Abstract
Growth differentiation factor 15 (GDF15) is a divergent member of the transforming growth factor–β superfamily and has been identified in different contexts as a hypoxia-inducible gene product and as a molecule involved in hepcidin regulation. The biology of iron and oxygen is closely related, and known regulatory pathways involving hypoxia-inducible factor (HIF) and iron-regulatory proteins (IRPs) are responsive to both these stimuli. We therefore sought to characterize the regulation of GDF15 by iron and oxygen and to define the involvement or otherwise of HIF and IRP pathways. Here we show that GDF15 is strongly up-regulated by stimuli that deplete cells of iron and that this response is specifically antagonized by the reprovision of iron. GDF15 exhibits greater sensitivity to iron depletion than hypoxia, and responses to hypoxia and iron depletion are independent of HIF and IRP activation, suggesting a novel mechanism of regulation. We also report significant induction of serum GDF15 in iron-deficient subjects and after administration of an iron chelator to normal subjects. These findings indicate that GDF15 can be induced by pathophysiologic changes in iron availability, raising important questions about the mechanism of regulation and its role in iron homeostasis.
Introduction
Originally identified as factors important for regulating development, differentiation, and tissue repair, the transforming growth factor–β (TGF-β) superfamily of proteins comprises more than 40 members, broadly divided into 2 branches, defined by sequence homology and the signaling pathways that they activate. The first branch contains TGF-β, activins, and the nodal family, and the second encompasses bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs), and Mullerian-inhibiting substance.1 Despite sharing a 7-cysteine domain that results in a characteristic “cysteine knot,” sequence identity between subfamilies is low, accounting for the diverse range of biologic functions. GDF15 is one of the most divergent members of the TFG-β family, showing only 15% to 29% identity to other family members, suggesting a unique biologic role.
GDF15 is also known as macrophage inhibitory cytokine 1 (MIC-1),2 nonsteroidal anti-inflammatory drug-regulated protein 1,3 prostate differentiation factor,4 placental bone morphogenic protein,5 and placental TGF-β.6 GDF15 exerts diverse biologic functions in distinct cellular contexts. It inhibits the late phase of macrophage activation,2 inhibits proliferation of immature hematopoietic progenitors,5 inhibits growth of assorted tumor cell lines,6,7 and is involved in embryonic, osteogenic, and hematopoietic development.4,8 The gene comprises 2 exons, and in common with other TGF-β family members, GDF15 is synthesized as a 62-kDa intracellular proprotein that contains a conserved diarginine motif (RXXR), which, after cleavage by a furin-like protease, is secreted as a 25-kDa disulfide-linked dimeric protein.2
Bioinformatic and functional promoter analysis has revealed several putative and confirmed transcription factor–binding sites in the 5′-flanking region of the GDF15 gene, including several sites common to other TGF-β family members, such as AP-1, AP-2, Sp1, Sp3, and Nkx-2.3,9 In addition, the GDF15 promoter contains 2 binding sites for p53 that mediate GDF15 response to several stimuli, including DNA damage.6,7
In the basal state, GDF15 is strongly expressed in the placenta5 and during erythroblast maturation,10 with more modest levels also observed in the colon, prostate, and kidney. However, GDF15 is present to some degree in most tissues2,9 and is highly inducible during macrophage activation in response to phorbol myristate acetate and several proinflammatory cytokines,11 after tissue injury,12,13 in response to ionizing radiation,14 and during tumorigenesis. GDF15 mRNA has also been identified in gene expression profiling experiments as an oxygen-regulated transcript responding both to hypoxia and anoxia.15,16
Although essential for many biologic processes, inappropriately high oxygen concentrations lead to the generation of excess oxygen-free radicals that are damaging to biologic molecules. Because of its ability to act as both electron donor and electron acceptor, iron promotes the production of such reactive oxygen species in the presence of oxygen. In keeping with this interaction, several homeostatic systems for iron and oxygen are responsive to both stimuli. For example, the hypoxia-inducible factor (HIF) hydroxylases, responsible for regulation of HIF by oxygen, require iron as a cofactor,17,18 rendering this major transcriptional pathway sensitive to changes in iron concentration as well as oxygen tension.19 In addition, the intracellular iron-regulatory pathway mediated by the RNA-binding iron-regulatory proteins 1 and 2 (IRP1 and IRP2) also exhibits dual sensitivity to iron and oxygen.20,21
Physiologic iron levels are determined predominantly by its intestinal absorption and recycling from senescent red blood cells, processes that are regulated by the liver-derived iron-regulatory hormone hepcidin.22 In healthy persons, hepcidin production appears to respond to physiologic iron demand, although little is known about the mechanisms through which hepcidin production is regulated. Several members of the TGF-β superfamily (BMP2, BMP4, and BMP9) modulate hepcidin expression through Smad second messenger pathways.23-25 Recent reports have also described GDF15-dependent suppression of hepcidin in β-thalassemia patients, suggesting a role in iron metabolism.10
Taken together, these findings prompted us to determine whether GDF15 might itself be responsive to cellular iron levels. We present data demonstrating that expression of both GDF15 mRNA, and protein is strongly and specifically responsive to intracellular iron depletion, generated through a variety of mechanisms, in a number of human cell lines. We further demonstrate that this up-regulation is independent of IRP1 and IRP2, and of the HIF pathway. Finally, we report that disruption of iron homeostasis in vivo, by administration of a single dose of intravenous desferrioxamine (DFO), or in iron-deficient persons leads to up-regulation of serum GDF15 levels.
Methods
The cell-penetrant iron chelator 2,2′-bipyridyl (BIP) and human holotransferrin were purchased from Sigma-Aldrich (St Louis, MO).
Cell lines and cell culture
The human embryonic kidney cell line 293T (number CRL-11268; ATCC, Manassas, VA), the breast adenocarcinoma cell line MCF-7 (ATCC number HTB-22), the hepatocellular carcinoma cell line HEP3B (ATCC number HB-8064), and the cervical adenocarcinoma cell line HeLa (ATCC number CCL-2) were cultured at 37°C in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 2.5 mM l-glutamine, 50 U/mL penicillin, and 50 μg/mL streptomycin (Sigma-Aldrich). The renal cell carcinoma cell line RCC4 stably transfected with either empty vector or a plasmid encoding hemagglutinin (HA)–tagged wild-type von Hippel–Lindau protein (VHL-HA) have been previously described.26 Cells were seeded at 2 × 105 cells/mL in 6-well plates for all experiments.
RNA extraction and cDNA synthesis
Adherent cells were washed twice with phosphate-buffered saline, then lysed in TRIzol (Sigma-Aldrich), and mRNA was extracted by phase separation. Equal amounts of the mRNA template were used for cDNA synthesis by Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA).
