To the editor:
With particular attention we read the article by Wu et al1 dealing with regulation of activation-induced cytidine deaminase (AID) by alternative splicing. We have also studied 3 alternative AID splice forms identical to the variants AID-ΔE4, AID-ΔE4a, and AID-ivs3 described by the authors. In accordance with Wu et al, we found that these splice variants did not reconstitute class switch recombination (CSR) in AID knockout mouse splenocytes. In the same assay, dominant-negative regulation was tested but not found. We studied somatic hypermutation (SHM) as well, measuring pGFP* revertants in NIH-3T3 cells,2 initially with results comparable with those reported by Wu et al.
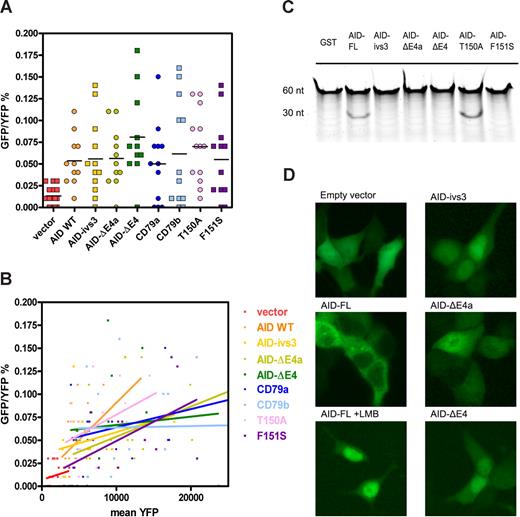
However, we noticed that the occurrence of revertants correlated with the transcription levels, as measured by mean fluorescence of the internal ribosome entry site–yellow fluorescent protein (IRES-YFP). Linear regression analysis of the green fluorescent protein (GFP)–positive frequency in relation to the mean YFP (Figure 1B) shows that all transductants exhibit a similar correlation, except for the empty vector control (LZRS-IRES-YFP), which never reaches a mean YFP intensity alike the others. We concluded that the empty vector is not a suitable control for this assay. Additional controls with the cDNAs of CD79a and CD79b inserted in the vector as mock yielded a comparable number of GFP+ cells as the full-length AID (AID-FL) and splice variants (Figure 1A). Similarly, 2 AID mutants, T150A and F151S, both gave rise to GFP+ cells. As T150A was able to induce CSR it was likely to have SHM activity, but F151S is a mutant known to have caused HIgM syndrome with a defect in SHM3 and should, therefore, lack GFP+ cells.
Deaminase activity and cytoplasmic localization of AID splice variants. (A) Dot plot of the somatic hypermutation assay depicting the frequency of GFP+ revertants in YFP+ NIH-3T3 cells. Each dot represents the measurement of one culture well. Virus stocks were applied in serial dilutions, resulting in a spread of the measurements (also visible in Figure 1B). (B) The same measurements shown in panel A depicted as a function of expression levels of the variants (mean IRES-YFP). Linear regression analysis shows the correlations between mutation frequency and expression levels. (C) In vitro deaminase activity: 100 fmol of a 60-nt FAM-labeled oligo, containing one cytidine located in a hot-spot motif, was incubated with 2 μg recombinant glutathione S-transferase (GST)–AID fusion protein. Subsequent recombinant UDG and NaOH heat treatments resulted in a 30-nt product, which was detected in AID-FL and T150A mutant, but not in the GST control, F151S AID mutant, or with any of the splice variants. Input and appropriate size of the recombinant proteins were verified by Coomassie staining (data not shown). (D) HEK293 cells were transfected with C-terminally tagged AID-GFP fusion proteins, and photographed alive at day 2 after transfection. AID-FL was located in the cytoplasm, but LMB incubation with 10 ng/mL for 3 hours resulted in nuclear accumulation. The AID splice variants display a predominantly nuclear or diffuse localization. Images were acquired with a Leica DM5000B microscope and a Leica DFC500 camera (Leica Microsystems, Rijswijk, The Netherlands) at the original magnification of 200×, and were further processed using Adobe Photoshop 7.
