Abstract
Understanding mechanisms controlling expression of the α-spectrin gene is important for understanding erythropoiesis, membrane biogenesis, and spectrin-linked hemolytic anemia. We showed previously that a minimal α-spectrin promoter directed low levels of expression only in early erythroid development, indicating elements outside the promoter are required for expression in adult erythrocytes. Addition of noncoding exon 1′ and intron 1′ conferred a 10-fold increase in activity in reporter gene assays. In this report, we used a transgenic mouse model to show that addition of exon 1′ and intron 1′ to the α-spectrin promoter conferred tissue-specific expression of a linked Aγ-globin gene in erythroid cells at all developmental stages. Expression was nearly position-independent, as 21 of 23 lines expressed the transgene, and γ-globin protein was present in 100% of erythrocytes, indicating uniform expression. Additional in vivo studies revealed that exon 1′ functions as an insulator with barrier-element activity. Chromatin immunoprecipitation assays demonstrated that this region was occupied by the upstream stimulatory factors 1/2 (USF1/USF2), similar to the well-characterized chicken HS4 insulator. These data identify the first barrier element described in an erythrocyte membrane protein gene and indicate that exon 1′ and intron 1′ are excellent candidate regions for mutations in patients with spectrin-linked hemolytic anemia.
Introduction
Spectrin is the major structural component of the erythrocyte membrane skeleton that maintains cellular shape, regulates the lateral mobility of integral membrane proteins, and provides structural support for the lipid bilayer.1 It is composed of 2 subunits, α- and β-spectrin, encoded by separate genes.1,2 Throughout erythropoiesis, there are significant changes in the synthesis, expression, and membrane assembly of spectrin. Early in erythropoiesis, α-spectrin is synthesized in great excess,3,4 a process controlled at the transcriptional level.3,5,6 The molecular mechanisms that regulate the erythroid tissue-specific and developmental stage-specific expression of α-spectrin, including the mechanisms that control the increase in α-spectrin gene transcription to high levels during the early stages of erythropoiesis, are unknown.
In the mature erythrocyte, quantitative and qualitative disorders of α-spectrin have been associated with inherited hemolytic anemias, including hereditary spherocytosis (HSp), hereditary elliptocytosis (HE), and hereditary pyropoikilocytosis (HPP).7-12 In most recessive HSp and many HPP patients, there is a defect in α-spectrin mRNA accumulation associated with spectrin deficiency.7,13,14 With a few rare exceptions, the cause of the defect in α-spectrin expression in erythrocytes of these patients is unknown, even after nucleotide sequence analysis of the exons corresponding to the α-spectrin coding region and the minimal promoter region.15,16
Our previous studies demonstrated that a minimal α-spectrin promoter directed low levels of expression only in the early stages of erythroid development, indicating elements outside the promoter are required for expression in adult erythroid cells.17 Additional studies identified a 183-bp region 3′ of the α-spectrin gene promoter composed of noncoding exon 1′ and intron 1′ of the α-spectrin gene, which directed high levels of expression in reporter gene/transfection assays.18 Both exon 1′ and intron 1′ in their proper genomic orientation relative to the α-spectrin minimal promoter were required for full activity in reporter gene assays, in part due to GATA-1–dependent positive expression directed by intron 1′. The chromatin in this 183-bp region contains a DNase I–hypersensitive site and exhibits hyperacetylation of histones H3 and H4.
We hypothesize that both cis sequences and trans factors modulate chromatin structure and control α-spectrin gene regulation. Identifying these elements will provide important insights into the role of α-spectrin synthesis in early erythropoiesis, the pathogenesis of decreased α-spectrin expression in spectrin-deficient patients with inherited hemolytic anemia, and may provide a tool to direct high-level, tissue-specific expression of other erythroid-specific genes in gene transfer applications. To address this hypothesis in vivo, we examined the 183-bp exon 1′ and intron 1′ region of the α-spectrin gene fragment in a transgenic mouse assay. When added to the minimal α-spectrin promoter, the region directed expression of a linked Aγ-globin gene in erythroid cells at all developmental stages. Both exon 1′ and intron 1′ were required for transgene expression in vivo. Additional in vivo studies revealed that exon 1′ functions as an insulator with barrier-element activity. Chromatin immunoprecipitation demonstrated binding of the upstream stimulatory factors 1/2 (USF1/USF2) in this region, similar to the chicken HS4 barrier element.19,20 These data identify the first barrier element described in an erythrocyte membrane protein gene and indicate this is an excellent candidate region for mutations in patients with spectrin-linked hemolytic anemia.