Real-time polymerase chain reaction for quantification of mRNA
mRNA measurement, by quantitative real-time polymerase chain reaction (qPCR), was achieved using Sybrgreen Fluorescein qPCR (Bio-Rad, Hercules, CA), with the following primer pairs: GDF15 forward (for), GTGTTGCTGGTGCTCTCGTG; GDF15 reverse (rev), CGGTGTTCGAATCTTCCCAG; CA9for, TTGCCAGAGTTGACGAGGC; CA9rev, CGATTTCTTCCAAGCGAGAC; TfR1for, ACCATTGTCATATACCCGGTTCA; TfR1rev, GGCCTTTGTGTTATTGTCAGCAT; IRP1for, AACCCATTCGCACACCTTG; IRP1rev, ATGGTAAGCGCCCATATCTT; IRP2for, GCAAAGCCAAACTCGAATCAATG; IRP2rev, CAGATTAATCTGGATCACCTGGG; β-actin for, ACCATGGATGATGATATCGCC; β-actin rev, GCCTTGCACATGCCGG. Expression levels were normalized to β-actin expression in the respective sample.
Quantification of human GDF15 protein by ELISA
Human GDF15 protein levels in cell supernatants and human sera were determined by enzyme-linked immunosorbent assay (ELISA) using a DuoSet Sandwich ELISA Kit (R&D Systems, Minneapolis, MN). Briefly, tissue-culture supernatants and serum samples were centrifuged to remove residual cells; 96-well plates were coated with 2 μg/mL monoclonal mouse anti–human GDF15 capture antibody and blocked with 1% bovine serum albumin in phosphate-buffered saline. After incubation with tissue-culture supernatants or serum, the wells were washed and bound GDF15 was detected using a biotinylated goat anti–human GDF15 antibody. Recombinant human GDF15 protein was used to generate a standard curve.
IRP and HIF RNA interference
HeLa cells were transfected in serum-free Optimem medium (Invitrogen) with small interfering RNA (siRNA) oligonucleotides (Dharmacon RNA Technologies, Lafayette, CO) at a final concentration of 200 nM, using Oligofectamine transfection reagent (Invitrogen). Cells were transfected on days 1 and 2 and harvested on day 3. Immediately before harvest, medium was collected for analysis of secreted GDF15 by ELISA. Drosophila HIF (dHIF) siRNA was used as an irrelevant siRNA control in all transfections.
Ferroportin and HFE-transient transfections
HeLa cells were transfected with 1 μg/well of the mammalian expression vector pQCXIX (Clontech, Mountain View, CA) encoding wild-type (WT) human ferroportin (FPN)27 or the inactive mutant V162Δ FPN28 fused to enhanced green fluorescent protein (EGFP) as described.29 Transient transfections were performed using Fugene transfection reagent (Invitrogen). pQCXIX-encoding human β2 microglobulin–hereditary hemochromatosis protein precursor (HFE) (WT) heavy chain fusion or β2 microglobulin–HFE (V100A) heavy chain fusion with EGFP (A.R.M.T., oral communication, February 2008) were similarly transfected into HeLa cells. The V100A mutation (numbering from the initiating methionine) abrogates HFE binding to transferrin receptor–1.30 Transfection efficacy was determined at different time intervals by counting EGFP-expressing cells with fluorescence microscopy.
Human studies
Eight healthy volunteers took part in the DFO study. Informed consent was obtained from subjects before participation. The study conformed to the principles of the Declaration of Helsinki and had approval from the Oxfordshire Clinical Research Ethics Committee.
DFO and saline treatments were administered by intravenous infusions, over a period of 8 hours, using a controlled-rate intravenous infusion pump. The DFO dose was 4 g/70 kg body weight. Separate infusions of DFO and saline were administered to the same person on different days. Blood was taken immediately before infusion (t = 0), at 4 hours after commencement of the infusion (t = 4), at the end of the infusion (t = 8), and again 4 and 16 hours later (t = 12 and t = 24). Plasma was frozen at −20°C, pending analysis by GDF15 ELISA. No difference in GDF15 immunoreactivity was seen between frozen and unfrozen samples.
Random samples of iron-deficient and normal sera were identified from routine analysis by the hematology laboratory at the John Radcliffe Hospital (Oxford, United Kingdom) to estimate the population characteristics for GDF15 levels in each group. Iron-deficient sera were taken as having an iron concentration of less than or equal to 8 μM (mean ± SD, 5.0 ± 2.0 μM), and iron-replete sera had an iron concentration more than or equal to 14 μM (mean ± SD, 19.6 ± 3.7 μM). Serum was stored at 4°C, for up to 7 days. No difference in GDF15 immunoreactivity was seen between fresh serum and serum stored for this time. Samples were each tested by ELISA twice, and the means of duplicate measurements were used to plot the data.
Statistical analysis
Data are mean plus or minus SEM. Statistical analysis of in vitro data was performed using the SPSS (version 16.0.0) software package. Analysis of mRNA data was performed on the normalized threshold cycles (ΔCT) before antilog transformation. Comparison of means of multiple groups was by one-way analysis of variance with post-hoc analysis performed using the Dunnett method (2-sided) for multiple comparisons with a single control group. A value of P less than .05 was deemed significant. In human studies, comparisons of GDF15 concentrations in blood samples were performed with the nonparametric Mann-Whitney test. Correlations were calculated using Spearman rank correlation.
Results
Transcription of the GDF15 gene is induced by iron chelation in human cell lines
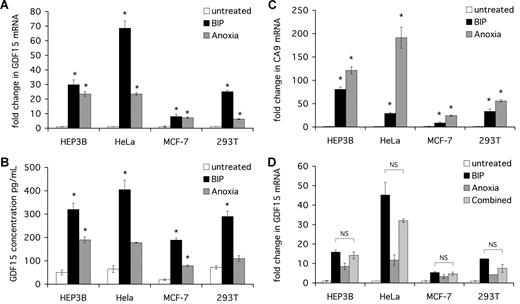
We first studied the regulation of GDF15 mRNA and protein by oxygen and iron in vitro in a range of cell lines: HEP3B, HeLa, MCF-7, and 293T. Cells were exposed for 16 hours to 21% oxygen (untreated), 0% oxygen (anoxia), or 100 μM bipyridyl (BIP), a cell-penetrant iron chelator, and fold change in GDF15 mRNA and protein determined. Similar results were obtained when BIP was substituted by the iron-chelator DFO (data not shown). Expression of the iron and oxygen-regulated gene carbonic anhydrase 9 (CA9) was measured to permit comparison with an established HIF-responsive gene.
In all cell lines, GDF15 mRNA was markedly up-regulated by anoxia and BIP treatment, reaching 60-fold induction in BIP-treated HeLa cells (Figure 1A). The concentrations of GDF15 protein, measured by ELISA in cell supernatants, reflected the changes in mRNA levels, confirming that both GDF15 mRNA and protein are up-regulated by anoxia and iron chelation (Figure 1B).