Deaminase activity and cytoplasmic localization of AID splice variants. (A) Dot plot of the somatic hypermutation assay depicting the frequency of GFP+ revertants in YFP+ NIH-3T3 cells. Each dot represents the measurement of one culture well. Virus stocks were applied in serial dilutions, resulting in a spread of the measurements (also visible in Figure 1B). (B) The same measurements shown in panel A depicted as a function of expression levels of the variants (mean IRES-YFP). Linear regression analysis shows the correlations between mutation frequency and expression levels. (C) In vitro deaminase activity: 100 fmol of a 60-nt FAM-labeled oligo, containing one cytidine located in a hot-spot motif, was incubated with 2 μg recombinant glutathione S-transferase (GST)–AID fusion protein. Subsequent recombinant UDG and NaOH heat treatments resulted in a 30-nt product, which was detected in AID-FL and T150A mutant, but not in the GST control, F151S AID mutant, or with any of the splice variants. Input and appropriate size of the recombinant proteins were verified by Coomassie staining (data not shown). (D) HEK293 cells were transfected with C-terminally tagged AID-GFP fusion proteins, and photographed alive at day 2 after transfection. AID-FL was located in the cytoplasm, but LMB incubation with 10 ng/mL for 3 hours resulted in nuclear accumulation. The AID splice variants display a predominantly nuclear or diffuse localization. Images were acquired with a Leica DM5000B microscope and a Leica DFC500 camera (Leica Microsystems, Rijswijk, The Netherlands) at the original magnification of 200×, and were further processed using Adobe Photoshop 7.
In this SHM assay, false-positive results may arise for several reasons. Klasen et al previously described the occurrence of GFP reversion due to the action of viral reverse transcriptase.4 In addition, we found mutations in YFP, and, as YFP is a GFP derivative, this might have contributed to the GFP signal. Induction of endogenous AID may be another complicating factor. Mouse AID was detectable in all samples at very low levels. Of note, additional induction due to signaling of the CD79a and CD79b-ITAM motifs was not observed (data not shown). The absence of “aspecific” GFP+ cells in the empty vector control may be related to the altered distance of YFP to the promotor, or to the lower transcription/transcript levels.
To circumvent the problems with the SHM assay mentioned and to settle this issue by a more robust biochemical method, we then used an in vitro deaminase assay as described previously.5,6 The results, depicted in Figure 1C, demonstrate that the 3 splice variants are not able to deaminate the cytidine in the oligonucleotide substrate and are therefore to be regarded as catalytically inactive.
We also studied the cellular localizations of the AID variants, at first using amino-terminally tagged proteins, which all located to the cytoplasm like the authors described. However, treatment with leptomycin B (LMB; Sigma-Aldrich, Zwijndrecht, The Netherlands), an inhibitor of the nuclear exporter CRM1, was not able to trap AID-FL in the nucleus. In contrast, carboxyl-terminal tagging resulted in nucleocytoplasmic shuttling for AID-FL as was described previously,7,8 and a disturbed cellular localization for the splice variants (Figure 1D). This result is in agreement with the expectations, since AID-ivs3 and AID-ΔE4 both lack the nuclear export signal and deletion of the α-helix might have dislocated it in AID-ΔE4a, as was also discussed by the authors.
Altogether, these results led us to the conclusion that the AID splice variants are nonfunctional.
Authorship
We thank A. Martin and M. Larijani for their help with the in vitro deaminase assay, H. Spits for the LZRS-IRES-YFP vector, M. Wabl for pGFP*, and T. Honjo for the AID knockout mice.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carel J. M. van Noesel, Department of Pathology, Academic Medical Center, Meibergdreef 9, 1105 AZ, Amsterdam, The Netherlands; e-mail: c.j.vannoesel@amc.uva.nl.