Methods
Preparation of α-spectrin gene promoter/Aγ-globin reporter transgenes
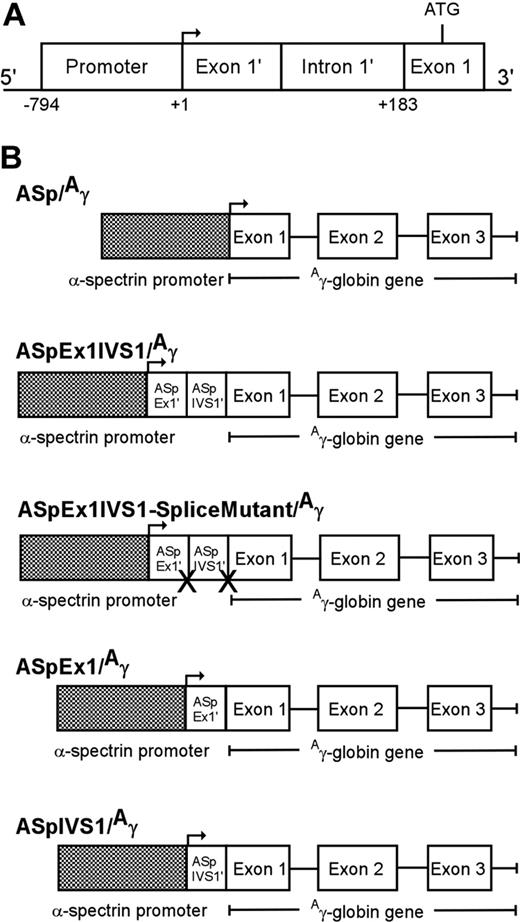
To generate α-spectrin/Aγ-globin transgenes, an 183-bp α-spectrin gene fragment encoding exon 1′ and intron 1′ was cloned between a 794-bp minimal α-spectrin gene promoter fragment (Figure 1A) and a 2266-bp fragment containing the human Aγ-globin gene to generate AspEx1IVS1/Aγ-globin (Figure 1B). This plasmid construct was sequenced to confirm fragment location and orientation. The 2877 α-spectrin promoter plus exon 1′ plus intron 1′/Aγ-globin gene fragment was excised from this plasmid with KpnI and HindIII for microinjection. Similarly, a 61-bp α-spectrin gene fragment encoding exon 1′ or a 122-bp α-spectrin fragment encoding intron 1′ was cloned between a 794-bp minimal α-spectrin gene promoter fragment and a 2266-bp fragment containing the human Aγ-globin gene, sequenced, and excised for microinjection (Figure 1B). Finally, the 183-bp α-spectrin gene fragment encoding exon 1′ and intron 1′ was manipulated via PCR mutagenesis to render the splice sites at the 5′ and 3′ ends of intron 1′ nonfunctional, AG:gt to AA:tt and AG:gt to TT:gt, respectively. Similar to wild-type, this mutant fragment was cloned between the minimal α-spectrin gene promoter and the human Aγ-globin gene to generate AspEx1IVS1-splice mutant/Aγ-globin (Figure 1B). Sequence integrity, position, and orientation of all transgenes were confirmed prior to excision and microinjection.
Generation of transgenic mice
Transgenic mice were generated as described by Hogan et al.21,22 Founder animals were identified by Southern blot analysis of DNA extracted from tail biopsies by probing with α-spectrin/Aγ-globin probe. Founder animals were crossed to FVB/N mice for propagation. Copy number determination and statistical analyses were performed as described.21,23 The Animal Care and Use committees of Yale University and the National Institutes of Health (NIH) have approved the animal studies described in this report.
Expression of human γ-globin mRNA and protein in erythrocytes of transgenic mice
Total cellular RNA was extracted from adult reticulocytes, obtained by collecting 200 μL of blood from phlebotomized animals, using TRIzol reagent (Invitrogen, Carlsbad, CA).
Three different riboprobes were used to analyze mRNA reporter gene expression in ribonuclease protection assays (RPA). Riboprobe 1 contains sequences for both the human Aγ-globin gene and the murine α2-globin gene.23 This riboprobe ensures that both the human Aγ-globin and murine α-globin sequences are labeled to equal specific activity, allowing direct comparison of human Aγ-globin and murine α-globin mRNA levels. The dual riboprobe α2-globin gene fragment, derived from a SKIVE/EiJ strain, contains a single nucleotide polymorphism that leads to detection of 2 α-globin bands in RPA analyses in FVB/N mice. Riboprobe 2 includes exon 2 of the human Aγ-globin gene and riboprobe 3 contains exon 2 of the murine α2-globin gene. Riboprobes 2 and 3 were always used in combination. Linear DNA templates for the RNase protection assay were prepared by BglII digestion of the dual riboprobe, EcoRI digestion of the Aγ-globin riboprobe, or HindIII digestion of the murine α-globin riboprobe. Templates were purified by agarose gel electrophoresis and a Geneclean II Kit (Bio101, Vista, CA).
32P-labeled RNA probes were transcribed using the MAXIscript In Vitro Transcription kit (Ambion, Austin, TX). Probe-RNA (0.1 to 0.25 μg) hybridization was carried out overnight according to standard procedures (RPA II; Ambion). RNase digestion was performed using an RNase A/RNase T1 mixture and the protected fragments separated on an 8% nondenaturing polyacrylamide gel. To quantitate levels of mRNA, the gel was exposed to a PhosphorImager screen and scanned on a Molecular Dynamics PhosphorImager (Amersham Pharmacia Biotech, Chalfont St Giles, United Kingdom). At least 3 independent RPA reactions were performed on each RNA sample followed by scanning quantitation. The relative amounts of the bands (ie, human Aγ-globin and mouse α-globin) were estimated by the following formula: (Aγ-globin RNA/transgene copy number) × (1/mouse α-globin RNA).
Detection and measurement of γ-globin protein in erythrocytes was performed as described by Thorpe et al.24
Insulator assays
Barrier-element activity was analyzed in a gene-silencing/position-effect assay.25,26 Test plasmids containing the enhanced green fluorescent protein (EGFP) as reporter gene were cotransfected into K562 cells with a pRSV-neomycin plasmid. After selection, individual clones were isolated and expanded in G418-containing medium and switched to nonselective medium before analysis of GFP expression via fluorescence-activated cell sorting (FACS). Southern blot analysis was used to confirm selected clones had an intact GFP construct. The negative control plasmid, HS2-β-GFP, consisted of the mouse β-globin HS2 enhancer, the human β-globin gene promoter, and the EGFP reporter gene. The positive control plasmid consisted of the HS2-β-GFP cassette flanked with hypersensitive site 4 from the chicken β-globin cluster locus control region (cHS4) insulator element.