GDF15 mRNA and protein are induced by iron chelation in human cell lines. Cells were seeded at 2 × 105 cells/mL and then cultured for 16 hours alone (untreated), in the presence of 100 μM BIP or at 0% oxygen (anoxia). Cells were then harvested for mRNA extraction and cDNA synthesis. Supernatants were collected and stored at −20°C for use in ELISA. (A) qPCR quantification of GDF15 expression. GDF15 signal was normalized to β-actin signal in the respective sample, and fold change in GDF15 calculated relative to untreated cells. (B) ELISA measurement of secreted GDF15 protein in cell supernatants. Serial dilutions of the recombinant human GDF15 were used as standards. (C) qPCR quantification of CA9 expression. CA9 signal was normalized to β-actin signal in the respective sample, and fold change in expression calculated relative to untreated cells. (D) Cells were seeded at 2 × 105 cells/mL and then cultured for 16 hours alone (untreated) with 100 μM BIP, or at 0% oxygen (anoxia), or in the presence of both BIP and anoxia (combined). Cells were then harvested for mRNA extraction and cDNA synthesis, GDF15 signal was normalized to β-actin signal in the respective sample, and fold change in GDF15 calculated relative to untreated cells. *P < .05 compared with untreated cells. NS indicates not significant.
GDF15 mRNA and protein are induced by iron chelation in human cell lines. Cells were seeded at 2 × 105 cells/mL and then cultured for 16 hours alone (untreated), in the presence of 100 μM BIP or at 0% oxygen (anoxia). Cells were then harvested for mRNA extraction and cDNA synthesis. Supernatants were collected and stored at −20°C for use in ELISA. (A) qPCR quantification of GDF15 expression. GDF15 signal was normalized to β-actin signal in the respective sample, and fold change in GDF15 calculated relative to untreated cells. (B) ELISA measurement of secreted GDF15 protein in cell supernatants. Serial dilutions of the recombinant human GDF15 were used as standards. (C) qPCR quantification of CA9 expression. CA9 signal was normalized to β-actin signal in the respective sample, and fold change in expression calculated relative to untreated cells. (D) Cells were seeded at 2 × 105 cells/mL and then cultured for 16 hours alone (untreated) with 100 μM BIP, or at 0% oxygen (anoxia), or in the presence of both BIP and anoxia (combined). Cells were then harvested for mRNA extraction and cDNA synthesis, GDF15 signal was normalized to β-actin signal in the respective sample, and fold change in GDF15 calculated relative to untreated cells. *P < .05 compared with untreated cells. NS indicates not significant.
For comparison, we also measured the HIF-regulated CA9 mRNA in the same experiments. CA9 mRNA was markedly up-regulated in all cell lines by both anoxia and iron chelation (Figure 1C). However, significant differences in the response patterns were observed, most markedly in HeLa cells, where GDF15 was more sensitive to iron chelation than to anoxia, whereas CA9 was less responsive to iron chelation than to anoxia, suggesting that the regulation of these genes by iron might be mediated by distinct pathways.
We next examined for synergy between the 2 stimuli. The combination of anoxia and BIP did not enhance the GDF15 mRNA response compared with BIP alone in any of the cell lines (Figure 1D).
The effects of BIP treatment on viable cell number and apoptotic activity were assessed by trypan blue and 4′,6′-diamidino-2-phenylindole staining, respectively. Treatment with 100 μM BIP over 16 hours did not significantly reduce viable cell number or increase apoptotic cell numbers in any of the cell lines (data not shown).
GDF15 induction by iron depletion is independent of IRP
HeLa cells showed the greatest magnitude of GDF15 response to iron chelation and were therefore selected for further mechanistic analyses. We next examined for the involvement of known iron- and oxygen-dependent transcriptional pathways in the GDF15 response to iron chelation. The GDF15 promoter contains 2 functional p53-binding sites, and the p53 transcriptional pathway is known to be up-regulated by hypoxia. However, we observed the most marked up-regulation of GDF15 mRNA and protein after iron chelation in HeLa cells, which are functionally deficient in p53, indicating that an intact p53 pathway is not required for the iron-responsiveness of GDF15.
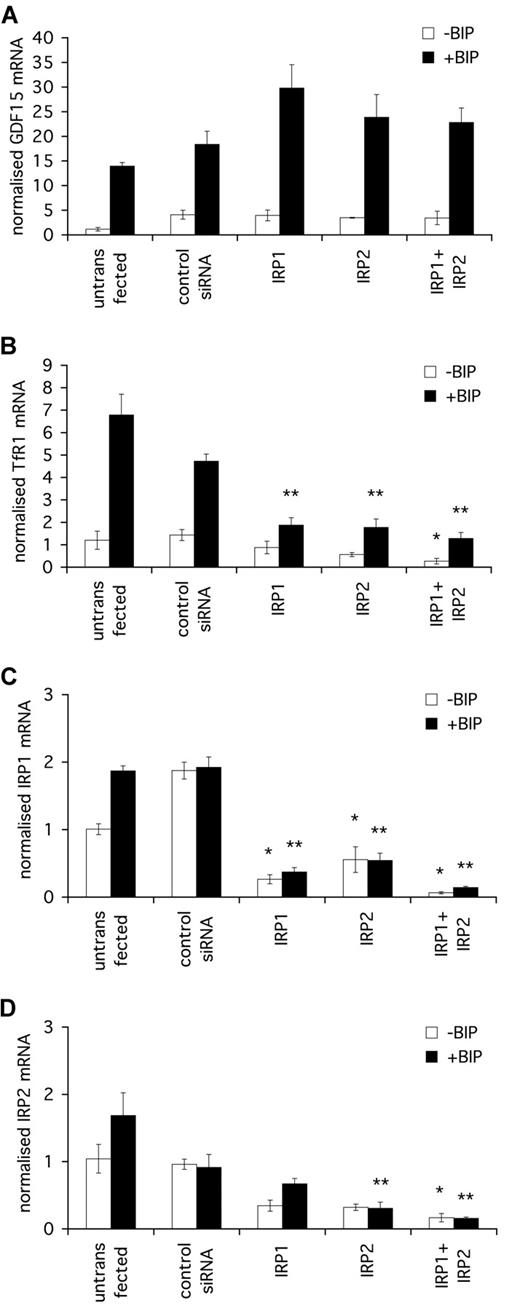
Regulating gene expression through control of mRNA stability and protein translation, the major iron-regulatory pathway IRP1/IRP2 could potentially mediate GDF15 iron-responsiveness, directly by modulating mRNA stability or indirectly through secondary effects on the GDF15 transcriptional machinery. We therefore determined whether the iron-responsiveness of GDF15 is dependent on IRP1 and/or IRP2. HeLa cells were transfected with IRP1 or IRP2 siRNA, individually or in combination, and the responsiveness of GDF15 mRNA expression to iron chelation was assessed (Figure 2A). dHIF siRNA was used as an irrelevant control siRNA. For comparison, the responsiveness to iron chelation of transferrin receptor 1 (TfR1), an established IRP-regulated gene, was also determined (Figure 2B). A small, but significant, increase in basal GDF15 level was seen in response to transfection with all siRNAs, including control siRNA, possibly reflecting a nonspecific response to transfection with siRNA or incubation in serum-free medium before transfection. For this reason, all further comparisons of the effects of gene-specific siRNAs are made relative to the dHIF control siRNA transfected in an identical manner.
