We developed a real-time copy number polymerase chain reaction assay for deletions on chromosome 20q (del20q), screened peripheral blood granulocytes from 664 patients with myeloproliferative disorders, and identified 19 patients with del20q (2.9%), of which 14 (74%) were also positive for JAK2-V617F. To examine the temporal relationship between the occurrence of del20q and JAK2-V617F, we performed colony assays in methylcellulose, picked individual burst-forming units–erythroid (BFU-E) and colony-forming units–granulocyte (CFU-G) colonies, and genotyped each colony individually for del20q and JAK2-V617F. In 2 of 9 patients, we found that some colonies with del20q carried only wild-type JAK2, whereas other del20q colonies were JAK2-V617F positive, indicating that del20q occurred before the acquisition of JAK2-V617F. However, in colonies from 3 of 9 patients, we observed the opposite order of events. The lack of a strict temporal order of occurrence makes it doubtful that del20q represents a predisposing event for JAK2-V617F. In 2 patients with JAK2-V617F and 1 patient with MPL-W515L, microsatellite analysis revealed that del20q affected chromosomes of different parental origin and/or 9pLOH occurred at least twice. The fact that rare somatic events, such as del20q or 9pLOH, occurred more than once in subclones from the same patients suggests that the myeloproliferative disorder clone carries a predisposition to acquiring such genetic alterations.
Introduction
Deletions of the long arm of chromosome 20 (del20q) have been observed frequently in patients with myeloproliferative disorders (MPDs), myelodysplastic syndromes (MDSs), and acute myeloid leukemia (AML).1,,–4 A common deleted region (CDR) of 2.7 megabases (Mb) was mapped for patients with MPD, and a 2.6-Mb CDR was defined for MDS/AML with an overlap of 1.7 Mb between the 2 CDRs.5,,,,–10 However, to date, it was not possible to identify a gene mutation within the CDR that is functionally linked to the expansion of the del20q clone.11 In MPD patients, del20q was found in the bone marrow from approximately 1% of patients with essential thrombocythemia (ET), up to 9% of polycythemia vera (PV) and up to 12% of primary myelofibrosis (PMF).10,12 The del20q occurred preferentially together with the JAK2-V617F mutation in 28 of 29 MPD patients studied.13 We previously analyzed 2 such patients with del20q and JAK2-V617F and found that the size of the del20q clone by far exceeded the size of the clone carrying JAK2-V617F,14 suggesting that del20q preceded the acquisition of the JAK2-V617F mutation. Here we expanded these studies by screening for del20q in 664 patients with MPD and examined the temporal order of the acquisition of JAK2-V617F and del20q in patients carrying both mutations by analyzing single colonies grown in methylcellulose
Methods
Patients
The collection of blood samples was performed at the study centers in Basel, Switzerland, Vienna, Austria, and Dijon and Nantes, France and was approved by the local Ethics Committees (Ethik Kommission Beider Basel, the Ethik Kommission der Universität Wien und des Allgemeinen Krankenhauses der Stadt Wien-AK, and the institutional boards of Dijon and Nantes). Written informed consent was obtained from all patients in accordance with the Declaration of Helsinki. The diagnosis of MPD was established according to the criteria of the World Health Organization.4
Cells and DNA analysis
Purification of granulocytes and peripheral mononuclear cells (PBMCs) and extraction of DNA were performed as described.15,16 The granulocyte DNA was assayed using a real-time polymerase chain reaction (PCR)–based copy number assay as previously published.16 The following primers were used to detect the copy number of 2 different exon of L3MBTL: 1736-GATCCCAATCAGGACCCCC, 1737-CGCCTGGCACTGACAGGT for exon 2 and 1995-CATGAAGCTGGAGGCTGTTG, 1996-GCCACGCAGACAAGGGAC for exon 9. Primers for LOC221154 (1283-CCATGGACGACGGGTTTCT, 1284-TGTACAGGACGTAGGAGGGTGA) were used as a diploid reference. Samples that showed a decrease in copy number in both exons were validated using primers in the neighboring genes PTPRT(1740-TGCCCCGGAACCATGATA, 1741-GGTCCAGAGGCAGCACGT) and TOX2 (1949-CCTGCCTACTCCTATCAGGCC, 1950-GTTGGACACCATGATGGCTG). The colony assays were performed using PBMCs from patients as published.17 Methylcellulose-based media (no. 4431) containing erythropoietin from StemCell Technologies (Vancouver, BC) was used to grow burst-forming units–erythroid (BFU-E) and colony-forming units–granulocyte (CFU-G). To ensure that plates with an optimal density were obtained so that colonies can be picked without contamination by cells from neighboring colonies, PBMCs were plated at 3 different concentrations (50 000, 100 000, and 200 000 per mL).
Comparative genomic hybridization
The custom oligo comparative genomic hybridization (CGH) array was designed using the eArray platform from Agilent Technologies (Palo Alto, CA). The chips were processed following the instructions of the supplier. Either a pool of male whole blood DNAs (Promega, Madison, WI) or T-cell DNAs from the respective patients were used as reference DNA. The data were analyzed using Capweb software (http://bioinfo-out.curie.fr/CAPweb/).18
Results
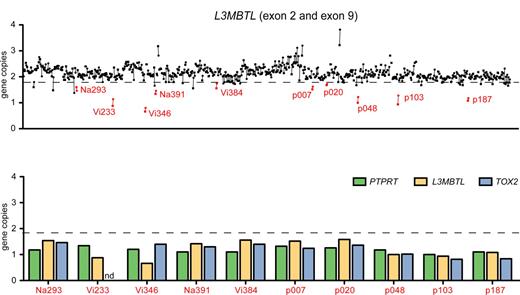
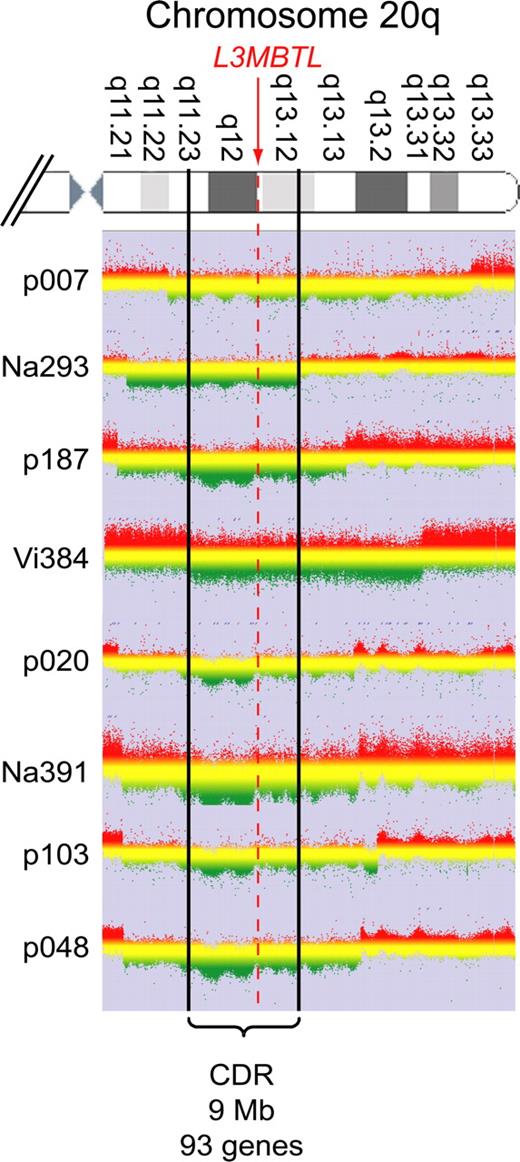
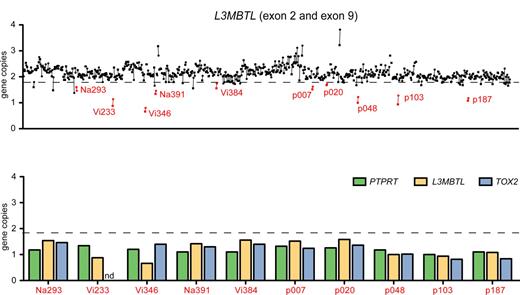
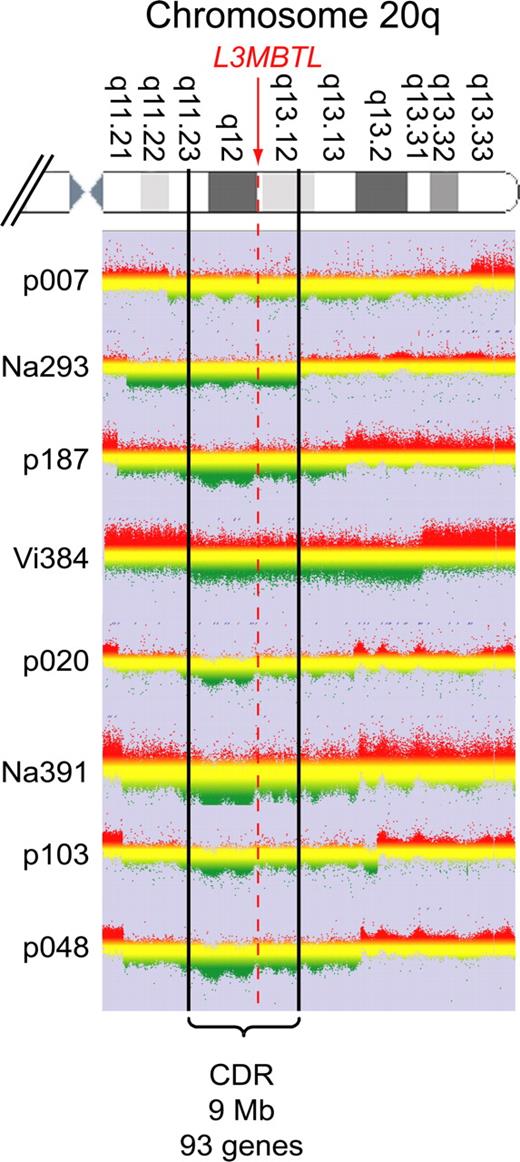
Purified peripheral blood granulocyte DNAs of 664 MPD patients (320 ET, 262 PV, 82 PMF) were screened for del20q by monitoring the copy number of L3MBTL, a gene located in the common deleted region on chromosome 20q. Two real-time PCR assays with primers placed in exons 2 and 9 of L3MBTL were used in parallel (Figure 1A). Patients with decreased gene copy numbers in both assays were further examined with real-time PCR assays for PTPRT and TOX2, 2 genes flanking L3MBTL (Figure 1B). The del20q was confirmed in 19 of 664 (2.9%) MPD patients, or specifically in 4 of 320 (1.3%) ET, 7 of 262 (2.7%) PV, and 8 of 82 (9.8%) PMF. The JAK2-V617F mutation was present in 14 of 19 (74%) patients with del20q, whereas the MPL-W515L was present in 1 patient without JAK2-V617F. No mutations in JAK2 exon 12 were found among the patients with del20q (Table 1). Cytogenetic analysis was available for 8 of the 19 patients with del20q and confirmed the results obtained by real-time PCR in all 8 cases. To map the breakpoints of the individual deletions, we performed custom high-density oligonucleotide CGH arrays on 8 del20q patients from whom DNA of sufficient quality was available. A CDR of 8.98 Mb (35.95-44.93 Mb) containing 93 genes was defined (Figure 2).
Screening for del20q by real-time PCR. (A) The copy number of the L3MBTL gene was determined in DNA from purified peripheral blood granulocytes using 2 SYBR real-time PCR assays with primer pairs located in L3MBTL exon 2 (quadrangles) and exon 9 (circles). Samples with gene copy values less than a threshold of 1.8 in both assays are marked in red and identified by unique patient numbers. (B) Patients with decreased L3MBTL gene copy number were further examined with real-time PCR assays for the neighboring genes PTPRT and TOX2. The decreased copy number was confirmed in all cases. nd indicates not determined.
Screening for del20q by real-time PCR. (A) The copy number of the L3MBTL gene was determined in DNA from purified peripheral blood granulocytes using 2 SYBR real-time PCR assays with primer pairs located in L3MBTL exon 2 (quadrangles) and exon 9 (circles). Samples with gene copy values less than a threshold of 1.8 in both assays are marked in red and identified by unique patient numbers. (B) Patients with decreased L3MBTL gene copy number were further examined with real-time PCR assays for the neighboring genes PTPRT and TOX2. The decreased copy number was confirmed in all cases. nd indicates not determined.
CGH. Granulocyte DNA from patients and a reference DNA were used to perform custom high-density oligonucleotide CGH arrays. The graphs show the log ratios for all probes located on chromosome 20q. Green represents regions deleted in the patient's DNA. The boundaries are marked by a clear drop in the ratio between the patient's DNA and the reference. Patient Vi384 delineates the centromeric and patient Na293 the telomeric border of the CDR, which spans 9 Mb and includes 93 genes.
CGH. Granulocyte DNA from patients and a reference DNA were used to perform custom high-density oligonucleotide CGH arrays. The graphs show the log ratios for all probes located on chromosome 20q. Green represents regions deleted in the patient's DNA. The boundaries are marked by a clear drop in the ratio between the patient's DNA and the reference. Patient Vi384 delineates the centromeric and patient Na293 the telomeric border of the CDR, which spans 9 Mb and includes 93 genes.
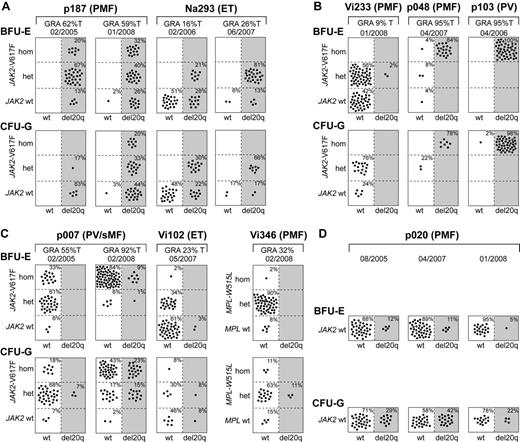
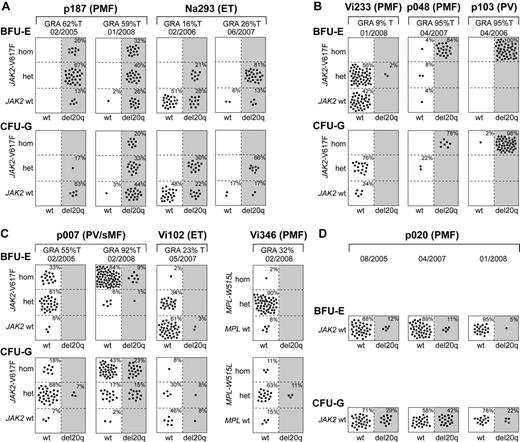
To determine the temporal relationship between the occurrence of del20q and JAK2-V617F, we performed colony assays in methylcellulose, picked single BFU-E and CFU-G colonies grown in the presence of erythropoietin, and genotyped each colony individually for del20q, JAK2-V617F, or in patient Vi346 for MPL-W515L (Figure 3). We observed 3 different patterns: First, in patients p187 and Na293, we found that some del20q-positive colonies carried the wild-type JAK2, whereas other del20q-positive colonies were positive for JAK2-V617F, indicating that the del20q clone is larger than the JAK2-V617F clone (Figure 3A). Because all colonies positive for JAK2-V617F also displayed del20q, we infer that del20q occurred before the JAK2 mutation. Second, in 3 patients, we observed the reverse order, with JAK2-V617F preceding del20q, as illustrated by the presence of JAK2-V617F–positive colonies with and without del20q (Figure 3B). Patient Vi233 appears to have acquired del20q in a cell heterozygous for JAK2-V617F, whereas in patients p048 and p103 the transition to del20q occurred in a cell homozygous for JAK2-V617F. Third, in patients p007, Vi102, and Vi346, we found a more complex pattern (Figure 3C). In the first sample of p007 from February 2005, only a few CFU-G heterozygous for JAK2-V617F carried del20q, whereas colonies homozygous for JAK2-V617F already existed. Two years later, del20q was also present in a subset of colonies homozygous for JAK2-V617F. This pattern could be explained either by postulating 2 independent del20q events or 2 independent 9pLOH events. The del20q-positive BFU-E colonies in patient Vi102 were wild-type for JAK2, and the JAK2-V617F–positive colonies were all negative for del20q, which is compatible with originating from 2 independent clones. However, the finding of a CFU-G colony positive for both JAK2-V617F and del20q suggests that either del20q or JAK2-V617F occurred twice independently. Patient Vi346 was negative for JAK2-V617F but carried the MPL-W515L mutation. The del20q was only detectable in CFU-G colonies, and all of these colonies were also heterozygous for MPL-W515L. Some colonies were homozygous for MPL-W515L but negative for del20q.
Single colony assays for del20q and JAK2-V617F. PBMCs were grown in methylcellulose in the presence of erythropoietin. Single erythroid colonies (BFU-E) and granulocytic colonies (CFU-G) were picked and analyzed individually for the presence of del20q and JAK2-V617F by microsatellite PCR and allele-specific PCR, respectively. Each colony is represented by a dot that is placed into one of 6 quadrangles representing the 6 possible genotypes: wild-type (wt), heterozygous (het), and homozygous (hom) for JAK2-V617F or MPL-W515L on the vertical axis, and absence (open quadrangles) or presence of del20q (gray quadrangles) on the horizontal axis. Results for BFU-Es are shown in the upper part and CFU-Gs in the lower part of the panels. The unique patient numbers, the diagnoses (PMF, ET, PV, sMF indicates secondary myelofibrosis), the allelic ratio of JAK2-V617F or MPL-W515L in purified granulocytes (%T), and the date of the sample drawing are shown above the corresponding boxes. (A) This group of patients has acquired the del20q before JAK2-V617F, as demonstrated by the presence of JAK2 wt colonies carrying del20q. For both patients shown, 2 sequential samples were analyzed. (B) In this group of patients, del20q was acquired in a cell already carrying JAK2-V617F, as indicated by the presence of JAK2-V617F-positive colonies with or without del20q. (C) Patients with a complex pattern of del20q acquisition that does not comply with a linear temporal order of events. In patient Vi346, the del20q coexists with MPL-W515L. (D) A patient with del20q that is negative for the JAK2-V617F mutation.
Single colony assays for del20q and JAK2-V617F. PBMCs were grown in methylcellulose in the presence of erythropoietin. Single erythroid colonies (BFU-E) and granulocytic colonies (CFU-G) were picked and analyzed individually for the presence of del20q and JAK2-V617F by microsatellite PCR and allele-specific PCR, respectively. Each colony is represented by a dot that is placed into one of 6 quadrangles representing the 6 possible genotypes: wild-type (wt), heterozygous (het), and homozygous (hom) for JAK2-V617F or MPL-W515L on the vertical axis, and absence (open quadrangles) or presence of del20q (gray quadrangles) on the horizontal axis. Results for BFU-Es are shown in the upper part and CFU-Gs in the lower part of the panels. The unique patient numbers, the diagnoses (PMF, ET, PV, sMF indicates secondary myelofibrosis), the allelic ratio of JAK2-V617F or MPL-W515L in purified granulocytes (%T), and the date of the sample drawing are shown above the corresponding boxes. (A) This group of patients has acquired the del20q before JAK2-V617F, as demonstrated by the presence of JAK2 wt colonies carrying del20q. For both patients shown, 2 sequential samples were analyzed. (B) In this group of patients, del20q was acquired in a cell already carrying JAK2-V617F, as indicated by the presence of JAK2-V617F-positive colonies with or without del20q. (C) Patients with a complex pattern of del20q acquisition that does not comply with a linear temporal order of events. In patient Vi346, the del20q coexists with MPL-W515L. (D) A patient with del20q that is negative for the JAK2-V617F mutation.
In one patient (Na391) with 50% of erythroblasts in the peripheral blood, all colonies were positive for both del20q and JAK2-V617F, which precluded us from determining the order of events (not shown), and one patient (p020) had del20q without a mutation in the JAK2 or MPL gene (Figure 3D). We analyzed p020 at 3 different time points, and we were able to detect an increase in del20q-positive colonies in CFU-Gs in 2007. However, one year later, the percentage of del20q-positive colonies was again comparable with the first time point. An increase in the percentage of del20q-positive colonies was observed in serial samples from Na293 and p007, whereas in p187 the colonies were all del20q-positive and remained so during follow-up, except for one BFU-E and one CFU-G (Figure 3A,C).
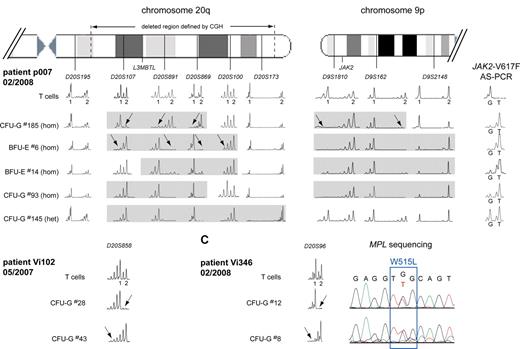
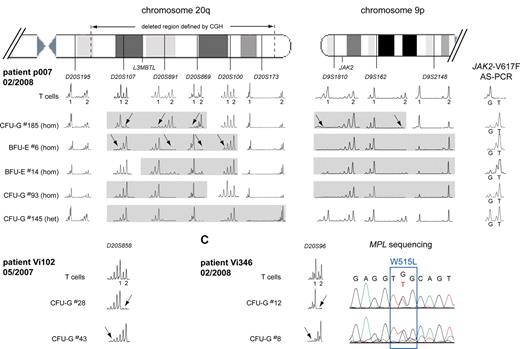
To examine the complex pattern in patients p007, Vi102, and Vi346, we performed a more detailed microsatellite analysis for regions on chromosomes 9 and 20 (Figure 4). In p007, the deletion in colony no. 185 affected the chromosome 20q of a different parental origin than in colony no. 6, indicating that 2 independent deletion events must have occurred (Figure 4A left panel). Furthermore, 4 different sizes of the latter del20q haplotype were detected when additional colonies were analyzed. This pattern could arise when a small del20q event was followed by sequential events that increased the size of the deleted region in the same cell, or alternatively, each of the del20q regions could represent a separate de novo deletion event. At present, we cannot distinguish between these possibilities. In addition, colony no. 185 also differed in the size of the 9pLOH region, suggesting the presence of 2 different 9pLOH events (Figure 4A right panel). In contrast, analysis of the deleted regions in colonies from other patients showed a unique size for each of the del20q regions (not shown), making it doubtful that the pattern observed in p007 is the result of artifacts that occurred during the methylcellulose culture. Evidence for 2 independent del20q events was also found in patients Vi102 (Figure 4B) and Vi346 (Figure 4C). In both cases, colonies were found that showed loss of heterozygosity affecting 2 different parental chromosomes 20.
Multiple del20q and 9pLOH events in colonies from 3 MPD patients. (A) The deleted region on chromosome 20q was mapped in individual colonies from patient p007 using 6 microsatellites distributed along chromosome 20q. T-cell DNA was used to define the 2 alleles for each informative microsatellite. The gray boxes mark the deleted region for each individual colony. del20q in colony no. 185 created a different haplotype than the deletion in colony no. 6 (marked by arrows), indicating that 2 del20q events occurred independently and affected the chromosome 20q of different parental origin. Furthermore, 4 different sizes of the del20q haplotype were detected when additional colonies were analyzed. The results from the mapping of the 9pLOH region are shown on the right side. The 9pLOH region is smaller in colony no. 185, as shown by the heterozygosity of D9S2148 in this colony. (B) Two separate del20q events affecting the chromosome 20q of different parental origin occurred in patient Vi102. (C) Two separate del20q events affecting the chromosome 20q of different parental origin occurred in patient Vi346. The chromatograms for the MPL-W515L mutation are shown for the 2 colonies analyzed.
Multiple del20q and 9pLOH events in colonies from 3 MPD patients. (A) The deleted region on chromosome 20q was mapped in individual colonies from patient p007 using 6 microsatellites distributed along chromosome 20q. T-cell DNA was used to define the 2 alleles for each informative microsatellite. The gray boxes mark the deleted region for each individual colony. del20q in colony no. 185 created a different haplotype than the deletion in colony no. 6 (marked by arrows), indicating that 2 del20q events occurred independently and affected the chromosome 20q of different parental origin. Furthermore, 4 different sizes of the del20q haplotype were detected when additional colonies were analyzed. The results from the mapping of the 9pLOH region are shown on the right side. The 9pLOH region is smaller in colony no. 185, as shown by the heterozygosity of D9S2148 in this colony. (B) Two separate del20q events affecting the chromosome 20q of different parental origin occurred in patient Vi102. (C) Two separate del20q events affecting the chromosome 20q of different parental origin occurred in patient Vi346. The chromatograms for the MPL-W515L mutation are shown for the 2 colonies analyzed.
Discussion
In the present study, we assessed the potential connection between JAK2-V617F and del20q by analyzing the temporal order of acquisition of the 2 events. Our copy number assay allowed us to screen for del20q by real-time PCR in peripheral blood from a large number of MPD patients. The L3MBTL gene, which we have chosen for the copy number assay, is located within the CDR shared between patients with MPD and MDS/AML.9 This assay requires that the majority of granulocytes carry del20q and therefore selects for del20q events that dominate in granulocytes in the peripheral blood. Nevertheless, our observed frequencies in patients with ET, PV, and PMF of del20q are comparable with results obtained with cytogenetic studies of bone marrow.10,12 Furthermore, real-time PCR and cytogenetic analysis were in agreement in all 8 patients in whom both analyses have been performed. We found no difference in the frequencies of JAK2-V617F in PV and PMF patients with and without del20q. Our results do not confirm a previously reported preferential association of del20q with JAK2-V617F-positive cases of PMF (Table 2).13 The reason for this discrepancy is currently unclear. In ET, 4 of 4 patients with del20q were JAK2-V617F–positive, similar to 5 of 5 del20q-positive ET cases reported previously (Table 2).13 Thus, by combining the studies, 9 of 9 ET patients with del20q were also JAK2-V617F–positive. However, this association should be examined in a larger series of patients with del20q. Mapping of the del20q region by CGH revealed that in most patients large deletions have occurred (Figure 2) and that the CDR derived from analyzing granulocytes overlaps with the published region obtained by mapping bone marrow samples.9
Disparity between the size of the JAK2-V617F–positive clone and the clone determined by analysis of the X-chromosome inactivation pattern,14,19 presence of endogenous erythroid colonies that are negative for JAK2-V617F,17,20,21 and coexistence of JAK2-V617F and MPL-W515L/K in the same patients,22,23 suggested that clonal events preceding the acquisition of JAK2-V617F exist in patients with MPD. We previously found 2 patients in whom the clone carrying del20q was larger than the clone positive for JAK2-V617F, and we hypothesized that JAK2-V617F preferentially occurs on the background of clonal hematopoiesis, which in some cases may be caused by del20q.14 Our single clone analysis confirmed the previously observed temporal order of events in 2 patients (Figure 3A), but in 3 additional patients we detected the inverse order, ie, JAK2-V617F preceding del20q (Figure 3B). Thus, there appears to be no strict temporal order of acquisition, making it doubtful that del20q represents a predisposing event for JAK2-V617F. At present, we cannot formally exclude the possibility that del20q may represent a “passenger mutation,” ie, a genetic alteration without functional consequence.24,25 The fact that in serial samples of patients Na293 and p007 we observed an expansion of the del20q subclone (Figure 3) could be interpreted in favor of a growth advantage provided by del20q and thus for a role as a “driver mutation.” Similarly, in patients p048 and p103, nearly all colonies were del20q-positive, suggesting a growth advantage of the cells harboring del20q.
Unexpectedly, in 3 patients (p007, Vi102, and Vi346), we found evidence for several independent del20q events and in p007 for 2 independent 9pLOH events (Figure 4). Patient p007 was diagnosed with PV 22 years ago and for 11 years showed signs of a progression to secondary myelofibrosis. The long disease duration and/or progression to spent phase may favor accumulation of de novo mutation events. However, the disease duration in ET patient Vi102 (4 years) and PMF patient Vi346 (6 years) was shorter. Genomic instability as a consequence of JAK2-V617F was recently reported.26 This could explain the findings in p007. However, in patient Vi102, one of the del20q events occurred in a JAK2-V617F–negative cell (Figure 3C) and patient Vi346 was negative for JAK2-V617F, suggesting that genomic instability in these patients is independent of JAK2.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Michael Medinger for contributing clinical data, Jean-Luc Harousseau (Nantes) and Bruno Villemagne (La Roche sur Yon) for providing patient samples, and Jürg Schwaller and Alex Theocharides for comments on the manuscript.
This work was supported by the Swiss National Science Foundation (grants 310000-108006/1) and the Swiss Cancer League (OCS-01 742-08-2005) to R.C.S. and from the Initiative for Cancer Research of the Medical University of Vienna and the Austrian Science Fund FWF (P20033-B11) to R.K.
Authorship
Contribution: F.X.S. performed research, analyzed data, and wrote the paper; R.J., R.L., and H.H.-S. performed research; S.H., F.G., A.T., H.G., and R.K. provided essential reagents and analyzed data; and R.C.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Radek C. Skoda, Department of Biomedicine, Experimental Hematology, University Hospital Basel, Hebelstrasse 20, 4031 Basel, Switzerland; e-mail: radek.skoda@unibas.ch.