Enhancer blocking activity was analyzed in a well-defined assay where elements under study are assessed for their ability to act as an enhancer blocker when placed between an enhancer and a promoter.27 α-Spectrin sequences were used to separate the mouse β-globin HS2 enhancer28 from a γ-globin promoter linked to a neomycin resistance gene. In the positive control plasmid, cHS4 sequences separated the HS2 enhancer from the γ-globin promoter-neomycin cassette. Plasmids were cotransfected with a neomycin resistance gene into K562 cells, plated in semisolid medium to allow growth of individual clones, and cultured under G418 selection for 16 weeks. Colony number was normalized to the number of colonies promoted by a HS2 γ-globin neomycin plasmid.
Quantitative chromatin immunoprecipitation assay
Anti-CTCF (07-729) antibody was obtained from Upstate Biotechnology (Lake Placid, NY). Anti-USF1 (H-86, sc-8983) and USF2 (C-20, sc-862) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Chromatin immunoprecipitation (ChIP) analyses were performed as described.18 After elution and extraction, immunoprecipitated DNA was analyzed by quantitative real-time PCR (iCycler; Bio-Rad, Hercules, CA) using primers shown in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Samples from at least 3 independent immunoprecipitations were each analyzed at least in triplicate, as described.18 The amount of product was determined relative to a standard curve of input chromatin. Amplification of a single product was confirmed by dissociation curve analysis and agarose gel electrophoresis with ethidium bromide staining. Parallel controls for each experiment included samples of no chromatin, no antibody, and nonimmune rabbit immunoglobulin G (IgG). Positive controls for USF and CTCF were also performed.29-31
Electrophoretic mobility shift assays
Results
Transgenic mice express a minimal α-spectrin promoter-exon 1′-intron-1′/Aγ-globin transgene in reticulocytes
In previous transgenic mouse studies, the human α-spectrin gene promoter directed expression of a linked Aγ-globin reporter gene only in early stages of erythroid development, with no expression in reticulocytes of lines transmitting the ASp/γ transgene (Table S3). These results indicated that elements outside the minimal α-spectrin gene promoter are required for expression in adult erythroid cells.17 Our previous studies identified a region 3′ of the core α-spectrin gene promoter, encoding exon 1′ and intron 1′, which directed high-level expression in reporter gene/transfection assays (Figure 1A). We created transgenic mice with a minimal human α-spectrin gene promoter fragment plus the region encoding exon 1′ and intron 1′ linked to a human Aγ-globin reporter gene (ASpEx1IVS1/Aγ, Figure 1B). To assess transgene expression, we used either a dual riboprobe that detects sequences from both the human γ-globin gene and the murine α-globin gene23 and/or a set of riboprobes with one detecting exon 2 of the human Aγ-globin gene and the other detecting exon 2 of the murine α2-globin gene. Twenty-three transgenic lines containing the ASpEx1IVS1/Aγ transgene were analyzed.
Human α-spectrin gene/Aγ-globin transgenes. (A) A map of the erythrocyte α-spectrin promoter region. The minimal promoter (−794 to +1), untranslated exon 1′ (exon 1′), and intron 1′ (IVS1′), which is spliced out in 50% of α-spectrin transcripts, are shown. (B) A human α-spectrin (ASp) fragment containing the minimal α-spectrin promoter with or without downstream regions (ASp/Aγ), exon 1′ + IVS 1 (ASpEx1IVS1/Aγ), exon 1′ alone (ASpEx1/Aγ), or IVS 1 alone (ASpIVS1/Aγ), was fused to the human Aγ-globin gene to create the transgene constructs shown. One transgene, ASp promoter-exon 1-IVS1-splice mutant/Aγ-globin-splice mutant (ASpEx1IVS1 splice mutant/Aγ) was created with the conserved splice junction sequences of intron 1′ abolished.
Human α-spectrin gene/Aγ-globin transgenes. (A) A map of the erythrocyte α-spectrin promoter region. The minimal promoter (−794 to +1), untranslated exon 1′ (exon 1′), and intron 1′ (IVS1′), which is spliced out in 50% of α-spectrin transcripts, are shown. (B) A human α-spectrin (ASp) fragment containing the minimal α-spectrin promoter with or without downstream regions (ASp/Aγ), exon 1′ + IVS 1 (ASpEx1IVS1/Aγ), exon 1′ alone (ASpEx1/Aγ), or IVS 1 alone (ASpIVS1/Aγ), was fused to the human Aγ-globin gene to create the transgene constructs shown. One transgene, ASp promoter-exon 1-IVS1-splice mutant/Aγ-globin-splice mutant (ASpEx1IVS1 splice mutant/Aγ) was created with the conserved splice junction sequences of intron 1′ abolished.
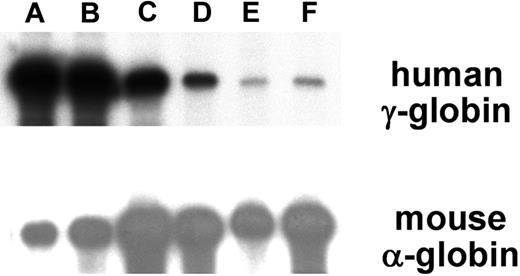
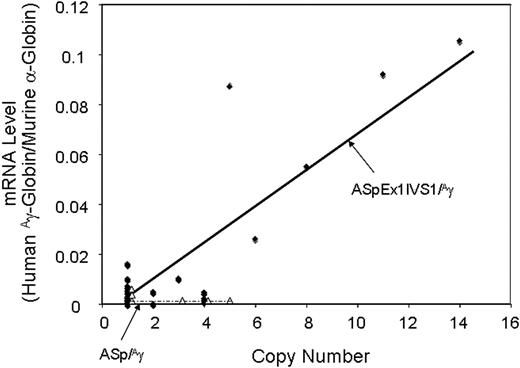
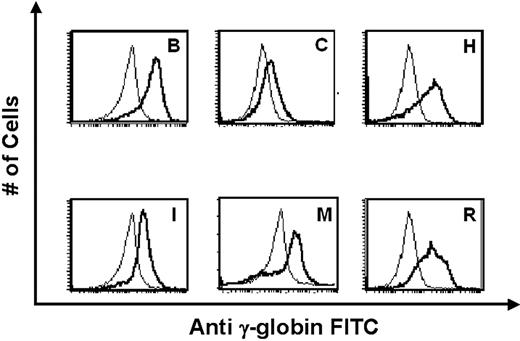
RNase protection demonstrated that 21 of 23 ASpEx1IVS1/Aγ-transgenic lines expressed detectable levels of mRNA from the linked Aγ-globin gene in adult erythroid cells (reticulocytes), (Figure 2, Tables 1, S3), indicating nearly position-independent expression. ASpEx1IVS1/Aγ-transgenic mice had mean levels of Aγ-globin mRNA of approximately 0.42% per mouse α-globin mRNA/transgene copy (Table 1, Table S3). The number of transgenes in each line was estimated by Southern blot analyses to be between 1 and 14 copies per expressing animal. Mice with the ASpEx1IVS1/Aγ transgene expressed the Aγ-globin transgene in a copy number-dependent fashion (copy number correlation with mRNA level r2 = 0.7659, P < .001 for all 23 strains, linear relationship, Figure 3, Tables 1, S3). To determine the distribution of Aγ-globin protein in erythrocytes of transgenic animals, we used an anti-human γ-globin monoclonal antibody for FACS analyses. ASpEx1IVS1/Aγ-transgenic mice expressed human γ-globin in a uniform pattern (ie, in 100% of erythrocytes; Figure 4).
Detection of α-spectrin gene/Aγ-globin mRNA in reticulocytes of transgenic mice. RNA from adult reticulocytes was hybridized to 32P-labeled antisense riboprobes for the human Aγ-globin gene (top panel) and the mouse α-globin gene (bottom panel) and digested with RNase. Protected fragments were separated by polyacrylamide gel electrophoresis followed by radiography. Representative transgenic lines are shown, (A-F) with the letters above the lanes corresponding to Table S3. Autoradiography time of the exposure shown was 20 hours for the γ-globin probe and 5 hours for the α-globin probe, respectively.
Detection of α-spectrin gene/Aγ-globin mRNA in reticulocytes of transgenic mice. RNA from adult reticulocytes was hybridized to 32P-labeled antisense riboprobes for the human Aγ-globin gene (top panel) and the mouse α-globin gene (bottom panel) and digested with RNase. Protected fragments were separated by polyacrylamide gel electrophoresis followed by radiography. Representative transgenic lines are shown, (A-F) with the letters above the lanes corresponding to Table S3. Autoradiography time of the exposure shown was 20 hours for the γ-globin probe and 5 hours for the α-globin probe, respectively.
Expression of α-spectrin/Aγ-globin mRNA in transgenic mice
| Transgene construct . | No. of expressing lines/total no. of lines . | Copy number range . | Copy number correlation . | Mean human Aγ-globin/murine α-globin/copy . |
|---|---|---|---|---|
| ASp/γ | 0/7 | 1-5 | Not detected | |
| ASpEx1IVS1/γ | 21/23 | 1-14 | r2 = 0.48 | 0.0042 + 0.004 |
| P = .001 | P = .047* | |||
| ASpEx1IVS1 splice mutant/γ | 9/9 | 1-10, 45 | r2 = 0.963 | 0.0047 + 0.003 |
| P = .001 | P = .002* | |||
| ASpIVS1/γ | 4/6 | 1-6 | 0.0055 + 0.0067 | |
| ASpEx1/γ | 2/7 | 1-15 | 0.001 + 0.0006 |
| Transgene construct . | No. of expressing lines/total no. of lines . | Copy number range . | Copy number correlation . | Mean human Aγ-globin/murine α-globin/copy . |
|---|---|---|---|---|
| ASp/γ | 0/7 | 1-5 | Not detected | |
| ASpEx1IVS1/γ | 21/23 | 1-14 | r2 = 0.48 | 0.0042 + 0.004 |
| P = .001 | P = .047* | |||
| ASpEx1IVS1 splice mutant/γ | 9/9 | 1-10, 45 | r2 = 0.963 | 0.0047 + 0.003 |
| P = .001 | P = .002* | |||
| ASpIVS1/γ | 4/6 | 1-6 | 0.0055 + 0.0067 | |
| ASpEx1/γ | 2/7 | 1-15 | 0.001 + 0.0006 |
P values from comparison versus mean human Aγ-globin/murine α-globin/copy of ASp/γ-transgenic mice.
Correlation of transgene copy number with the levels of α-spectrin gene/Aγ-globin mRNA in ASpEx1IVS1/Aγ-transgenic mice. ●, Linear regression analysis of the transgene copy number with the corrected mRNA expression level was performed with ASpEx1IVS1/Aγ mice; △, ASp/Aγ mice. There is a linear relationship in ASpEx1IVS1/Aγ mice, indicating copy number–dependent expression (r2 = 0.7659, P < .001 for all 23 strains). The lines shown were generated via a “best fit” algorithm with each point weighted equally.
Correlation of transgene copy number with the levels of α-spectrin gene/Aγ-globin mRNA in ASpEx1IVS1/Aγ-transgenic mice. ●, Linear regression analysis of the transgene copy number with the corrected mRNA expression level was performed with ASpEx1IVS1/Aγ mice; △, ASp/Aγ mice. There is a linear relationship in ASpEx1IVS1/Aγ mice, indicating copy number–dependent expression (r2 = 0.7659, P < .001 for all 23 strains). The lines shown were generated via a “best fit” algorithm with each point weighted equally.
Expression of human γ-globin protein in erythrocytes of ASpEx1IVS1/Aγ-transgenic mice. Fluorescence intensity from a FITC-conjugated monoclonal antibody against human γ-globin was correlated with the number of erythrocytes counted in 6 representative transgenic lines chosen for high Aγ-globin mRNA expression. In each panel, the thick line represents a transgenic mouse and the thin line represents a nontransgenic, littermate control. The single peaks observed represent uniform expression of human γ-globin in these cells, whereas a bimodal peak represents nonuniform or variegated expression.23 Representative transgenic lines are shown, with the letters corresponding to Table S3.
Expression of human γ-globin protein in erythrocytes of ASpEx1IVS1/Aγ-transgenic mice. Fluorescence intensity from a FITC-conjugated monoclonal antibody against human γ-globin was correlated with the number of erythrocytes counted in 6 representative transgenic lines chosen for high Aγ-globin mRNA expression. In each panel, the thick line represents a transgenic mouse and the thin line represents a nontransgenic, littermate control. The single peaks observed represent uniform expression of human γ-globin in these cells, whereas a bimodal peak represents nonuniform or variegated expression.23 Representative transgenic lines are shown, with the letters corresponding to Table S3.
RNA was collected from 10.5 dpc (days postcoitum) of yolk sac–derived peripheral blood cells, 13.5 dpc of fetal livers, and adult reticulocytes from 3 ASpEx1IVS1/Aγ-transgenic lines. RNase protection analysis demonstrated that the transgene was expressed in yolk sac, fetal liver, and adult erythroid cells in all 3 lines analyzed. The average levels of γ-globin mRNA during development ranged between 0.05 and 0.75% that of the endogenous mouse α-globin per transgene copy (Table S4).
Three transgenic lines expressing the ASpEx1IVS1/Aγ transgene in reticulocytes were chosen for further analyses of expression in various tissues. In these 3 lines, RNase protection analysis of γ-globin mRNA levels was performed on RNA extracted from 10 different tissues after systemic perfusion with saline immediately prior to sacrificing and tissue harvest. RNase protection did not detect γ-globin mRNA in kidney, brain, heart, liver, lung, or skeletal muscle. Expression was detected in the hematopoietic tissues, spleen, bone marrow, and thymus (Table S5).
Splicing of intron 1′ is not required for expression of the α-spectrin promoter-exon 1′-intron-1′/Aγ-globin transgene in reticulocytes
The 183-bp region encodes exon 1′ and an alternately spliced intron 1′ (Figure 1A), which is excised in approximately 50% of α-spectrin mRNA transcripts.32 To test the hypothesis that alternate splicing of intron 1′ is responsible for the increase in expression due to transcriptional or posttranscriptional regulation of α-spectrin mRNA,33,34 the conserved splice junctions of intron 1′ were mutated in the context of the transgene to abolish its excision during splicing. ASpEx1IVS1–splice mutant/Aγ-transgenic mice demonstrated levels of Aγ-globin mRNA expression in reticulocytes in 9 of 9 lines at levels comparable to the wild type ASpEx1IVS1/Aγ transgene (Tables 1, S3). Reverse-transcription polymerase chain reaction of random-primed reticulocyte mRNA isolated from ASpEx1IVS1/Aγ and ASpEx1IVS1-splice mutant/Aγ-transgenic mice, using primers in exon 1′ of α-spectrin and exon 1 of Aγ-globin (Table S1), only a single product of 285 bp was obtained from both ASpEx1IVS1/Aγ and ASpEx1IVS1-splice mutant/Aγ-transgenic mouse RNA from 3 expressing lines (not shown). Sequence analysis demonstrated that intron 1′ was not spliced out of either transgene, a result that is not surprising, as the corresponding exon 2 and intron 2 sequences of the α-spectrin gene required for splicing are not present in the transgenes. Together, these data indicate that splicing of intron 1′ is not required for α-spectrin–directed Aγ-globin expression in reticulocytes of these transgenic mice, insuring that expression directed by the ASpEx1IVS1/Aγ transgene was not due to a functional link between transcription and splicing.35-37
Intron 1′ and exon 1′ are both required for expression in vivo
Our previous studies demonstrated that both exon 1′ and intron 1′ in their proper genomic orientation relative to the α-spectrin minimal promoter were required for full activity in reporter gene assays. This activity was predominantly due to GATA-1–dependent positive expression directed by intron 1′, while exon 1′ alone had no activity in reporter gene assays. We separated exon 1′ and intron 1′ and evaluated them in our transgenic mouse assay by linking the minimal human α-spectrin gene promoter was to either exon 1′ or intron 1′ and the Aγ-globin reporter gene to create ASpEx1/γ or ASpIVS1/γ, respectively. Four of 6 transgenic lines with the ASpIVS1/γ transgene expressed Aγ-globin mRNA at levels approximately comparable to the ASpEx1IVS1/Aγ transgene (Tables 1, S3). The other 2 lines transmitting the ASpIVS1/γ transgene were silenced and did not express Aγ-globin mRNA. Two of 7 transgenic lines with ASpEx1/γ transgene expressed Aγ-globin mRNA at low levels, whereas no expression was detected in the other 5 lines, similar to the minimal α-spectrin promoter alone (Tables 1, S3). These data confirm our previous studies that both exon 1′ and intron 1′ together in their proper genomic orientation relative to the α-spectrin minimal promoter are required for reporter gene expression in vivo.
α-Spectrin exon 1′ but not intron 1′ exhibits properties of an insulator with barrier-element activity
Insulators are DNA sequences and their associated binding proteins that establish and/or maintain the boundaries between euchromatin and heterochromatin. They are associated with DNase I–hypersensitive sites, often flank gene clusters or loci, or are tightly associated with gene promoters, but do not themselves cause an increase in the rate of transcription.38-43 One type of insulator creates a barrier to protect against heterochromatin-mediated gene silencing (barrier insulators), whereas another type of insulator establishes chromatin domains to separate enhancers and promoters and prevent their interaction (enhancer-blocking insulators).39 Some insulators perform both functions, such as the Drosophila Gypsy insulator and the well-characterized chicken 5′HS4 insulator element from the β-globin locus control region (cHS4).20,39,44-48
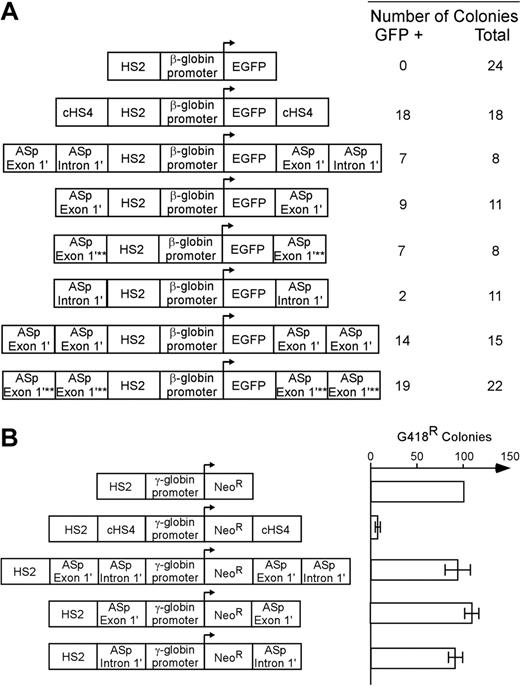
Flanking promoter/reporter genes with insulator elements that convey barrier activity in vivo prevents gene silencing and reduces position effect variegation (PEV; barrier function).25,49,50 We used a gene silencing/position-effect assay to determine whether the regions of the α-spectrin gene encoding exon 1′ and intron 1′ possess these barrier-element functions. A mouse β-globin HS2 enhancer/human β-globin gene promoter/enhanced green fluorescent protein reporter gene plasmid (HS2-β-GFP, negative control) was cotransfected with a neomycin resistance gene into K562 cells and cultured under G418 selection (Figures 5, S1).26,48 All 24 clones containing the HS2-β–GFP cassette were silenced within 21 days (Figure 5A). As shown previously,48 when the HS2-β–GFP cassette was flanked with the cHS4 insulator element as a positive control, 18 of 18 lines expressed GFP in 100% of cells for more than 16 weeks (Figure 5A; P < .001). When the HS2-β–GFP cassette was flanked with exon 1′ plus intron 1′ of the α-spectrin gene, 7 of 8 clones expressed GFP after 16 weeks (P < .001). When the HS2-β–GFP cassette was flanked with exon 1′ of the α-spectrin gene, or exon 1′ of the α-spectrin gene with a mutation that prevents GATA-1 binding, 10 of 12 and 7 of 8 clones, respectively, expressed GFP in 100% of cells for more than 16 weeks (P < .001; P < .001). When the HS2-β–GFP cassette was flanked with intron 1′ of the α-spectrin gene, only 2 of 11 clones expressed GFP after 16 weeks (P > .05). Flanking the HS2-β–GFP cassette with 2 copies of exon 1′ or 2 copies of exon 1′ with a mutant GATA-1 site, 14 of 15 and 19 of 22 clones, respectively, expressed GFP for more than 16 weeks (Figure 5A; P < .001, P < .001). These data indicate that exon 1′ of the α-spectrin gene functions as a barrier-element function in erythroid cells.
Insulator functions of the α-spectrin gene 183-bp exon 1′ + intron 1′ region. (A) Analysis of barrier-element function. The indicated constructs were cotransfected into K562 cells along with a pRSV-neomycin plasmid for selection of individual clones. Individual clones were isolated and expanded in G418 containing medium and switched to nonselective medium before analysis of GFP expression by FACS. The total number of clones analyzed and the number of GFP-expressing (+) clones are shown at the right. Southern blot analysis was used to confirm that all clones had an intact GFP construct. χ2 analysis of the observed versus expected number of GFP+ clones demonstrated that gene silencing was significantly inhibited (P < .001) for all test fragments except for α-spectrin intron 1′ (P > .05). Abbreviations are: HS2: Hypersensitive site 2 from the mouse β-globin cluster locus control region; EGFP: the coding sequence for the enhanced green fluorescent protein gene; cHS4: Hypersensitive site 4 from the chicken β-globin cluster locus control region; ASp: α-spectrin; asterisks denote mutation of the exon 1′ GATA site that disrupts GATA binding to DNA. (B) Analysis of enhancer blocking activity. The indicated constructs were transfected into K562 cells and plated in semisolid medium to allow growth of individual clones. The relative number of colonies was normalized to HS2 γ-Neo. The cHS4 insulator element was used as positive control. HS2, Hypersensitive site 2 from the mouse β-globin gene locus control region; γ, the human γ-globin promoter; Neo, the coding sequence for the neomycin-resistance gene. Error bars represent numbers of normalized colonies obtained from multiple experiments.
Insulator functions of the α-spectrin gene 183-bp exon 1′ + intron 1′ region. (A) Analysis of barrier-element function. The indicated constructs were cotransfected into K562 cells along with a pRSV-neomycin plasmid for selection of individual clones. Individual clones were isolated and expanded in G418 containing medium and switched to nonselective medium before analysis of GFP expression by FACS. The total number of clones analyzed and the number of GFP-expressing (+) clones are shown at the right. Southern blot analysis was used to confirm that all clones had an intact GFP construct. χ2 analysis of the observed versus expected number of GFP+ clones demonstrated that gene silencing was significantly inhibited (P < .001) for all test fragments except for α-spectrin intron 1′ (P > .05). Abbreviations are: HS2: Hypersensitive site 2 from the mouse β-globin cluster locus control region; EGFP: the coding sequence for the enhanced green fluorescent protein gene; cHS4: Hypersensitive site 4 from the chicken β-globin cluster locus control region; ASp: α-spectrin; asterisks denote mutation of the exon 1′ GATA site that disrupts GATA binding to DNA. (B) Analysis of enhancer blocking activity. The indicated constructs were transfected into K562 cells and plated in semisolid medium to allow growth of individual clones. The relative number of colonies was normalized to HS2 γ-Neo. The cHS4 insulator element was used as positive control. HS2, Hypersensitive site 2 from the mouse β-globin gene locus control region; γ, the human γ-globin promoter; Neo, the coding sequence for the neomycin-resistance gene. Error bars represent numbers of normalized colonies obtained from multiple experiments.
The other activity of insulator elements is the ability to act as an enhancer blocker when placed between an enhancer and a promoter.27 To determine whether exon 1′ and intron 1′ of the α-spectrin gene conferred this activity, these sequences were studied in a well-defined enhancer blocking assay. The α-spectrin sequences or cHS4 (positive control) were used to separate the mouse β-globin HS2 enhancer28 from a γ-globin promoter linked to a neomycin resistance gene. Plasmids were cotransfected with a neomycin resistance gene into K562 cells and cultured under G418 selection (Figure 5B). In the absence of an enhancer blocker (negative control), the HS2 enhancer promoted the growth of numerous G418-resistant colonies. The cHS4 element blocked this activity, as shown previously.51 None of the α-spectrin fragments could block the effects of mouse β-globin HS2 enhancer elements in this assay (Figure 5B). These data indicate that neither exon 1′ not intron 1′ have enhancer blocking activity in erythroid cells.
USF but not CTCF proteins occupy the exon 1′-intron 1′ region
Previous study of histone modifications using a ChIP assay demonstrated that the core histones H3 and H4 were hyperacetylated in the 183-bp region of the α-spectrin gene in erythroid cells, a finding associated with insulator elements conferring barrier function, but not enhancer blocking activity.48,52-54 The region of the cHS4 insulator that functions as a barrier element binds the upstream stimulatory factor (USF) proteins, thought to be necessary for recruitment of histone modifications associated with active chromatin and, by inference, barrier activity.19,20,50 In contrast, vertebrate insulator elements that exhibit enhancer-blocking activity have been shown to bind the 11-zinc finger protein CTCF.27,49 USF1 and USF2 are basic helix-loop-helix proteins involved in regulation of many hematopoietic genes including HoxB4, β-globin, and glycophorin.50,55-57
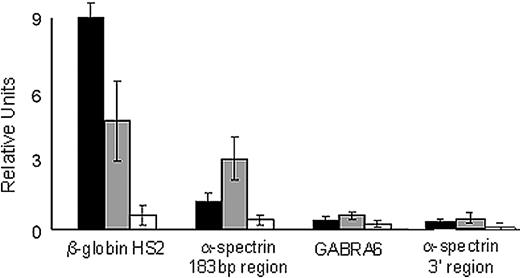
We performed ChIP analyses of this region in native K562 chromatin using anti-USF and anti-CTCF antibodies. In K562 cell chromatin, both USF1 and USF2 occupied the 183-bp region of the α-spectrin gene (Figure 6). USF1 and USF2 occupancy was found in the positive control, the HS2 region of the β-globin gene locus control region,29 but not in either of 2 negative controls, a region of the cerebellar-specific GABRA6 gene, and an unrelated region of the α-spectrin gene downstream of exon 1′/intron 1′ that does not contain any USF consensus-binding sites (Figure 6).
USF proteins interact with the α-spectrin gene 183-bp exon 1′ + intron 1′ region. Quantitative ChIP analyses of the 183-bp region of the α-spectrin gene encoding exon 1′ and intron 1′ were performed with K562 cell chromatin and antibodies against USF1 and USF2. The hypersensitive site 2 of the locus control region of the human β-globin locus was included as a positive control. Negative controls include GABRA6 and an unrelated region of the α-spectrin gene downstream of the 183-bp region. ■, USF1-precipitated chromatin; ▩, USF2-precipitated chromatin; □, IgG-precipitated chromatin. Standard error bars represent results from multiple (≥ 3) experiments.
USF proteins interact with the α-spectrin gene 183-bp exon 1′ + intron 1′ region. Quantitative ChIP analyses of the 183-bp region of the α-spectrin gene encoding exon 1′ and intron 1′ were performed with K562 cell chromatin and antibodies against USF1 and USF2. The hypersensitive site 2 of the locus control region of the human β-globin locus was included as a positive control. Negative controls include GABRA6 and an unrelated region of the α-spectrin gene downstream of the 183-bp region. ■, USF1-precipitated chromatin; ▩, USF2-precipitated chromatin; □, IgG-precipitated chromatin. Standard error bars represent results from multiple (≥ 3) experiments.
In HeLa cell chromatin, occupancy by USF1 and USF2 was found only in the positive control, a known USF binding site in the ubiquitously expressed DOLK gene,30 but not in the 183-bp region of the α-spectrin gene, the GABRA6 gene, or the unrelated region of the α-spectrin gene downstream of exon 1′/intron 1′ (Figure S2).
In other ChIP assays with native K562 cell chromatin, CTCF occupancy was found in the positive control, a validated CTCF-binding site on chromosome one (chr1:147764685-147765334),31 but not in the GABRA6 gene negative control. CTCF did not occupy the exon 1′/intron 1′ region of the α-spectrin gene in chromatin from K562 cells (not shown), correlating with the enhancer blocking studies.
Double-stranded oligonucleotide probes corresponding to the 2 potential USF-binding sites in exon 1′, the region with barrier-element activity, intron 1′, exon 1′ and intron 1′ with flanking sequence, or a control USF binding sequence from the HoxB4 promoter were prepared (Table S2) and used in EMSA with K562 cell nuclear extracts. None of the α-spectrin probes yielded a complex on EMSA that migrated at the same location as a complex obtained with the control USF probe and only the control USF probe yielded a complex that was shifted by both USF-1 and USF-2 antiserum (data not shown). These data indicate that USF proteins interact with other DNA-binding proteins at this site in the α-spectrin gene or are brought to the α-spectrin exon 1′ region from a distance (eg, by formation of a chromatin loop).
Discussion
The major erythrocyte membrane protein genes, including α-spectrin, β-spectrin, ankyrin, and band 3, share the common feature of using alternate first exons and likely alternate promoters to direct tissue-specific isoform expression.17,21,51,58-60 In erythroid cells, all 4 genes use GATA-1–dependent, tissue-specific promoters to direct expression of their respective transcripts. In contrast to the other promoters, which direct high levels of expression at all stages of erythroid development, the core α-spectrin gene promoter directs low levels of expression, and then only in the early stages of erythroid development.17 This observation is surprising, as previous studies have demonstrated that the α-spectrin gene is expressed at high levels in erythroid cells and that expression is controlled at the transcriptional level.3-6 The data in this report indicate that a region downstream of the core α-spectrin gene promoter is required for expression in adult erythroid cells (reticulocytes), and, based on the low levels of Aγ-globin reporter gene expression in the reticulocytes of transgenic mice, suggest that additional regulatory elements are required for high-level expression in erythroid cells.
The spectrin family of genes, including α-spectrin, β-spectrin, and dystrophin, serve as excellent models of complex gene loci structure and function. These loci encode multiple tissue, developmental, and stage-specific isoforms, arising from alternative splicing, alternate polyadenylation, and usage of alternate promoters.1,61 Because of the great diversity and complexity of the spectrin family of genes, it is expected that insulator elements with barrier-element and/or enhancer blocker activity participate in their regulation. Such activity could participate in the regulation of the heterogeneous isoforms of α-spectrin found in spleen, heart, and reticulocytes.62
The mechanisms controlling barrier-element structure and function in vertebrates are poorly understood. Studies of the cHS4 barrier element have shown that the transcription factors USF1 and USF2 recruit histone methyltransferase activity, histone acetyltransferase, and ATP-dependent nucleosome remodeling complexes,19,20 supporting the hypothesis that recruitment of enzymes associated with activating histone modifications blocks the mechanism(s) that lead to spreading of gene-silencing–associated chromatin alterations into regions of active chromatin.19,52,54,63 Transgenic studies in both Drosophila and mice have used barrier elements to flank reporter genes to prevent gene silencing and reduce position effect variegation.64-67 Barrier elements are found flanking genes or gene clusters or, as in the case of α-spectrin, tightly associated with gene promoters.68-71 Much of the data available on barrier-element structure and function has come from model organisms or study of the chicken, mouse, or human β-globin gene loci.70,72,73 The barrier element in α-spectrin is the first barrier element described in an erythrocyte membrane protein gene and suggests that other membrane gene loci will require barrier elements to maintain the proper chromatin configuration for expression in erythroid cells as well.
Our previous studies of the 183-bp region of the α-spectrin gene demonstrated characteristics associated with barrier elements, including the inability of exon 1′ to act as an enhancer in transfection assays,39 presence of a broad DNase I–hypersensitive site indicating a nucleosome-free region, and histone H3 and H4 hyperacetylation.48,54,74 The barrier function contributed by exon 1′ accounts for the nearly position-independent, uniform expression of the transgene, an activity confirmed in the gene silencing/position effect variegation barrier assay. Barrier elements mark the boundary between euchromatin and heterochromatin and, although they do not directly affect the level of expression of a given gene, they are thought to be responsible for preventing the spread of heterochromatin into the gene, thereby maintaining the open chromatin structure needed for optimal expression.75 Occupancy of the region of the chicken β-globin HS4 and α-spectrin 183-bp regions with barrier-element activity by USF proteins suggests that USF binding is a mechanism also used by mammalian barriers, but, as suggested by our studies, not necessarily via DNA binding.
We propose a model of α-spectrin gene regulation that includes exon 1′ engaging several proteins, some as yet unknown, including USF and GATA-1 factors, which recruit proteins that provide chromatin modifications to protect the locus from encroachment of heterochromatin into the α-spectrin gene and direct erythroid-specific expression. It is possible that another barrier 5′ or 3′ of this region participates in protection of euchromatin from the encroachment of heterochromatin into this region in erythroid cells.
Studies of patients with α-spectrin linked hereditary hemolytic anemia have shown that erythroid cells of many patients exhibit decreased α-spectrin mRNA levels and diminished α-spectrin synthesis leading to abnormal, spectrin-deficient erythrocytes.7,13-15 The precise genetic basis(es) of decreased spectrin mRNA accumulation in these cases is not known, even after mutation screening of the minimal promoter and 52 coding region exons of the α-spectrin gene.16 Thus, it is critical to identify cis-elements in the α-spectrin gene outside the coding region as potential sites of mutation in these patients. This region of the α-spectrin gene is an excellent candidate region for mutations associated with decreased α-spectrin gene expression in patients with spectrin-linked hemolytic anemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by a grant from the National Heart, Lung, and Blood Institute, NIH (HL65448), the National Institute of Child Health and Human Development, NIH (HD000850), and intramural funds from the National Human Genome Research Institute, NIH.
National Institutes of Health
Authorship
Contribution: P.G.G. designed and interpreted experiments and prepared the manuscript; D.G.N., J.Y. L., L.A.S., and Y.D.M. designed and performed experiments and reviewed the manuscript; and D.M.B. designed and interpreted experiments, prepared figures, and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Patrick G. Gallagher, Department of Pediatrics, Yale University School of Medicine, 333 Cedar Street, PO Box 208064, New Haven, CT 06520-8064; e-mail: patrick.gallagher@yale.edu.