GDF15 induction by iron depletion is independent of IRP. HeLa cells were seeded at 2 × 105 cells/mL and transfected at 24 hours and 48 hours with 200 nM IRP1 siRNA, IRP2 siRNA, or a combination of both. dHIF siRNA was used as an irrelevant control siRNA. After the second transfection, cells were treated with 100 μM BIP for a further 16 hours, after which cells were harvested for RNA extraction. (A) qPCR quantification of GDF15 induction by BIP in the presence of various IRP siRNA treatments. (B) qPCR quantification of transferrin receptor 1 (TfR1) induction by BIP. TfR1 was used as a control for an IRP1- and IRP2-dependent iron-responsive gene. (C) qPCR quantification of IRP1 expression in cells treated with IRP1 siRNA alone, IRP2 siRNA alone, or with a combination of siRNAs. (D) qPCR quantification of IRP2 expression in cells treated with IRP1 siRNA alone, IRP2 siRNA alone, or with a combination of siRNAs. *P < .05 compared with control siRNA without BIP. **P < .05 compared with control siRNA with BIP.
GDF15 induction by iron depletion is independent of IRP. HeLa cells were seeded at 2 × 105 cells/mL and transfected at 24 hours and 48 hours with 200 nM IRP1 siRNA, IRP2 siRNA, or a combination of both. dHIF siRNA was used as an irrelevant control siRNA. After the second transfection, cells were treated with 100 μM BIP for a further 16 hours, after which cells were harvested for RNA extraction. (A) qPCR quantification of GDF15 induction by BIP in the presence of various IRP siRNA treatments. (B) qPCR quantification of transferrin receptor 1 (TfR1) induction by BIP. TfR1 was used as a control for an IRP1- and IRP2-dependent iron-responsive gene. (C) qPCR quantification of IRP1 expression in cells treated with IRP1 siRNA alone, IRP2 siRNA alone, or with a combination of siRNAs. (D) qPCR quantification of IRP2 expression in cells treated with IRP1 siRNA alone, IRP2 siRNA alone, or with a combination of siRNAs. *P < .05 compared with control siRNA without BIP. **P < .05 compared with control siRNA with BIP.
GDF15 mRNA was induced by BIP in cells treated with either siRNA alone or the combination of IRP1 and IRP2 siRNAs. The magnitude of GDF15 induction by BIP was not reduced by any of the siRNA treatments compared with control siRNA, confirming that GDF15 regulation by iron was independent of both proteins. Although slightly higher levels of GDF15 mRNA were seen in response to BIP after IRP1 siRNA, this was not statistically significant compared with control siRNA and was in the opposite direction to that predicted if the GDF15 response to iron was mediated by IRPs (Figure 2A). In contradistinction, both basal and BIP-induced levels of TfR1 mRNA expression were markedly reduced by IRP1 and IRP2 siRNA compared with control siRNA, confirming sufficient functional knockdown of IRP1 and IRP2 (Figure 2B).
The efficacy of IRP1 and IRP2 siRNA was further confirmed by qPCR and demonstrated 90% knockdown of IRP1 mRNA (Figure 2C) and IRP2 mRNA (Figure 2D) in cells treated with the combination of both siRNAs. Partial but significant knockdown of IRP1 mRNA by IRP2 siRNA and of IRP2 mRNA by IRP1 siRNA was also observed, probably reflecting cross-reactivity of the siRNAs with their target sequences.
GDF15 induction by iron depletion is independent of HIF
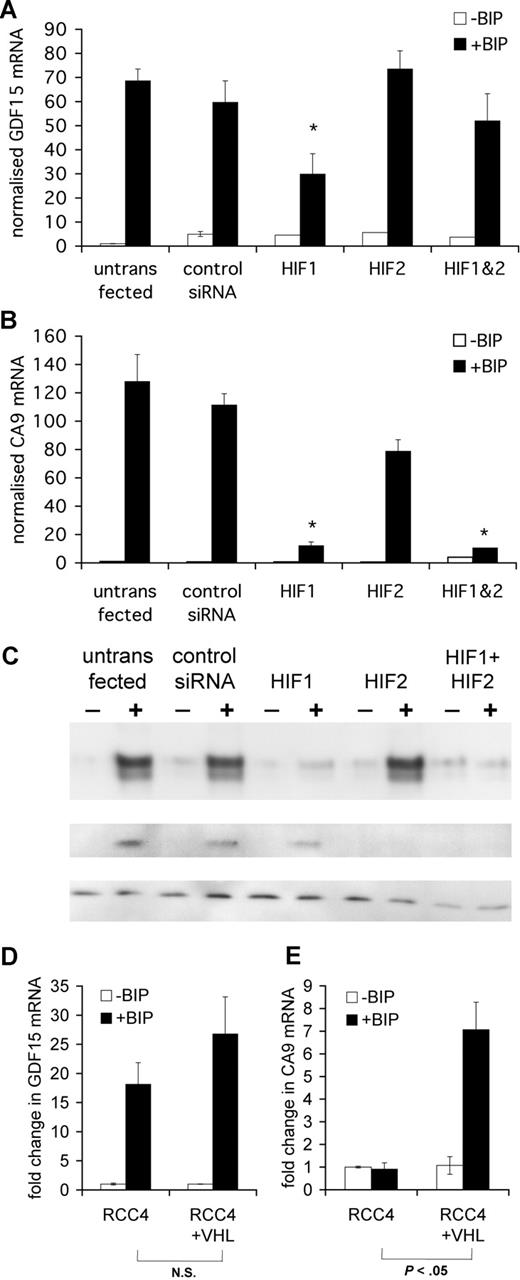
Although GDF15 and the HIF-target gene CA9 both respond to iron chelation and anoxia, the differing patterns of sensitivity to the 2 stimuli suggested that GDF15 might be regulated by a distinct, HIF-independent pathway. Furthermore, although the GDF15 promoter does contain DNA sequence motifs corresponding to the core HIF recognition consensus RCGTG, none is conserved across mammalian species, and none binds HIF-1α or HIF-2α proteins in chromatin immunoprecipitation experiments (D.R.M., unpublished data, October 2008). To determine directly whether the GDF15 response to iron chelation is affected by the integrity of the HIF pathway, we tested responses after transfection of HeLa cells with HIF-1α or HIF-2α siRNA individually or in combination. dHIF siRNA was used as an irrelevant control siRNA.
GDF15 mRNA was induced by BIP after treatment with either HIF-1α or HIF-2α siRNA alone or the combination of both. Although, in some experiments, a reduction in GDF15 responses to BIP was seen with HIF-1α siRNA, this was inconsistent and was not seen when combined HIF-1α and HIF-2α siRNAs were used, suggesting that GDF15 induction by BIP was not mediated by HIF (Figure 3A).
GDF15 induction by iron depletion is independent of HIF. HeLa cells were seeded at 2 × 105 cells/mL and transfected at 24 hours and 48 hours with 200 nM HIF-1α siRNA, HIF-2α siRNA, or a combination of both. dHIF siRNA was used as an irrelevant control siRNA. After the second transfection, cells were treated with 100 μM BIP for a further 16 hours, after which cells were harvested for mRNA and protein extraction. (A) qPCR quantification of GDF15 induction by BIP in the presence of various HIF siRNA treatments. (B) qPCR quantification of CA9 induction by BIP. (C) Western blotting for human HIF-1α and HIF-2α. Cell lysates were electrophoresed on SDS-polyaccrylamide gel, transferred onto cellulose acetate membrane, and stained with human monoclonal HIF-1α or HIF-2α antibodies. The housekeeping protein β-tubulin was stained as a loading control. (D,E) RCC4 cells stably transfected with empty vector or wild-type VHL-HA were seeded at 2 × 105 cells/mL and then cultured for 16 hours in the presence of 100 μM BIP. GDF15 and CA9 fold-induction by BIP compared with untreated cells was determined by qPCR after normalization to β-actin levels. In contrast with CA9, GDF15 was strongly induced by BIP irrespective of VHL status. *P < .05 compared with control siRNA with BIP.
GDF15 induction by iron depletion is independent of HIF. HeLa cells were seeded at 2 × 105 cells/mL and transfected at 24 hours and 48 hours with 200 nM HIF-1α siRNA, HIF-2α siRNA, or a combination of both. dHIF siRNA was used as an irrelevant control siRNA. After the second transfection, cells were treated with 100 μM BIP for a further 16 hours, after which cells were harvested for mRNA and protein extraction. (A) qPCR quantification of GDF15 induction by BIP in the presence of various HIF siRNA treatments. (B) qPCR quantification of CA9 induction by BIP. (C) Western blotting for human HIF-1α and HIF-2α. Cell lysates were electrophoresed on SDS-polyaccrylamide gel, transferred onto cellulose acetate membrane, and stained with human monoclonal HIF-1α or HIF-2α antibodies. The housekeeping protein β-tubulin was stained as a loading control. (D,E) RCC4 cells stably transfected with empty vector or wild-type VHL-HA were seeded at 2 × 105 cells/mL and then cultured for 16 hours in the presence of 100 μM BIP. GDF15 and CA9 fold-induction by BIP compared with untreated cells was determined by qPCR after normalization to β-actin levels. In contrast with CA9, GDF15 was strongly induced by BIP irrespective of VHL status. *P < .05 compared with control siRNA with BIP.
In contrast, the magnitude of the response of CA9 mRNA to BIP was greatly attenuated after suppression of HIF-1α or combined suppression of both HIFα isoforms, consistent with CA9 being predominantly regulated by the HIF-1 isoform31 (Figure 3B). Induction of HIF proteins by iron chelation and efficient knockdown by their respective siRNA were confirmed by Western blot (Figure 3C).
To further determine the dependence or otherwise of the iron-responsiveness of GDF15 on HIF, we determined the magnitude of the GDF15 response to iron chelation in the VHL-deficient renal carcinoma cell line RCC4 and the same cell line stably transfected with wild-type VHL-encoding vector. RCC4 cells have previously been shown to express constitutively high levels of HIF-1α and HIF-2α that are unresponsive to iron chelation, whereas the VHL-reconstituted cell line used has a normal HIF response.26,32 Both the VHL-deficient cells (stably transfected with empty vector) and the VHL-reconstituted cells showed a significant GDF15 response to iron chelation (P < .05). No significant difference was seen in the magnitude of GDF15 mRNA induction by BIP between the 2 cell lines (Figure 3D). In contrast, the fold induction of CA9 mRNA was significantly reduced in the VHL-deficient cell line (P < .05) (Figure 3E).
Taken together, these results indicate that, unlike CA9, GDF15 responsiveness to iron depletion is independent of the HIF/VHL pathway.
GDF15 is induced by overexpression of wild-type ferroportin and HFE
The observation that GDF15 expression is up-regulated by iron chelation in a manner that is independent of p53, IRP, and HIF pathways suggested the involvement of a novel iron-regulatory pathway. However, although they are potent chelators of iron, BIP and DFO are also able to bind other metal ions, raising the possibility that GDF15 might be responding to changes in nonferrous metal concentrations. We therefore wished to investigate the specificity of the observed GDF15 response to intracellular iron using alternative methods to reduce iron concentration.
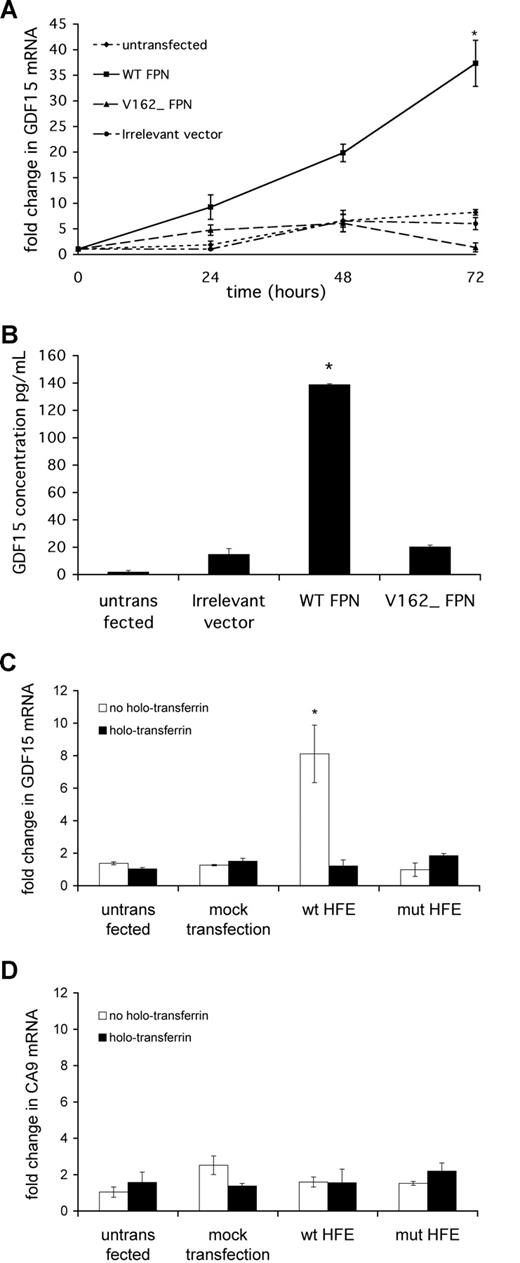
We first determined the effect of reducing intracellular iron levels by overexpressing the iron-exporting protein FPN.27,33,34 HeLa cells were transfected with 1 μg/well of the mammalian expression vector pQCXIX (Clontech) encoding WT human FPN, the inactive mutant V162Δ, or an irrelevant insert fused to EGFP. Comparable transfection efficiency was determined by fluorescence microscopy of the GFP-fusion protein in each case (data not shown). GDF15 mRNA was assayed by qPCR immediately before transfection and at 24, 48, and 72 hours after transfection. The effect of FPN overexpression on the iron-responsive HIF target gene CA9 was also assessed for comparison.
Untransfected cells and cells transfected with the irrelevant vector showed a moderate and steady 5-fold increase in GDF15 mRNA during the time course of the study, possibly reflecting basal iron depletion from the medium as a result of cell growth.19 In contrast, GDF15 mRNA increased markedly and progressively in WT FPN-transfected cells, achieving a 35-fold increase after 72 hours, whereas cells transfected with the inactive mutant V162Δ FPN showed changes in GDF15 expression comparable with background (Figure 4A). A similar pattern of induction was seen for CA9 (data not shown). The concentrations of GDF15 in cell supernatants reflected mRNA levels, with GDF15 protein secretion being significantly induced by WT FPN overexpression relative to V162Δ FPN and irrelevant vector (Figure 4B).
GDF15 is induced by intracellular iron depletion. HeLa cells were seeded at 2 × 105 cells/mL 8 hours before transfection. Cells were transfected with 1 μg/well PQCXIX mammalian expression vector encoding wild-type ferroportin (WT FPN), inactive mutant V162Δ ferroportin (V162Δ FPN), or an irrelevant insert fused to EGFP (irrelevant vector). (A) Cells were harvested at 24, 48, and 72 hours after transfection and GDF15 mRNA were determined by qPCR. *P < .05 compared with irrelevant vector. (B) ELISA measurement of secreted GDF15 protein in cell supernatants collected 48 hours after transfection. *P < .05 compared with irrelevant vector. (C,D) HeLa cells were transfected with 1 μg/well PQCXIX mammalian expression vector encoding β2M-HFE (WT), inactive mutant β2M-HFE V100Δ (mut HFE), or FuGene transfection reagent alone (mock transfection). Cells were harvested 72 hours after transfection and GDF15 mRNA and CA9 mRNA determined by qPCR. For 36 hours before harvest, cells were treated with or without 2 mg/mL holotransferrin. *P < .05 compared with mock transfection. Plotted data are mean plus or minus SD of 3 independent biologic replicates.
GDF15 is induced by intracellular iron depletion. HeLa cells were seeded at 2 × 105 cells/mL 8 hours before transfection. Cells were transfected with 1 μg/well PQCXIX mammalian expression vector encoding wild-type ferroportin (WT FPN), inactive mutant V162Δ ferroportin (V162Δ FPN), or an irrelevant insert fused to EGFP (irrelevant vector). (A) Cells were harvested at 24, 48, and 72 hours after transfection and GDF15 mRNA were determined by qPCR. *P < .05 compared with irrelevant vector. (B) ELISA measurement of secreted GDF15 protein in cell supernatants collected 48 hours after transfection. *P < .05 compared with irrelevant vector. (C,D) HeLa cells were transfected with 1 μg/well PQCXIX mammalian expression vector encoding β2M-HFE (WT), inactive mutant β2M-HFE V100Δ (mut HFE), or FuGene transfection reagent alone (mock transfection). Cells were harvested 72 hours after transfection and GDF15 mRNA and CA9 mRNA determined by qPCR. For 36 hours before harvest, cells were treated with or without 2 mg/mL holotransferrin. *P < .05 compared with mock transfection. Plotted data are mean plus or minus SD of 3 independent biologic replicates.
To further test GDF15 responses to specific reduction in cellular iron, we went on to block cellular iron uptake by the overexpression of HFE. HFE is a major histocompatibility complex class I–type protein that associates with beta2-microglobulin (β2M) and regulates iron absorption by blocking the interaction of surface transferrin receptor with transferrin.35-37 This intervention also allowed for specific manipulation of intracellular iron through the reprovision of iron in the form of iron-laden transferrin. pQCXIX vector encoding human β2M-HFE (WT) heavy chain fusion or β2M-HFE (V100A) heavy chain fusion with EGFP was transfected into HeLa cells. The V100A mutation (numbering from the initiating methionine) abrogates HFE binding to transferrin receptor-1.30 Fluorescence microscopy was used to ensure comparable transfection efficiency in all cases (data not shown).
Thirty-six hours after transfection, qPCR showed an 8-fold induction of GDF15 mRNA in cells transfected with β2M-HFE (WT) but not with β2M-HFE (V100Δ) or mock transfection. When iron-laden holotransferrin (2 mg/mL) was added to compete the effect of HFE on the transferrin receptor, holotransferrin reversed the effect of HFE on GDF15 expression, indicating that the effect of HFE was mediated through its actions on cellular iron uptake (Figure 4C). Interestingly, no appreciable effect was seen on CA9 expression, suggesting that the pathway regulating GDF15 is distinct and possibly more sensitive to changes in iron levels than HIF (Figure 4D).
Taken in combination, the effects of iron chelation, ferroportin, or HFE overexpression and competition by holotransferrin demonstrate that GDF15 is robustly and specifically induced by intracellular iron depletion. Furthermore, the magnitude of the in vitro response suggests a high degree of sensitivity, with regulation of up to 60-fold observed in HeLa cells treated with iron chelator.
Serum GDF15 is increased in vivo by DFO treatment
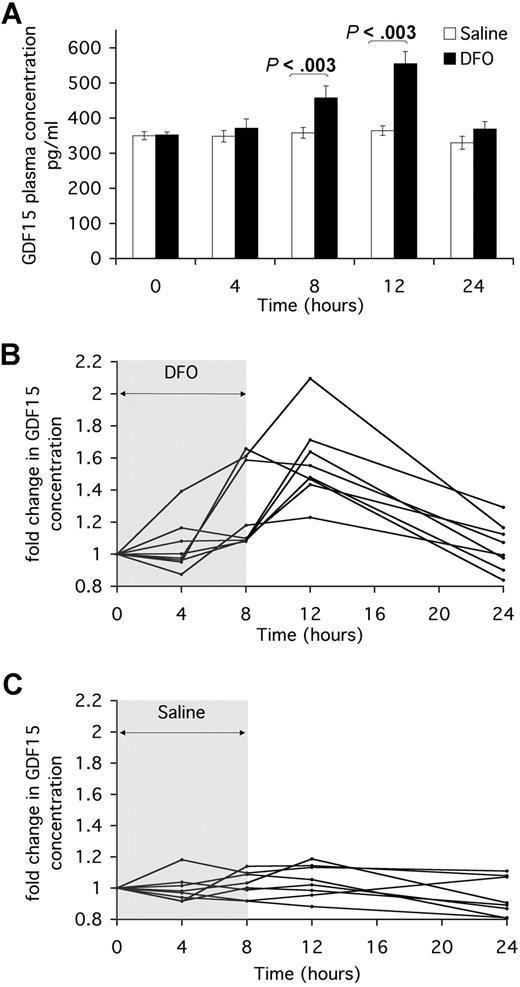
We therefore aimed to determine whether GDF15 was similarly induced in vivo by the iron chelator DFO. Eight persons were each subjected to an 8-hour infusion with either saline or DFO on separate days. Blood was taken from each person at the start of the infusion (t = 0 hours), 4 hours into the infusion (t = 4 hours), at the end of the infusion (t = 8 hours), and 4 hours or 16 hours after the end of the infusion (t = 12 hours and t = 24 hours, respectively). GDF15 protein was measured by ELISA in duplicate aliquots from each stored plasma sample.
Mean levels of constitutive GDF15 in the 8 persons were comparable at the start of the saline and DFO infusions (349 ± 11 pg/mL and 352 ± 8 pg/mL, respectively). Saline infusion induced no change in GDF15 plasma levels, which remained steady over time in all persons. DFO treatment induced a significant increase in GDF15 plasma levels at t = 8 hours (457 ± 34 pg/mL, P < .003) and up to 4 hours (t = 12 hours) after the end of the infusion (555 ± 43 pg/mL, P < .003). Sixteen hours after the end of the infusion (t = 24 hours), plasma GDF15 levels had decreased again in all persons (Figure 5A). Plotting the fold change in GDF15 concentration over time relative to the initial concentration, for each person, demonstrated that GDF15 was induced by DFO treatment, but not by saline, with similar temporal patterns of induction in all but one person (Figure 5B,C). Although of smaller magnitude than the GDF15 response to iron chelation observed in vitro, the observed induction of GDF15 serum concentration, after a single dose of iron chelator, was statistically significant (P < .003 at 8 hours and 12 hours compared with saline control).
Serum GDF15 is increased by DFO treatment in vivo. Eight persons were infused with saline or 4 g/70 kg DFO for 8 hours on 2 separate days. On each day, blood was taken from each person at the start of the infusion (t = 0 hours), 4 hours into the infusion (t = 4 hours), at the end of the infusion (t = 8 hours), and 4 and 16 hours after the end of the infusion (t = 12 hours and t = 24 hours, respectively). Serum samples were stored at −20°C for use in ELISA. (A) Serum concentration of GDF15 in saline and DFO treatment were measured by ELISA. Data for each time point are plotted as the mean plus or minus SE of GDF15 serum levels in all 8 persons. Mann-Whitney test was used to calculate the significance of difference between GDF15 levels in DFO and saline infusions at t = 8 hours and t = 12 hours. (B,C) Fold change in GDF15 serum concentration over time was calculated relative to time t = 0 in all 8 DFO- and saline-treated persons. Plotted data are the means of duplicate ELISA measurements.
Serum GDF15 is increased by DFO treatment in vivo. Eight persons were infused with saline or 4 g/70 kg DFO for 8 hours on 2 separate days. On each day, blood was taken from each person at the start of the infusion (t = 0 hours), 4 hours into the infusion (t = 4 hours), at the end of the infusion (t = 8 hours), and 4 and 16 hours after the end of the infusion (t = 12 hours and t = 24 hours, respectively). Serum samples were stored at −20°C for use in ELISA. (A) Serum concentration of GDF15 in saline and DFO treatment were measured by ELISA. Data for each time point are plotted as the mean plus or minus SE of GDF15 serum levels in all 8 persons. Mann-Whitney test was used to calculate the significance of difference between GDF15 levels in DFO and saline infusions at t = 8 hours and t = 12 hours. (B,C) Fold change in GDF15 serum concentration over time was calculated relative to time t = 0 in all 8 DFO- and saline-treated persons. Plotted data are the means of duplicate ELISA measurements.
Serum GDF15 is elevated in iron deficiency
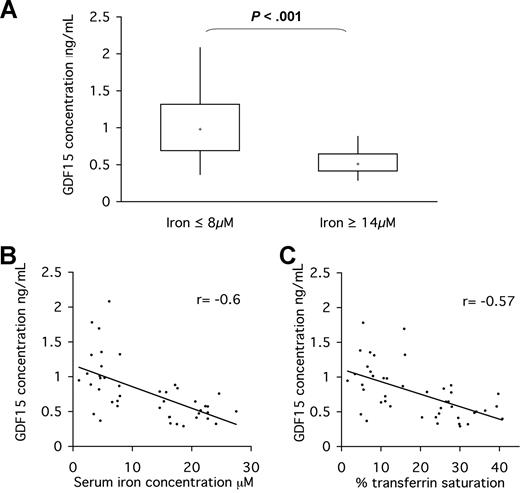
Having observed an induction of serum GDF15 concentration in response to iron chelation, we next determined whether differences in iron-loading status were associated with altered serum GDF15 levels. We conducted a comparative population study of serum from persons undergoing routine analysis of iron-loading indices. Samples were divided into 2 groups: those with serum iron concentration more than or equal to 14 μM (iron-replete) and those with serum iron concentration less than or equal to 8 μM (iron-deficient). Twenty-one iron-replete and 22 iron-deficient samples were randomly selected from each group. Duplicate aliquots of each sample were analyzed for GDF15 concentration by ELISA.
The mean concentration of GDF15 in iron-replete sera was 542 plus or minus 39 pg/mL, whereas the mean concentration of GDF15 in iron-deficient persons was 1048 plus or minus 91 pg/mL (Figure 6A). The difference in GDF15 levels between the 2 groups was found to be statistically significant by the Mann-Whitney U test (P < .001).
Serum GDF15 is elevated in iron-deficient persons relative to iron-replete controls. Persons with iron level less than or equal to 8 μM (n = 21) and iron level more than or equal to 14 μM (n = 22) were selected from a serum bank available at the Hematology and Biochemistry Unit, John Radcliffe Hospital. Serum samples had been stored at 4°C for up to 5 days. (A) Serum concentration of GDF15 in iron-deficient and iron-replete groups was measured by ELISA. Data are presented in a box and whisker plot showing the median, upper value, lower value, upper quartile, and lower quartile for each group. The Mann-Whitney test was used to determine the significance of difference in GDF15 serum levels between the 2 groups (P < .001). (B) Correlation between iron blood levels and serum GDF15 as determined by Spearman correlation (n = 43). (C) Correlation between transferrin saturation and serum GDF15 as determined by Spearman correlation (n = 43). Plotted data are the means of duplicate ELISA measurements. Regression lines are plotted.
Serum GDF15 is elevated in iron-deficient persons relative to iron-replete controls. Persons with iron level less than or equal to 8 μM (n = 21) and iron level more than or equal to 14 μM (n = 22) were selected from a serum bank available at the Hematology and Biochemistry Unit, John Radcliffe Hospital. Serum samples had been stored at 4°C for up to 5 days. (A) Serum concentration of GDF15 in iron-deficient and iron-replete groups was measured by ELISA. Data are presented in a box and whisker plot showing the median, upper value, lower value, upper quartile, and lower quartile for each group. The Mann-Whitney test was used to determine the significance of difference in GDF15 serum levels between the 2 groups (P < .001). (B) Correlation between iron blood levels and serum GDF15 as determined by Spearman correlation (n = 43). (C) Correlation between transferrin saturation and serum GDF15 as determined by Spearman correlation (n = 43). Plotted data are the means of duplicate ELISA measurements. Regression lines are plotted.
To determine whether GDF15 levels in these samples correlated to serum iron indices, GDF15 serum concentrations were plotted against either serum iron concentration or transferrin saturation. Both correlations showed a negative trend between markers of iron loading and GDF15 (r = −0.6 and r = −0.57, respectively; Figure 6B,C). Our data thus indicate that (1) iron deficiency is associated with elevated serum GDF15 concentration; and (2) iron-mediated regulation of GDF15 concentration occurs at pathophysiologic levels of iron.
Discussion
We have demonstrated robust and sensitive up-regulation of GDF15 mRNA and secreted protein in response to iron depletion in a range of human cell lines. The specificity of GDF15 responsiveness to iron was further confirmed by GDF15 induction in HeLa cells by specific and physiologically relevant iron-depleting stimuli (ferroportin and HFE overexpression) that could be antagonized by addition of excess holotransferrin.
The regulation of GDF15 by iron and oxygen differs quantitatively from that of the HIF-target gene CA9, which exhibits a greater sensitivity to changes in oxygen tension than to depletion of iron. Earlier work had demonstrated that GDF15 mRNA induction by anoxia was independent of both p53 and HIF.16 Furthermore, we have shown GDF15 mRNA induction by iron depletion to be independent of HIF, IRP1, and p53, providing further support for the involvement of a novel iron and oxygen-sensing pathway.
The nature of this mechanism and the transcriptional and/or posttranscriptional pathways leading to the iron-sensitive regulation of GDF15 are currently undetermined. The diversity of iron-dependent processes in biologic systems raises many possibilities. The lack of synergy between anoxia and iron chelation is consistent with the possibility that these stimuli act either directly or indirectly on the same pathway to control GDF15 mRNA levels. Furthermore, it is interesting that, in addition to identifying GDF15 as an oxygen-sensitive gene, previous work has demonstrated induction of expression by the cell-penetrant, 2-oxoglutarate analog, dimethyloxalylglycine, which inhibits HIF hydroxylases. This compound is also known to inhibit many other members of the Fe(II) and 2-oxoglutarate dioxygenase superfamily, raising the possibility of involvement of an, as yet unidentified, Fe(II)– and 2-oxoglutarate–dependent dioxygenase in the regulation of GDF15.15
Recent bioinformatic predictions have indicated that the human genome encodes as many as 60 or more such enzymes, many of which have as yet no known function.38 Further work will be required to determine whether one or more of these enzymes is responsive for the iron-sensitive signal that regulates GDF15.
Although the induction of GDF15 by a single DFO infusion was modest in comparison to in vitro responses, the ability to follow the time course of changes in each person under tightly controlled conditions indicated that the response was robust. Moreover, we also observed a significant increase in GDF15 in sera of persons with reduced serum iron concentration. Mast et al have recently reported on GDF15 levels in high-intensity blood donors.39 Interestingly, in that study, GDF15 levels (males, 1261 ± 3162 pg/mL; females, 754 ± 945 pg/mL) were higher than the normal range reported by Tanno et al (450 ± 50 pg/mL),10 and in our own study (542 ± 39 pg/mL). Mast et al commented that, in their high-intensity blood donors, the distribution of GDF15 levels was skewed and did not correlate with ferritin levels.39 Although they suggest that this argues against a correlation of GDF15 with iron status, their finding of elevated GDF15 levels in certain apparently fit but iron-challenged persons would be consistent with our finding that GDF15 is responsive to pathophysiologic reduction in iron. The cellular source of elevated GDF15 in vivo is unclear; and in our tissue-culture studies, we observed cellular heterogeneity in the magnitude of induction of GDF15 by iron chelation. Thus, the more limited up-regulation of GDF15 in the in vivo studies (and the absence of correlation with ferritin observed by Mast et al39 ) could be the result of restriction of responses to particular tissues and/or a largely paracrine effect that is not reflected in circulating GDF15 concentrations.
It should be noted that the GDF15 levels we observed in vivo in the current study, as well as those reported by Mast et al,39 were substantially less than those used by Tanno et al10 to suppress hepcidin. Conceivably, while generating systemic iron overload, ineffective erythropoiesis and associated iron fluxes in β-thalassemia might generate an iron-deficiency signal in a relevant molecular or cellular context and consequent stimulation of GDF15 expression in a particular erythroid compartment. Therefore, further dissection of GDF15 regulation by iron and oxygen in erythroid progenitors should be of interest and may provide mechanistic explanations to elevated GDF15 levels in β-thalassemia patients.
Notably, erythroid and macrophage differentiation of precursor bone marrow cell lines are both associated with up-regulation of GDF15.2,8 GDF15 inhibits the proliferation of immature hematopoietic progenitors and suppresses macrophage differentiation, thereby potentially creating one or more feedback loops.2,5 Interestingly, we have previously shown that differentiation of macrophages is associated with a large reduction in chelatable intracellular iron that might be responsible for up-regulation of GDF15 as well as the observed increase in HIF.19
Many cancers, such as human prostate,40 pancreatic, and colorectal cancers,41,42 overexpress GDF15 where it has been implicated in increased tumor invasiveness. The mechanism of GDF15 up-regulation in these cancers has yet to be elucidated, although it is possible that local iron depletion and hypoxia in rapidly dividing cancer cells may contribute to this expression.19
Another potential role of GDF15 is in the regulation of hepcidin. This small peptide hormone, secreted by the liver, regulates iron absorption by blocking intestinal iron uptake and is in turn tightly regulated by iron levels. Although several signaling pathways are known to affect hepcidin transcription, the precise mechanism of hepcidin regulation by iron is poorly understood. Currently, much work focuses on the regulation of hepcidin by members of the TGF-β superfamily, specifically BMPs 2, 4, and 9 that act through phosphorylation of receptor SMADs 1, 5, and 8, which, on association with SMAD4, translocate to the nucleus and stimulate expression of hepcidin.1,24 GDF15 itself has been shown to induce SMAD2/3 transcriptional activity in cardiac myocytes.43 The elevated serum GDF15 levels seen in β-thalassemia patients, who have inappropriately low hepcidin, and the ability of GDF15, both recombinant and from β-thalassemia sera, to suppress hepcidin in vitro lead to the hypothesis that high GDF15 levels can suppress hepcidin production.10 Whether regulation of GDF15 by intracellular iron provides a mechanism by which intracellular iron might impinge on hepcidin production is unclear and requires further analyses. To this end, we also observed GDF15 induction by iron depletion in cultured murine cells (data not shown), suggesting that a mouse model will be useful in dissecting the physiologic role of GDF15 regulation by iron.
In conclusion, the data presented provide a new link between iron and gene expression control. Given the emerging role of GDF15 in diverse aspects of iron metabolism, it will be important to further define the relevance of this regulation to iron homeostasis. In addition, elucidation of the cellular pathways leading to the iron-responsiveness of GDF15 should provide valuable insights into cellular iron-sensing mechanisms, as well as the interplay between iron and oxygen-sensing pathways.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Wellcome Trust and European Union (Pulmotension grant).
Wellcome Trust
Authorship
Contribution: S.L., D.R.M., P.J.R., A.R.M.T., P.A.R., and C.W.P. designed the research; S.L., D.R.M., N.P.T., A.C., and C.S. performed experiments; A.R.M.T. contributed vital analytical reagents; S.L. and D.R.M. analyzed and interpreted the data and performed statistical analyses; and S.L., P.J.R., and D.R.M. wrote the manuscript.
Conflict-of-interest disclosure: P.J.R. and C.W.P. are scientific cofounders of ReOx. The remaining authors declare no competing financial interests.
Correspondence: David R. Mole, Henry Wellcome Building for Molecular Physiology, University of Oxford, Roosevelt Drive, Oxford, OX3 7BN, United Kingdom; e-mail: drmole@well.ox.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal