64 | 3.15 | 3.31 | 2.69 |
| rs7899137† | 76 338 468 | 138 | 3.0 | 2.86 | 2.58 | 2.27 | 1.62 | |
| D10S556 | 94.38 | 76 922 909 | 138 | −∞ | 1.38 | 2.33 | 2.37 | 1.81 |
| D10S195* | 94.38 | 77 289 176 | 138 | −∞ | −3.38 | −1.02 | −0.15 | 0.4 |
| D10S1136† | 95.77 | 78 009 813 | 138 | −∞ | 7.83 | 8.42 | 7.94 | 6.17 |
| D10S1730 | 96.85 | 78 600 935 | 41 | 0 | −0.4 | 1.69 | 2.03 | 1.86 |
| D10S2327* | 98.55 | 80 382 014 | 41 | −11.38 | −3.88 | −1.49 | −0.56 | 0.06 |
| D10S1419 | 92 354 576 | 41 | −∞ | −1.3 | −0.06 | 0.32 | 0.43 | |
| D10S677* | 114.71 | 95 954 428 | 41 | −∞ | −8.1 | −2.33 | −0.29 | 0.98 |
| D10S1239* | 121.96 | 103 186 224 | 41 | −∞ | −3.5 | −0.51 | 0.44 | 0.87 |
| D10S1237 | 133.64 | 116 109 968 | 41 | −∞ | 1.04 | 2.09 | 2.22 | 1.83 |
| D10S1230* | 145.08 | 122 732 623 | 41 | −∞ | −13.29 | −6.56 | −3.9 | −1.59 |
| D10S1213 | 150.11 | 125 396 627 | 41 | 0.26 | 0.24 | 0.15 | 0.06 | −0.02 |
| D10S1248* | 166.09 | 130 982 451 | 41 | −∞ | −7.09 | −3.82 | −2.06 | −0.53 |
| D10S212* | 174.19 | 134 299 489 | 41 | −8.49 | −2.61 | −0.27 | 0.51 | 0.85 |
Markers were evaluated for 41 selected individuals or all 138 QPD family members, as indicated. SNPs are designated by rs numbers, except for the newly identified SNP, QPD-1. PLAU indicates the location of the urokinase plasminogen activator gene. Rutgers map positions are in centimorgans, except for 6 markers whose genetic map positions are unknown. Build 36.1 positions indicate physical distances on chromosome 10.
Markers excluding linkage.
Markers giving the best evidence for linkage.
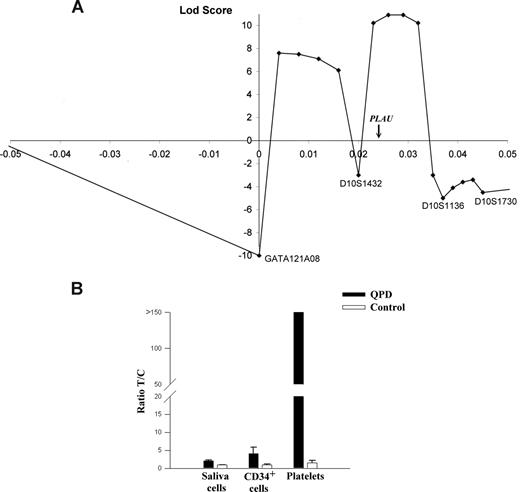
Multipoint linkage analysis with the markers closest to PLAU (Figure 1A) indicated that the most likely site for the disease gene was near PLAU (maximal LOD score of +11). Data for D10S195 excluded part of this region, and defined a 2-Mb region of chromosome 10q as the most likely location for the QPD mutation. This region contains 22 additional genes (Table S1), including 2 transcription factors (HSGT1 and MYST4) that are not expressed by megakaryocytes or known to regulate PLAU.10
Multipoint linkage and allelic expression analysis of PLAU in QPD. (A) Results of multipoint linkage using 4 microsatellite markers (positions are shown relative to GATA121A08). The genetic location of PLAU between markers D10S1432 and D10S1136 is indicated on the x-axis, which is the region with the highest LOD score for this analysis (+11). (B) Graph of reverse transcriptase–quantitative PCR (RT-qPCR) analyses of PLAU alleles for SNP rs4065 in different tissues and indicates the measured ratios of T/C alleles in saliva cells, CD34+ cells, and platelets in samples from 5 controls and 4 individuals with QPD. T/C ratios were significantly different for QPD compared with control samples for all tissues (P = .016).
Multipoint linkage and allelic expression analysis of PLAU in QPD. (A) Results of multipoint linkage using 4 microsatellite markers (positions are shown relative to GATA121A08). The genetic location of PLAU between markers D10S1432 and D10S1136 is indicated on the x-axis, which is the region with the highest LOD score for this analysis (+11). (B) Graph of reverse transcriptase–quantitative PCR (RT-qPCR) analyses of PLAU alleles for SNP rs4065 in different tissues and indicates the measured ratios of T/C alleles in saliva cells, CD34+ cells, and platelets in samples from 5 controls and 4 individuals with QPD. T/C ratios were significantly different for QPD compared with control samples for all tissues (P = .016).
At 13.7 kb upstream of the PLAU transcription start site, we identified a potentially informative marker for QPD (designated QPD-1)—a single nucleotide polymorphism (SNP; T/G; Build 36.1 position: 75 327 173). Genotypes of all 28 affected and 4 of 110 unaffected QPD family members were T/G, whereas other unaffected family members were T/T. Among 105 unrelated French-Canadians, the G allele was uncommon (3 individuals, T/G; 102, T/T). SNP QPD-1 was strongly linked with QPD (LOD score, +11.8; Table 1). This LOD score, and the maximal LOD score from multipoint analyses, were close to the maximum predicted by linkage simulations, excluding the possibility of another locus for QPD in this pedigree.
Southern blotting for a 25.2-kb region of chromosome 10 containing PLAU and its known regulatory elements excluded major alterations from QPD. DNA sequencing excluded the possibility that QPD results from mutations in the exons, introns, and characterized 3′ and 5′ regulatory elements of PLAU, or in the entire 24 kb upstream of the PLAU transcription start site and 2 kb downstream of the 3′ UTR. Complete sequencing of both PLAU alleles for these regions was confirmed, as the overlapping polymerase chain reaction (PCR) products demonstrated SNP heterozygosity in at least 1 QPD subject (Table S2).
To test for cis or trans effects of QPD on PLAU expression, the expressed exon 11 SNP rs4065 (T/C; T allele linked to QPD) was studied in heterozygous individuals. Unlike control platelets (T/C ratio, 1.5), QPD platelets contained an abundance of the T-allele transcript (all ratios greater than 150; P = .016; Figure 1B). In QPD CD34+ cells and saliva cells (which contained normal amounts of uPA mRNA; P = .36), the abundance of the T-allele transcript was more similar to controls (approximately 4-fold and 2-fold increases, respectively, compared with controls; P = .016; Figure 1B). As sequencing excluded mutated PLAU mRNA stability elements, these findings indicated that QPD results from a cis regulatory defect that distinctly increases transcription of the linked PLAU allele during megakaryocyte differentiation.
Genetic disorders with altered tissue-specific patterns of gene transcription appear to be rare.11 We had anticipated that QPD might result from mutations in conserved, 3′ message stability elements or in the conserved, 2.5-kb, 5′ regulatory region of PLAU that binds hematopoietic transcription factors, and other silencers and enhancers (reviewed in Diamandis et al2 and Nagamine et al12 ). As these possibilities were fully excluded, the search for the cis regulatory mutation that causes QPD must now extend beyond PLAU to identify the unique sequence changes that markedly increase transcription of the linked PLAU allele during megakaryocyte differentiation. In some disorders, the causative mutation that alters transcription is quite far (5′ and 3′) from the dysregulated gene,13,14 and it can be in an unrelated, neighboring gene.15,16 Regulatory elements with cis or trans effects have been identified as far as 1 Mb away from the genes transcribed.17-20 Interestingly, the nature of cis mutations that alter gene transcription in other blood disorders (eg, thalassemia) include deletions within locus control regions21 and point mutations that create promoter-like sequences for binding transcription factors.22
Future identification of the mutation in the linked region of chromosome 10 that increases PLAU transcription in QPD may be helpful to develop improved QPD therapy that targets the cause of uPA overexpression rather than limiting the consequences. The information may also suggest novel ways to up-regulate PLAU expression to protect against arterial and venous thrombosis.8
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Dr Bhupinder Bharaj and Barry Eng are gratefully acknowledged for technical help.
This work was supported by Heart and Stroke Foundation of Ontario grant 5888 and Career Investigator Award (C.P.M.H.), Bayer Canada (G.E.R.), grants from Genome Canada and Ontario Research Development Challenge Fund (D.E.B.), Ontario Graduate Student Scholarship (M.D.), Canadian Institute of Health Research/Heart and Stroke Foundation of Canada Focus on Stroke Doctoral Research Award (D.K.V.), Canada Research Chairs in Molecular Hemostasis (C.P.M.H.), and the Genetics of Complex Diseases (A.D.P.).
Authorship
Contribution: M.D. recruited subjects, designed and performed experiments, interpreted results, and wrote the manuscript; A.D.P. participated in study design, data analysis, and writing of the manuscript; J.M.R., D.E.B., J.S.W., and G.E.R. contributed to study design, interpretation of results, and manuscript writing; D.K.V. and J.B. performed experimental work and contributed to manuscript writing; F.D. and G.E.R. recruited subjects; and C.P.M.H. supervised the project, designed experiments, interpreted results, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine P. M. Hayward, McMaster University Health Sciences Center, Room 2N30, 1200 Main Street West, Hamilton, Ontario, L8N 3Z5 Canada; e-mail: haywrdc@mcmaster.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal