Abstract
The Src homology 2 domain–containing leukocyte phosphoprotein of 76 kilodaltons (SLP-76) is a cytosolic adaptor protein essential for thymocyte development and T-cell activation. It contains a sterile-α motif (SAM) domain, 3 phosphotyrosine motifs, a proline-rich region, and a Src homology 2 domain. Whereas the other domains have been extensively studied, the role of the SAM domain in SLP-76 function is not known. To understand the function of this domain, we generated SLP-76 knockin mice with the SAM domain deleted. Analysis of these mice showed that thymocyte development was partially blocked at the double-positive to single-positive transition. Positive and negative thymic selection was also impaired. In addition, we analyzed T-cell receptor (TCR)–mediated signaling in T cells from these mutant mice. TCR-mediated inositol 1,4,5-triphosphate production, calcium flux, and extracellular signal-regulated kinase activation were decreased, leading to defective interleukin-2 production and proliferation. Moreover, despite normal association between Gads and SLP-76, TCR-mediated formation of SLP-76 microclusters was impaired by the deletion of the SAM domain. Altogether, our data demonstrated that the SAM domain is indispensable for optimal SLP-76 signaling.
Introduction
The Src homology (SH)2 domain-containing leukocyte phosphoprotein of 76 kilodaltons (SLP-76) is a hematopoietic cell-specific adaptor protein that plays a critical role in thymocyte development1,2 and T-cell receptor (TCR) signaling.3 The N terminus of SLP-76 contains 3 tyrosine residues that are phosphorylated upon TCR engagement.4 These phosphotyrosines serve as docking sites for recruiting the Rac/Rho guanine nucleotide exchange factor Vav,5 the Tec-family protein tyrosine kinase Itk,6-8 and the adaptor protein Nck.9 The central proline-rich region of SLP-76 contains a specific sequence (amino acids 224-244) that constitutively binds to the adaptor protein Gads.10-12 An additional sequence (named P1 domain) in this proline-rich region also mediates a constitutive interaction with phospholipase C (PLC)-γ1.13,14 The C-terminal SH2 domain of SLP-76 provides TCR-dependent association with TCR-dependent association with adhesion- and degranulation-promoting adaptor protein (ADAP)15,16 and hematopoietic progenitor kinase 1 (HPK1).17,18 Upon TCR engagement, SLP-76 is phosphorylated by ζ-associated protein 70 (zap-70) and is recruited to the membrane-associated adaptor protein linker for activation of T cells (LAT) through the binding of Gads. Together, LAT and SLP-76 nucleate a large signaling complex, which couples TCR-proximal signaling to downstream biochemical events, such as calcium flux and mitogen-activated protein kinase (MAPK) activation.
SLP-76 is essential for the pre-TCR signaling that drives thymocyte development through the double-negative (DN)3 checkpoint. SLP-76−/− mice suffer from a profound block of thymocyte development at the DN3 stage, and completely lack double-positive (DP) thymocytes and mature T cells.1,2 Recent studies on CD4Cre/SLP-76 conditional knockout mice show that SLP-76 also plays an important role in mature TCR-mediated thymic selections, because absence of SLP-76 in DP thymocytes prevents them from further differentiating into single-positive (SP) thymocytes.19 The function of SLP-76 in mature TCR signaling was studied primarily in cell lines. Jurkat T cells deficient in SLP-76 (J14 cells) are defective in TCR-dependent calcium flux and extracellular signal-regulated kinase (ERK) activation, and are unable to activate the interleukin (IL)–2 nuclear factor of activated T cells (NFAT)/activator protein-1 (AP-1) promoter.3
The structural requirement of the SLP-76 domains for mediating thymopoiesis was studied using transgenic mice expressing various forms of mutant SLP-76 on a SLP-76−/− background. The SLP-76 Y3F mutant harboring Y112F, Y128F, and Y145F point mutations can partially rescue thymocyte development, as indicated by the accumulation of DN cells and the markedly reduced number of DP and SP cells.20 The SLP-76 Δ224-244 mutant, which fails to interact with Gads, is able to restore thymopoiesis in SLP-76−/− mice relatively better than the Y3F mutant, but not to wild-type levels.20,21 SLP-76 with a R448K point mutation in the SH2 domain, which prevents it from binding to ADAP, is able to efficiently reconstitute thymocyte development, suggesting that the SLP-76/ADAP association is largely dispensable for thymopoiesis.20 However, in the absence of the SH2 domain, the mutant SLP-76 can only partially rescue T-cell development.21 TCR signaling, including calcium flux and ERK activation, is impaired to various degrees in SLP-76−/− mice reconstituted with Y3F, Δ224-244, or ΔSH2 SLP-76 mutants. T-cell proliferation and IL-2 production are markedly defective as well.20,21 Consistent with the transgenic data, J14 Jurkat T cells expressing these SLP-76 mutants exhibit a partial reconstitution of TCR signaling by each mutant.13
Despite the extensive structure-function analysis of SLP-76, little is known about the role of the sterile-α motif (SAM) domain in the N terminus of SLP-76. SAM domains were identified more than a decade ago based on a conserved ∼70-amino-acid domain in 14 eukaryotic proteins.22 They were predicted to feature a high content of α-helices and to participate in protein-protein interactions.22,23 Since then, more than 1300 SAM-containing proteins have been identified in various organisms, ranging from yeast to humans. SAM domains can be found in all subcellular compartments and participate in a wide variety of cellular processes, from signal transduction to transcriptional/translational regulation.24 Most of the SAM domains studied to date are involved in protein interaction with different binding properties. Some SAM domains can interact with themselves25-27 or with other SAM domain-containing proteins26,28 to form oligomers. Others can interact with non-SAM–containing proteins.29 In addition to their role in mediating protein association, certain SAM domains also exhibit RNA-binding properties.30,31 Given such versatility, understanding the function of SAM domains relies heavily upon extensive experimental characterization in individual proteins.
The SAM domain of SLP-76 spans from amino acids 12 to 78 at the N terminus. Its requirement for SLP-76 function in mediating thymocyte development and TCR signaling has yet to be explored. Interestingly, transgenic expression of the Δ2-156 SLP-76 mutant, in which the entire N terminus, including both the SAM domain and 3 phosphotyrosine motifs, is deleted, fails to restore thymopoiesis in SLP-76−/− mice, as indicated by the complete block in thymocyte development at the DN3 stage.21 However, in SLP-76−/− mice reconstituted with the SLP-76 Y3F mutant, all subsets of thymocytes could be found, albeit reduced.20 These data suggested a potential role for the SAM domain in mediating SLP-76 functions. In this study, we generated SLP-76 knockin mice in which the SAM domain was successfully deleted. Our data showed for the first time that the SAM domain of SLP-76 plays a critical role in both thymic selection and TCR signaling.
Methods
Generation of ΔSAM–SLP-76 knockin mice
The slp-76 genomic fragments were amplified from embryonic stem (ES) cells by polymerase chain reaction (PCR), sequenced, and cloned into the targeting plasmid (Figure 1). The short arm contains a 1.5-kb sequence upstream of the slp-76 start codon and a modified exon 1 with the sequence, ATGGCCTTGAAGAATTCAAG, at its 3′ end. The long arm comprises a 5-kb sequence in the intron between exons 3 and 4. G418-resistant ES cells were screened by PCR and further confirmed by Southern blotting analysis of the genomic DNA (digested with XbaI). The genomic probe used in the Southern was amplified from ES cells using the following primers: 5′-CTC CCT GGT GAT TTA TCT GAG G-3′ and 5′-ACC AGG ACA ATG ACA ATG AAC A-3′. The correctly targeted ES cells were injected into blastocysts to generate chimeric mice. To delete the PGK-Neo fragment, chimeric mice were crossed with the β-actin Cre transgenic mice to generate SLP-76m/+ mice. SLP-76m/+ mice were subsequently backcrossed with C57BL/6 mice for at least 6 generations before analysis. Genotyping of littermates was done by PCR using 3 primers, as follows: 5′-GAA TCA GAA GAG CCA AGG ACA C-3′, 5′-ACA GTG GGT TGT GTC TGA CAA G-3′, and 5′-GGT CTC TCC CAT CCC TTT ATT T-3′. To confirm deletion of the sequence encoding the SAM domain, the slp-76 RNA was amplified by reverse transcription (RT)-PCR using the following 2 primers: 5′-AGA GCA TCT GGG AAT CAG AAG A-3′ and 5′-GGC TTT CTG TCT CCT CAA GAA-3′. C57BL/6 mice and β-actin Cre transgenic mice were purchased from The Jackson Laboratory. HY-TCR transgenic mice were purchased from Taconic Farms. Gads−/− mice were provided by A. Cheng. Mice were housed in specific pathogen-free conditions. All mice were used in accordance with National Institutes of Health guidelines. The experiments described in this study were reviewed and approved by the Duke University Institutional Animal Care Committee.
FACS analysis and antibodies
Fluorescence-conjugated antibodies used in flow cytometry, such as anti-CD4, CD8, CD25, CD44, CD62L, CD5, CD69, heat-stable antigen (HSA), TCR-β, and TCR-γδ, were all purchased from eBioscience. For cell surface marker staining, single-cell suspensions were prepared from mouse thymi, lymph nodes, or spleens, and were incubated with the 2.4G2 antibody (anti-Fcγ receptor) before staining with different antibody mixtures. For intracellular staining of SLP-76, cells were fixed, permeabilized, and then stained with anti-SLP-76 (Cell Signaling Technology; no. 4958), followed by Alexa Fluor 488 goat anti–rabbit immunoglobulin (Ig)G (Molecular Probes). Fluorescence-activated cell sorter (FACS) data were acquired on FACSDiva (BD Biosciences) and analyzed with the Flowjo software.
Cell separation
CD4+CD8+ DP thymocytes and CD4+CD44low T cells were sorted on FACSDiva (BD Biosciences). The postsort purity was greater than 99%. CD4+ T cells were purified using the EasySep Mouse CD4+ T Cell Enrichment Kit (StemCell Technologies). The purity was greater than 90%.
Western blotting and immunoprecipitation
Sorted DP thymocytes or purified CD4+ T cells before or after stimulation via CD3 were lysed in radioimmunoprecipitation assay buffer. For coimmunoprecipitation, thymocytes were lysed in 1% Brij lysis buffer; the lysates were then incubated with sheep anti–SLP-76 polyclonal antibody as well as protein G-coupled Sepharose beads for 1 hour. The protein lysates or immunoprecipitates were resolved on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes (Bio-Rad). The membranes were then blotted with different primary antibodies. The anti-pERK, pPLC-γ1, PLC-γ1, and SLP-76 antibodies were purchased from Cell Signaling Technology. Anti-pSLP-76 (Y128) was purchased from BD Biosciences. Anti-ERK2 was purchased from Santa Cruz Biotechnology. Anti-phosphotyrosine (4G10) and anti-Gads were purchased from Upstate Biotechnology. For secondary antibodies, Alexa Fluor 680 anti–mouse IgG (Molecular Probes) or IRDye 800 anti–rabbit IgG (Rockland) was used accordingly. The membranes were scanned by the LI-COR Odyssey infrared imaging system.
Cell proliferation and IL-2 production
Purified CD4+ T cells (for IL-2 production) or sorted CD4+CD44low T cells (for proliferation) were seeded in U-bottom 96-well plates (2 × 105 cells/150 μL) with the indicated concentration of plate-coated anti-CD3 plus soluble anti-CD28 (1 μg/mL), or with phorbol myristate acetate (PMA; 20 ng/mL) plus ionomycin (0.5 μg/mL). Triplicates were performed in each assay. For IL-2 production, after 8 hours, 50 μL of supernatant was collected from each well and subjected to IL-2 enzyme-linked immunosorbent assay (ELISA). For cell proliferation, after 36 hours, cells were pulsed with 1 μCi of [3H]-thymidine for an additional 6 hours, and were then harvested for scintillation counting. Cell proliferation was represented by the counted radioactivity (counts per minute [CPM]). For the IL-2 ELISA, anti–IL-2 capture antibody, biotinylated anti–IL-2 detection antibody, horseradish peroxidase–conjugated avidin, and recombinant mouse IL-2 standard were all purchased from eBioscience. The tetramethylbenzidine peroxidase substrate kit was purchased from Bio-Rad. The ELISA was performed according to the recommended protocol from eBioscience.
Calcium flux
Total thymocytes or splenocytes were first loaded with Indo-1 (Molecular Probes) in loading buffer (1× Hanks balanced salt solution [HBSS] with 10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] and 1% fetal bovine serum [FBS]) for 30 minutes and then stained with phycoerythrin anti-CD4 and fluorescein isothiocyanate anti-CD8 antibodies. Cells were then resuspended in loading buffer (107 cells/mL). Calcium flux was initiated by the addition of biotinylated anti-CD3 (5 μg/mL) and anti-CD4 (1 μg/mL) or anti-CD8 (1 μg/mL), followed by cross-linking with streptavidin (25 μg/mL; Sigma-Aldrich). Ionomycin (2 μg/mL; Sigma-Aldrich) was used to induce TCR-independent calcium flux. Calcium flux was assayed by monitoring the fluorescence emission ratio at 405/510 nm with FACStar (BD Biosciences) and analyzed using the Flowjo software. For the intracellular calcium release assay, a modified loading buffer (Ca2+-free 1 × HBSS with 10 mM HEPES, 1% FBS, and 5 mM ethyleneglycoltetraacetic acid [EGTA]) was used.
Inositol 1,4,5-triphosphate assay
Total thymocytes or purified CD4+ T cells were either left untreated or stimulated by CD3 cross-linking at 37°C for 2 minutes. The incubation was terminated with an equal volume of ice-cold 20% (wt/vol) trichloroacetic acid (Sigma-Aldrich). The soluble fraction was then washed 3 times with 10 volumes of water-saturated diethyl ether to eliminate the acid. Inositol 1,4,5-triphosphate (IP3) levels were measured using a GE Healthcare IP3 assay kit (TRK1000). The procedures are described in the manual provided by the manufacturer.
SLP-76 clustering and cellular imaging
The cDNA sequences encoding human wild-type SLP-76 or ΔSAM–SLP-76 (with amino acids 12-78 deleted) were cloned into pEGFP-N3 vector to express SLP with green fluorescent protein (GFP) fused at the C terminus. J14 cells were transfected with the pEGFP–SLP-76 or pEGFP–ΔSAM–SLP-76 plasmids by electroporation. GFP+ cells with comparable fluorescence intensity were sorted by FACS. The T-cell spreading assay was performed, as previously described,32 with some modifications. Coverslips were sequentially coated with biotinylated poly(L-lysine), streptavidin, and biotin anti–human CD3ϵ antibody (UCHT1). Such treated coverslips were then mounted onto a holder so that the stimulatory side served as the bottom of a chamber. Sorted GFP+ J14 cells were loaded into the chamber, and imaging data were collected every 20 seconds after the cells made contact with the coverslip. Alternatively, cells were added into the chamber, centrifuged onto the coverslips, and incubated at 37°C for 10 more minutes to allow maximal activation. The live cell imaging was performed on a Zeiss Observer D1 station equipped with a CoolSNAPHQ charge-coupled device camera (Roper Scientific) and a high-speed automatic objective stage for multiple Z stack recording. The images were collected with 40× objective lens and 2.5× camera zoom.
For each cell at each time point, GFP data were collected over 21 continuous vertical Z positions bracketing the cell/coverslip interface. Images corresponding to the interface were then identified within the stacks. To calculate the average intensity of the clusters, the Z stack data were first processed by 3D deconvolution using the AutoQuant X software (Media Cybernetics). Average fluorescence of the 2 Z positions immediately before and after the interface was subtracted from the image corresponding to the interface. The brightness of the clusters was represented as the average intensity per pixel. Unless otherwise indicated, all imaging manipulation and analysis were done using the MetaMorph software suite (Molecular Probes).
Results
Generation of SLP-76 knockin mice
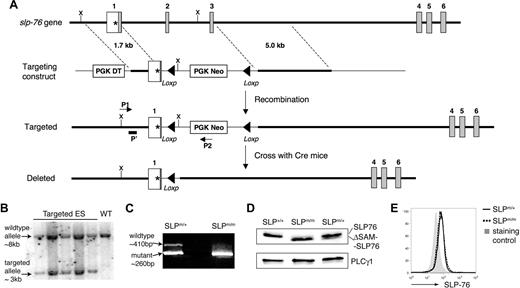
To understand the role of the SAM domain in SLP-76–mediated signaling, we generated knockin mice that express a SAM-domain–truncated form of SLP-76. The targeting strategy is illustrated in Figure 1A. The SAM domain of SLP-76 spans from amino acids 12 to 78, and its corresponding genomic sequence covers part of both exons 1 and 4, and the entirety of exons 2 and 3. The intron between exons 3 and 4 is approximately 14 kb long, making homologous recombination potentially difficult. We replaced the exons encoding the SLP-76 amino acids 6-63 with PGK-Neo and left exon 4 (encoding the remaining 15 amino acids of the SAM domain) intact. To avoid the reading frameshift after the deletion, the 3′ end of exon 1 was modified, resulting in the addition of 2 amino acids (Ser-Arg). We reason that deletion of most (51 of 66 amino acids) of the SAM domain should render it nonfunctional.
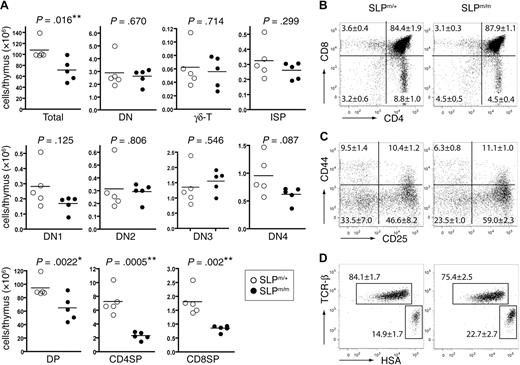
Generation of ΔSAM–SLP-76 knockin mice. (A) Illustration of the ΔSAM–SLP-76 knockin targeting strategy. The first 6 slp-76 exons are indicated. The initiation codon (*) is at exon 1. The SAM domain spans from exons 1 to 4. Part of exon 1, along with exons 2 and 3, is replaced by PGK-Neo, which is later deleted upon crossing with β-actin Cre transgenic mice. ▴, Represent the Loxp sites. P1 and P2 represent PCR primers used for ES clone screening. P′ represents a probe used in Southern blotting analysis to confirm correctly targeted ES clones. X = XbaΙ, the restriction endonuclease used to digest genomic DNA of ES cells for Southern blotting. (B) Southern blotting analysis of genomic DNA from 5 ES cell clones (lanes 1-5) and wild-type ES cells (lane 6). (C) RT-PCR products from SLP-76m/+ and SLP-76m/m thymocytes using 2 primers flanking the deleted region. (D) SLP-76 protein expression. DP thymocytes from SLP-76+/+, SLP-76m/+, and SLP-76m/m littermates were FACS sorted and subsequently lysed. Postnuclear lysates were subjected to Western blotting with anti-SLP-76. Anti-PLC-γ1 blot is used as a loading control. (E) Intracellular staining of SLP-76 in CD4+ splenic T cells from SLP-76m/+ (solid line) and SLP-76m/m (dotted line) littermates. The gray area represents staining control using B220+ lymphocytes.
Generation of ΔSAM–SLP-76 knockin mice. (A) Illustration of the ΔSAM–SLP-76 knockin targeting strategy. The first 6 slp-76 exons are indicated. The initiation codon (*) is at exon 1. The SAM domain spans from exons 1 to 4. Part of exon 1, along with exons 2 and 3, is replaced by PGK-Neo, which is later deleted upon crossing with β-actin Cre transgenic mice. ▴, Represent the Loxp sites. P1 and P2 represent PCR primers used for ES clone screening. P′ represents a probe used in Southern blotting analysis to confirm correctly targeted ES clones. X = XbaΙ, the restriction endonuclease used to digest genomic DNA of ES cells for Southern blotting. (B) Southern blotting analysis of genomic DNA from 5 ES cell clones (lanes 1-5) and wild-type ES cells (lane 6). (C) RT-PCR products from SLP-76m/+ and SLP-76m/m thymocytes using 2 primers flanking the deleted region. (D) SLP-76 protein expression. DP thymocytes from SLP-76+/+, SLP-76m/+, and SLP-76m/m littermates were FACS sorted and subsequently lysed. Postnuclear lysates were subjected to Western blotting with anti-SLP-76. Anti-PLC-γ1 blot is used as a loading control. (E) Intracellular staining of SLP-76 in CD4+ splenic T cells from SLP-76m/+ (solid line) and SLP-76m/m (dotted line) littermates. The gray area represents staining control using B220+ lymphocytes.
Successful targeting was confirmed by Southern blotting (Figure 1B), and the positive ES clones were used to generate chimeric mice, which were subsequently crossed with β-actin Cre transgenic mice to generate SLP-76m/+ mice. These heterozygous mice were backcrossed onto a B6 background for at least 6 generations. SLP-76m/m mice were generated from SLP-76m/+ breeding at an expected frequency. In contrast to SLP-76−/− mice, which frequently succumb to prenatal systemic hemorrhage, SLP-76m/m pups had no obvious signs of bleeding and appeared grossly healthy.
To confirm correct gene targeting and successful deletion of the sequence encoding the SAM domain, total RNAs were prepared from thymocytes from SLP-76m/+ and SLP-76m/m mice and were used in RT-PCR with 2 primers flanking the deleted region. As predicted, the PCR product amplified from the mutant sequence was approximately 150 base pairs shorter than that from the wild type (Figure 1C). Successful mRNA splicing between exon 1 (modified) and exon 4 was confirmed by sequencing the PCR products (data not shown). We also examined the expression of SLP-76 protein in sorted DP thymocytes from SLP-76+/+, SLP-76m/+, and SLP-76m/m littermates by Western blotting with polyclonal anti-SLP antisera. As expected, a truncated form of the protein was detected in SLP-76m/m cells (Figure 1D), and its expression level was comparable with that of the wild-type protein in SLP-76+/+ cells. Interestingly, SLP-76m/+ DP cells preferentially expressed the wild-type form of SLP-76 protein. Moreover, intracellular staining followed by FACS analysis revealed comparable expression levels of SLP-76 protein in peripheral T cells from SLP-76m/+ and SLP-76m/m littermates (Figure 1E). These data showed that the ΔSAM–SLP-76 protein is expressed in SLP-76m/m T cells at a level similar to that of wild-type SLP-76 in SLP-76+/+ cells.
Defective thymocyte development in SLP-76 knockin mice
Next, we analyzed thymocyte development in SLP-76+/+, SLP-76m/+, and SLP-76m/m mice. T-cell development appeared normal in SLP-76m/+ mice. Both percentages and total numbers of different thymocyte and mature T-cell subsets in SLP-76m/+ mice were similar to those observed in SLP-76+/+ littermates (data not shown).
Thymi from the 4-week-old SLP-76m/m mice appeared slightly smaller than those from SLP-76m/+ littermates (data not shown), and the total number of thymocytes was decreased (Figure 2A). The percentage of DN thymocytes in SLP-76m/m mice was slightly increased (Figure 2B), whereas the overall DN thymocyte numbers appeared relatively normal (Figure 2A). As shown by the CD25 versus CD44 profile in Figure 2C, the percentage of DN3 SLP-76m/m thymocytes increased moderately from 46.6% to 59.0%, accompanied by a mild decrease in the percentage of the DN4 subset (from 33.5%-23.5%). Yet the differences in the absolute numbers of the DN subsets between the mutant and control mice did not appear to be statistically significant (Figure 2A). Data obtained from a 5-bromo-2′-deoxyuridine incorporation assay showed that all 4 DN subsets in the SLP-76m/m mice proliferated at similar rates as their counterparts from control SLP-76m/+ mice (data not shown), suggesting that the relatively normal number of mutant DN thymocytes was not caused by any compensatory proliferation effect. We also found that the absolute number of CD4−CD8+HSAhighTCRlow intermediate SP (ISP) cells, which are immature precursors of DP thymocytes, remained relatively normal (Figure 2A). Altogether, these data suggested that the deletion of the SAM domain slightly impaired SLP-76 function in pre-TCR signaling, resulting in a minor developmental block at the DN3 to DN4 checkpoint.
Thymocyte development in ΔSAM–SLP-76 knockin mice. (A) Total numbers of different thymocyte populations from 4-week-old SLP-76m/+ and SLP-76m/m littermates. Five mice for each genotype were analyzed. Statistical analysis was performed using 2-tailed Student t test. *P < .05; **P < .01. (B-D) Thymocytes from 4-week-old SLP-76m/+ and SLP-76m/m littermates (5 mice for each genotype) were analyzed by flow cytometry. FACS plots shown are from 1 representative of each genotype. (B) Surface expression of CD4 versus CD8 on total thymocytes. (C) Expression of CD25 versus CD44 on CD4−CD8− DN thymocytes. (D) Expression of TCR-β versus HSA on CD4−CD8+ thymocytes. The numbers in each panel represent the average percentages of the gated populations.
Thymocyte development in ΔSAM–SLP-76 knockin mice. (A) Total numbers of different thymocyte populations from 4-week-old SLP-76m/+ and SLP-76m/m littermates. Five mice for each genotype were analyzed. Statistical analysis was performed using 2-tailed Student t test. *P < .05; **P < .01. (B-D) Thymocytes from 4-week-old SLP-76m/+ and SLP-76m/m littermates (5 mice for each genotype) were analyzed by flow cytometry. FACS plots shown are from 1 representative of each genotype. (B) Surface expression of CD4 versus CD8 on total thymocytes. (C) Expression of CD25 versus CD44 on CD4−CD8− DN thymocytes. (D) Expression of TCR-β versus HSA on CD4−CD8+ thymocytes. The numbers in each panel represent the average percentages of the gated populations.
We also examined the thymic production of γδTCR+ cells in SLP-76m/m mice. SLP-76 has been shown to play a critical role in γδ T-cell development because the number of γδTCR+ thymocytes is greatly decreased in SLP-76−/− mice.1 In contrast, the SLP-76m/m mice contained approximately the same number of γδTCR+ thymocytes as the heterozygous controls (Figure 2A), suggesting that the SAM domain is not required for SLP-76–mediated γδ T-cell development.
In contrast to SLP-76−/− mice, which completely lack DP and SP thymocytes, SLP-76m/m mice contained a slightly higher percentage of DP thymocytes than the heterozygous controls (Figure 2B). The total number of SLP-76m/m DP thymocytes was moderately decreased compared with SLP-76m/+ controls (Figure 2A). However, despite a largely normal DP subset, the percentage of CD4+SP thymocytes was significantly reduced (from 8.8% to 4.5%) in SLP-76m/m mice (Figure 2B), and their total number was decreased by more than 60% (Figure 2A). The relatively small decrease observed in the percentage of SLP-76m/m CD4−CD8+ thymocytes (from 3.6% to 3.1%; Figure 2B) was caused by an enrichment of ISP cells (Figure 2D). Therefore, the absolute number of mature CD8+ SP thymocytes was still reduced by approximately 50% in SLP-76m/m mice (Figure 2A). Together, our data suggested that deletion of the SAM domain in SLP-76 partially impaired the maturation of SP thymocytes, particularly the CD4+SP cells.
Defective positive and negative selection in SLP-76 knockin mice
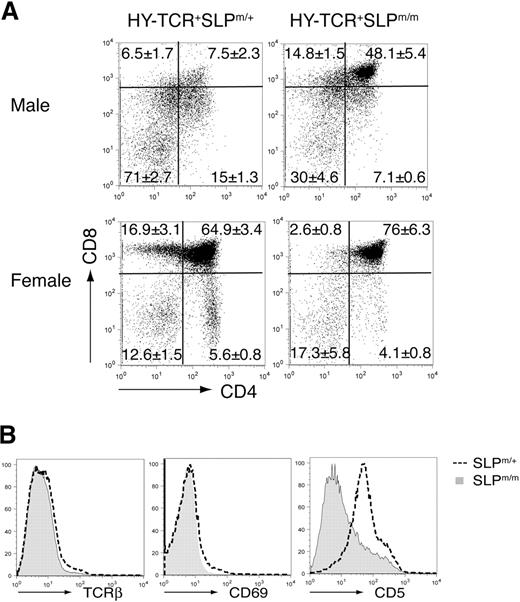
To study the function of the SAM domain of SLP-76 in thymic selection, we crossed these knockin mice with HY-TCR transgenic mice, which carry an αβTCR specific for the male antigen HY. The results are shown in Figure 3A. In the male HY-TCR+SLP-76m/+ mice, the HY-TCR+ thymocytes underwent extensive negative selection, resulting in a severe loss of DP thymocytes. In contrast, a substantial population of HY-TCR+ DP thymocytes persisted in the male HY-TCR+SLP-76m/m mice, indicating that thymic negative selection was severely impaired. We also examined positive selection in female HY-TCR transgenic mice, which lack the HY antigen. In female HY-TCR+SLP-76m/+ mice, the HY-TCR+ DP thymocytes underwent successful positive selection and matured into SP thymocytes (predominantly CD8+). On the contrary, despite a normal HY-TCR+ DP subset, the percentages of SP thymocytes were significantly reduced in the female HY-TCR+SLP-76m/m mice, suggesting that these mutant DP thymocytes have a defect in positive selection. These data showed that the deletion of the SAM domain from SLP-76 severely disrupted both positive and negative thymic selection in the HY-TCR transgenic mice.
Defective positive and negative thymic selections in ΔSAM–SLP-76 knockin mice. (A) ΔSAM–SLP-76 knockin mice were crossed with HY-TCR transgenic mice. HY-TCR+ gated thymocytes from 4-week-old male (top) and female (bottom) HY-TCR+SLP-76m/+ and HY-TCR+SLP-76m/m littermates were analyzed for their surface expression of CD4 and CD8. The numbers represent the average percentages of the gated populations. (B) Surface expression of TCR-β, CD69, and CD5 on DP thymocytes from 4-week-old SLP-76m/+ and SLP-76m/m littermates.
Defective positive and negative thymic selections in ΔSAM–SLP-76 knockin mice. (A) ΔSAM–SLP-76 knockin mice were crossed with HY-TCR transgenic mice. HY-TCR+ gated thymocytes from 4-week-old male (top) and female (bottom) HY-TCR+SLP-76m/+ and HY-TCR+SLP-76m/m littermates were analyzed for their surface expression of CD4 and CD8. The numbers represent the average percentages of the gated populations. (B) Surface expression of TCR-β, CD69, and CD5 on DP thymocytes from 4-week-old SLP-76m/+ and SLP-76m/m littermates.
We further examined the cell surface expression of TCR-β, CD5, and CD69, all of which are known markers for positive selection, on the DP thymocytes from SLP-76m/+ and SLP-76m/m mice. As shown in Figure 3B, CD5 expression on the SLP-76m/m DP cells was significantly decreased. Meanwhile, the percentages of TCR-β+ cells and CD69+ cells also declined among the SLP-76m/m DP thymocytes. Altogether, our data showed that the deletion of the SAM domain of SLP-76 affected both positive and negative thymic selection, resulting in a partial block during the DP to SP transition.
Fewer mature T cells in the periphery of the SLP-76 knockin mice
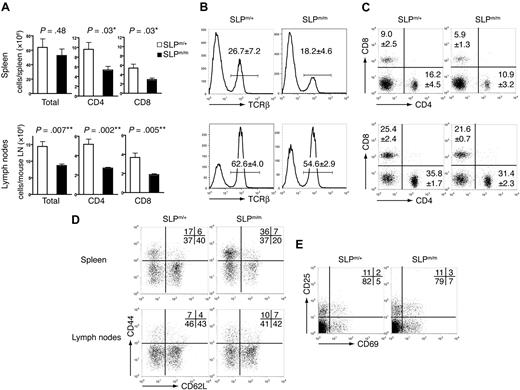
We next examined the peripheral lymphoid compartment of the SLP-76 knockin mice. The spleens in SLP-76m/m mice appeared grossly normal, and the total number of splenocytes did not appear significantly different from that of control mice (Figure 4A). However, consistent with the thymocyte data, TCR-β+ cells constituted a lower percentage of splenocytes in SLP-76m/m mice (Figure 4B); correspondingly, the numbers of both CD4+ and CD8+ splenic T cells were decreased by approximately 50% (Figure 4A). In contrast to the spleens, the percentage of TCR-β+ cells was only slightly decreased in the peripheral lymph nodes in SLP-76m/m mice (Figure 4B). However, the total number of peripheral lymph node cells in SLP-76m/m mice was significantly reduced, resulting in a markedly decreased amount of T cells (Figure 4A). The CD4:CD8 ratio in both spleen and peripheral lymph nodes of SLP-76m/m mice remained similar to that of the control mice (Figure 4C). Furthermore, despite their low numbers, the mutant T cells expressed normal levels of surface TCR on a single-cell basis (Figure 4B).
Peripheral T cells in ΔSAM–SLP-76 knockin mice. Splenocytes and peripheral lymph node cells from 5-week-old SLP-76m/+ and SLP-76m/m littermates were analyzed by flow cytometry. (A) Total numbers of splenocytes, lymph node cells, and CD4+ and CD8+ T cells. Five mice for each genotype were analyzed. Statistical analysis was performed using 2-tailed Student t test. *P < .05; **P < .01. (B) The percentages of TCR-β+ cells in spleens and lymph nodes. (C) Expression of CD4 versus CD8 on splenocytes and lymph node cells. (D) Expression of CD44 versus CD62L on CD4+ gated cells in spleens and lymph nodes. (E) Expression of CD25 versus CD69 on CD4+ gated cells in spleens. FACS plots shown are from 1 representative of 5 mice analyzed. The numbers represent the average percentages of the gated populations.
Peripheral T cells in ΔSAM–SLP-76 knockin mice. Splenocytes and peripheral lymph node cells from 5-week-old SLP-76m/+ and SLP-76m/m littermates were analyzed by flow cytometry. (A) Total numbers of splenocytes, lymph node cells, and CD4+ and CD8+ T cells. Five mice for each genotype were analyzed. Statistical analysis was performed using 2-tailed Student t test. *P < .05; **P < .01. (B) The percentages of TCR-β+ cells in spleens and lymph nodes. (C) Expression of CD4 versus CD8 on splenocytes and lymph node cells. (D) Expression of CD44 versus CD62L on CD4+ gated cells in spleens and lymph nodes. (E) Expression of CD25 versus CD69 on CD4+ gated cells in spleens. FACS plots shown are from 1 representative of 5 mice analyzed. The numbers represent the average percentages of the gated populations.
Analysis of surface activation markers showed that the frequency of effector-like CD44highCD62LlowCD4+ T cells increased in SLP-76m/m spleens, with a simultaneous reduction in the percentage of naive CD44lowCD62Lhigh CD4+ T cells (Figure 4D). A similar phenomenon was observed in splenic CD8+ T cells (data not shown), but not in lymph node T cells (Figure 4D). However, neither CD4+ nor CD8+ SLP-76m/m splenic T cells up-regulated surface expression of CD25 or CD69 (Figure 4E and data not shown), suggesting that basal activation of the mutant T cells was not disturbed. We reason that the SLP-76m/m splenic T cells might have undergone homeostatic proliferation upon entering the partially lymphopenic environment, which has been known to result in a similar CD44highCD62Llow phenotype.
Defective TCR-induced proliferation and IL-2 production by SLP-76m/m T cells
Next, we examined the role of the SLP-76 SAM domain in T-cell activation. Up-regulation of early activation markers, CD25 and CD69, upon anti-CD3 stimulation appeared normal in the SLP-76m/m T cells (Figure 5A). Proliferation of SLP-76m/m CD4+CD44low naive T cells in response to stimulation by various concentrations of plate-bound anti-CD3 and 1 μg/mL soluble anti-CD28 was significantly reduced compared with SLP-76m/+ CD4+CD44low T cells (Figure 5B). However, the mutant T cells proliferated at a comparable rate to the control T cells upon PMA plus ionomycin stimulation, which bypasses TCR engagement (Figure 5B). IL-2 production by SLP-76m/m CD4+ T cells was also diminished upon anti-CD3/CD28 stimulation (Figure 5C). In contrast, PMA plus ionomycin stimulation induced efficient production of IL-2 from the SLP-76m/m CD4+ T cells as well as the SLP-76m/+ controls (Figure 5C). These data demonstrated that the SAM domain is required for optimal T-cell proliferation and IL-2 production.
Defective T-cell activation in ΔSAM–SLP-76 knockin mice. (A) Splenocytes from SLP-76m/+ and SLP-76m/m littermates were either left untreated (gray area), stimulated with 3 μg/mL plate-bound anti-CD3 (solid line), or stimulated with 20 ng/mL PMA plus 0.5 μg/mL ionomycin (dotted line) for 6 hours and subsequently analyzed by flow cytometry. Surface expression of CD25 and CD69 on CD4+ gated cells is shown. (B) Purified SLP-76m/+ and SLP-76m/m CD4+CD44low T cells were either stimulated with various concentrations of plate-bound anti-CD3 plus 1 μg/mL soluble anti-CD28 (left), or with PMA plus ionomycin (right). After 36 hours, cells were pulsed with [3H]thymidine for an additional 6 hours before being harvested for scintillation counting. Cell proliferation is represented by the counted radioactivity (CPM). (C) Purified SLP-76m/+ and SLP-76m/m CD4+ T cells were stimulated the same as in panel B. Supernatants were harvested after 8 hours, and the IL-2 concentrations were determined by ELISA.
Defective T-cell activation in ΔSAM–SLP-76 knockin mice. (A) Splenocytes from SLP-76m/+ and SLP-76m/m littermates were either left untreated (gray area), stimulated with 3 μg/mL plate-bound anti-CD3 (solid line), or stimulated with 20 ng/mL PMA plus 0.5 μg/mL ionomycin (dotted line) for 6 hours and subsequently analyzed by flow cytometry. Surface expression of CD25 and CD69 on CD4+ gated cells is shown. (B) Purified SLP-76m/+ and SLP-76m/m CD4+CD44low T cells were either stimulated with various concentrations of plate-bound anti-CD3 plus 1 μg/mL soluble anti-CD28 (left), or with PMA plus ionomycin (right). After 36 hours, cells were pulsed with [3H]thymidine for an additional 6 hours before being harvested for scintillation counting. Cell proliferation is represented by the counted radioactivity (CPM). (C) Purified SLP-76m/+ and SLP-76m/m CD4+ T cells were stimulated the same as in panel B. Supernatants were harvested after 8 hours, and the IL-2 concentrations were determined by ELISA.
Impaired TCR signaling in SLP-76 knockin mice
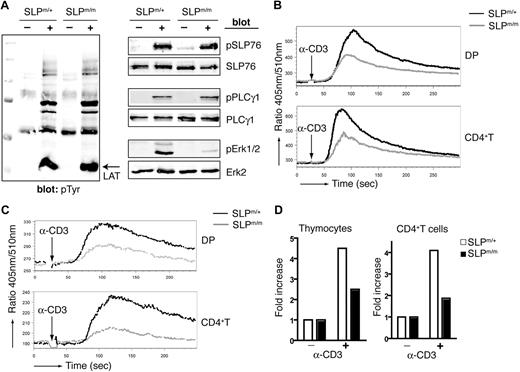
To further understand the TCR signaling events that led to impaired activation of SLP-76m/m T cells, we examined 2 major signaling pathways downstream of TCR engagement: ERK activation and calcium mobilization. In Figure 6A, purified CD4+ T cells from SLP-76m/m and SLP-76m/+ mice were stimulated by CD3 cross-linking. Total tyrosine phosphorylation of proteins in SLP-76m/m CD4+ T cells was comparable with that of control cells, and LAT phosphorylation was normal. It appears that deletion of the SAM domain does not affect TCR-proximal signaling events upstream of SLP-76. Phosphorylation of SLP-76 was also normal, suggesting that it is independent of the SAM domain. Phosphorylation of ERK1/2 was dramatically decreased, as shown in Figure 6A, but PLC-γ1 phosphorylation was surprisingly undisturbed. Similar results were observed in purified SLP-76m/m DP thymocytes (data not shown). These data showed that the SAM domain plays an essential role in SLP-76–mediated ERK-MAPK activation, but is dispensable for PLC-γ1 phosphorylation.
Defective TCR-mediated signaling in ΔSAM–SLP-76 T cells. (A) Purified SLP-76m/+ and SLP-76m/m CD4+ T cells were stimulated for 2 minutes by anti-CD3 cross-linking. Postnuclear lysates were analyzed by Western blotting with anti-pTyr, pSLP-76, pPLC-γ1, and pERK1/2 antibodies. SLP-76, PLC-γ, and ERK2 blots are shown as loading controls. (B-C) DP thymocytes and CD4+ splenic T cells from SLP-76m/+ and SLP-76m/m mice were loaded with Indo-1 and then stimulated by anti-CD3 cross-linking without (B) or with (C) the presence of 5 mM EGTA. Calcium concentration was monitored by flow cytometry and represented as the ratio of fluorescence at 405 nm and 510 nm. (D) Total thymocytes (left) and purified CD4+ T cells (right) from SLP-76m/+ and SLP-76m/m mice were stimulated for 2 minutes by anti-CD3 cross-linking. IP3 was extracted, and IP3 levels were measured.
Defective TCR-mediated signaling in ΔSAM–SLP-76 T cells. (A) Purified SLP-76m/+ and SLP-76m/m CD4+ T cells were stimulated for 2 minutes by anti-CD3 cross-linking. Postnuclear lysates were analyzed by Western blotting with anti-pTyr, pSLP-76, pPLC-γ1, and pERK1/2 antibodies. SLP-76, PLC-γ, and ERK2 blots are shown as loading controls. (B-C) DP thymocytes and CD4+ splenic T cells from SLP-76m/+ and SLP-76m/m mice were loaded with Indo-1 and then stimulated by anti-CD3 cross-linking without (B) or with (C) the presence of 5 mM EGTA. Calcium concentration was monitored by flow cytometry and represented as the ratio of fluorescence at 405 nm and 510 nm. (D) Total thymocytes (left) and purified CD4+ T cells (right) from SLP-76m/+ and SLP-76m/m mice were stimulated for 2 minutes by anti-CD3 cross-linking. IP3 was extracted, and IP3 levels were measured.
We next examined TCR-induced calcium mobilization in SLP-76m/m T cells. Upon CD3 cross-linking, SLP-76m/m DP thymocytes (Figure 6B), as well as SP thymocytes (data not shown), mounted a considerably weaker calcium flux compared with SLP-76m/+ controls. However, ionomycin treatment induced an equally strong Ca2+ response in both the mutant and control thymocytes, suggesting that the calcium mobilization machinery was intact in mutant cells and that the dye loading was equal in both samples (data not shown). The same phenomenon was also observed in both CD4+ (Figure 6B) and CD8+ (data not shown) peripheral T cells. The absence of the SAM domain most likely impaired the initial calcium release from endoplasmic reticulum (ER) stores, because the calcium flux in SLP-76m/m T cells appeared to be kinetically normal despite a lower peak concentration. To separate the intracellular calcium release from extracellular influx, EGTA was used to chelate free Ca2+ in the assay buffer. As shown in Figure 6C, in the presence of EGTA, CD3 cross-linking induced a weaker increase in Ca2+ levels in SLP-76m/+ T cells, reflecting calcium depletion from intracellular stores. Such elevation was much less noticeable in the SLP-76m/m T cells. Ionomycin, however, triggered a similar Ca2+ increase in both the mutant and control cells, suggesting comparable ER Ca2+ stores in these cells (data not shown). Furthermore, we assayed TCR-mediated production of the secondary messenger IP3. IP3 is generated from cleavage of 4,5-bis-phosphate by activated PLC-γ1 and subsequently interacts with IP3 receptors to trigger intracellular calcium release. As shown in Figure 6D, IP3 production was considerably lower in SLP-76m/m thymocytes as well as CD4+ T cells. Altogether, our data showed that deletion of the SLP-76 SAM domain caused defective TCR-induced IP3 generation, which subsequently led to impaired calcium flux and IL-2 production.
SAM domain in TCR-mediated formation of SLP-76 microclusters
Previous studies have analyzed SLP-76−/− mice reconstituted with SLP-76 Y3F, Δ224-244, ΔSH2, or R448K mutants.20,21 Among these mice, mice expressing the Δ224-244 mutant exhibited strikingly similar phenotypes to our SLP-76 ΔSAM knockin mice. The deletion of amino acids 224-244 of SLP-76 abolishes its interaction with Gads, through which SLP-76 can be recruited to LAT and lipid rafts upon TCR ligation. Accordingly, Gads−/− mice also share many gross similarities with SLP-76m/m mice.33 It is possible that, similar to the Gads-binding motif, the SAM domain plays a role in the recruitment of SLP-76 to the plasma membrane.
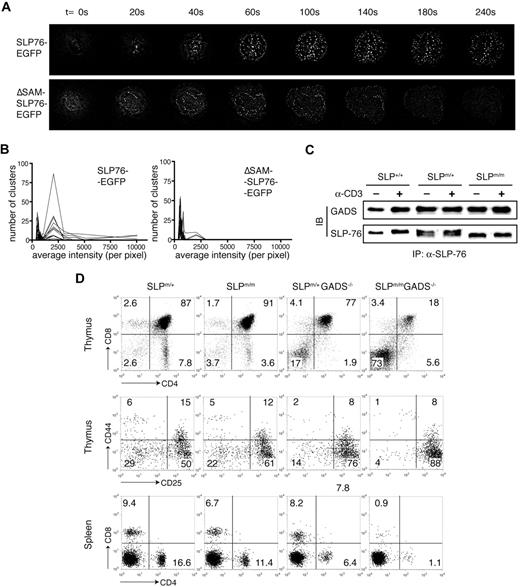
To investigate the subcellular localization of the ΔSAM-SLP-76 mutant after TCR stimulation, we reconstituted SLP-76–deficient J14 Jurkat cells with GFP-tagged wild-type or ΔSAM SLP-76 proteins at their C termini. These cells were then stimulated by anti-CD3ϵ antibody (UCHT1) coated onto coverslips. The cellular distribution of the GFP fluorescence was visualized every 20 seconds by live imaging. As previously reported,14,34 SLP-76–GFP began to form bright clusters and translocate to the plasma membrane within 20 seconds of TCR stimulation (Figure 7A). Such clusters were observed at the plane where the cells made contact with the coverslip. Very few, if any, clusters could be found in the interior of the cells or other areas of the plasma membrane that was not in contact with the coverslip (data not shown). These clusters were maintained at the interface over time and could still be observed after at least 15 minutes (Figure 7C and data not shown). In contrast, whereas the ΔSAM–SLP-76–GFP protein was capable of forming clusters at the coverslip surface, such clustering appeared much weaker in intensity and disappeared much faster (Figure 7B).
Deletion of the SAM domain inhibits TCR-mediated SLP-76 clustering. (A) J14 cells expressing SLP-76–GFP or ΔSAM–SLP-76–GFP fusion proteins were dropped onto coverslips coated with anti–human CD3ϵ antibody. Time point 0 represents the moment the cells made contact with the coverslip. GFP clustering at the cell/coverslip interface at indicated time points is shown. (B) Cells in panel A were activated on anti-CD3ϵ–coated coverslips for 10 minutes. Average intensity of the GFP clustering formed at the cell/coverslip interface was measured. The numbers of clusters within indicated intensity ranges were counted. Each line represents 1 analyzed cell. (C) Total thymocytes from SLP-76+/+, SLP-76m/+, and SLP-76m/m littermates were either stimulated with anti-CD3ϵ cross-linking or left untreated. Cell lysates were subjected to immunoprecipitation with anti-SLP-76 polyclonal antibody, followed by Western blotting with anti–SLP-76 and anti-Gads antibodies. (D) Thymocytes and splenocytes from 4-week-old SLP-76m/+, SLP-76m/m, Gads−/−, and SLP-76m/mGads−/− mice were analyzed by flow cytometry. (Top) Expression of CD4 versus CD8 on total thymocytes. (Middle) Expression of CD25 versus CD44 on CD4−CD8− DN thymocytes. (Bottom) Expression of CD4 versus CD8 on splenocytes.
Deletion of the SAM domain inhibits TCR-mediated SLP-76 clustering. (A) J14 cells expressing SLP-76–GFP or ΔSAM–SLP-76–GFP fusion proteins were dropped onto coverslips coated with anti–human CD3ϵ antibody. Time point 0 represents the moment the cells made contact with the coverslip. GFP clustering at the cell/coverslip interface at indicated time points is shown. (B) Cells in panel A were activated on anti-CD3ϵ–coated coverslips for 10 minutes. Average intensity of the GFP clustering formed at the cell/coverslip interface was measured. The numbers of clusters within indicated intensity ranges were counted. Each line represents 1 analyzed cell. (C) Total thymocytes from SLP-76+/+, SLP-76m/+, and SLP-76m/m littermates were either stimulated with anti-CD3ϵ cross-linking or left untreated. Cell lysates were subjected to immunoprecipitation with anti-SLP-76 polyclonal antibody, followed by Western blotting with anti–SLP-76 and anti-Gads antibodies. (D) Thymocytes and splenocytes from 4-week-old SLP-76m/+, SLP-76m/m, Gads−/−, and SLP-76m/mGads−/− mice were analyzed by flow cytometry. (Top) Expression of CD4 versus CD8 on total thymocytes. (Middle) Expression of CD25 versus CD44 on CD4−CD8− DN thymocytes. (Bottom) Expression of CD4 versus CD8 on splenocytes.
To quantitate the microcluster formation, we spun the GFP+ J14 cells onto anti-CD3–coated coverslips and incubated them at 37°C for 10 minutes before collecting imaging data. This procedure allowed these cells to be stimulated simultaneously. For each cell analyzed, the average intensity of microclusters was calculated by MetaMorph software, and the numbers of clusters within various ranges of intensity were also counted. As shown in Figure 7B, the number of clusters with high fluorescence intensity decreased dramatically in J14 cells expressing the ΔSAM–SLP-76–GFP compared with those cells expressing SLP-76–GFP.
Altogether, these data suggested that the SAM domain plays an important role in mediating or stabilizing the TCR-mediated membrane association of SLP-76, through mechanisms yet to be understood. The similarity between the ΔSAM knockin mice and the SLP-76−/− mice expressing the Δ224-244 mutant prompted us to speculate whether the SAM domain functions through Gads-dependent mechanisms. We first examined the interaction between ΔSAM–SLP-76 and Gads. Comparable amount of Gads protein was coimmunoprecipitated with wild-type or mutant SLP-76 from thymocyte lysates, suggesting that the constitutive interaction between SLP-76 and Gads was not affected by the deletion of the SAM domain (Figure 7C). Similar data were also obtained using J14 cells reconstituted with wild-type or mutant SLP-76 (data not shown). In addition, there were no obvious differences in the tyrosine-phosphorylated proteins that were coimmunoprecipitated with SLP-76, suggesting that SLP-76 association with Vav and PLC-γ1 is most likely normal (data not shown).
To further understand the relationship between the SAM domain and Gads in SLP-76 function, we generated SLP-76m/mGads−/− double-mutant mice and compared them with both SLP-76m/m and Gads−/− single-mutant littermates. Strikingly, the T-cell developmental defects observed in the SLP-76m/mGads−/− mice were much more severe than in either of the single-mutant mice. The combined effects of the SLP-76 SAM domain and Gads deletions caused a nearly complete arrest of thymocyte development at the DN3 stage. Only a few DN thymocytes managed to mature into DP cells, evidenced by the fact that only approximately 18% of total thymocytes were CD4+CD8+ in the double-mutant mice. Very few mature T cells could be detected in the periphery of the SLP-76m/mGads−/− mice (Figure 7D). These data suggested that the SAM domain most likely exerts its function in a Gads-independent fashion, and that both the SAM domain and the Gads–SLP-76 interaction are important in SLP-76 function.
Discussion
Upon TCR engagement, TCR-proximal signals are translated into distal biochemical events via the orchestration of a variety of adaptor proteins. Adaptor proteins are signaling molecules that lack intrinsic enzymatic activity, but act as protein scaffolds, nucleating multiprotein signaling complexes. They usually contain multiple domains that are capable of interacting with other signaling proteins either constitutively or inducibly. Therefore, to fully understand the functions of adaptor proteins, it is crucial to explore their protein interaction motifs and corresponding binding partners.
SLP-76 is a hematopoietic cell-specific adaptor protein critical for thymocyte development and mature TCR signaling. Extensive effort has been aimed at identifying the binding partners of SLP-76 and exploring the domain structures involved in such associations. Most structure-function analysis of SLP-76 has been focused on 3 motifs: the 3 N-terminal phosphotyrosines, the Gads-binding sequence in the central proline-rich region, and the C-terminal SH2 domain. Yet, more protein-binding motifs have been identified in SLP-76 over the years. A P1 domain within the proline-rich region of SLP-76 was found to mediate constitutive association with the SH3 domain of PLC-γ1.13,14 In fact, it was later found that at least 3 distinct sites within this proline-rich region of SLP-76 could bind to PLC-γ1.35 Moreover, the SH3 domain of the protein tyrosine kinase p56lck can associate with a 10-amino-acid–long sequence (amino acids 185-194) within SLP-76.36,37 SLP-76−/− mice reconstituted with the Δ185-194 mutant exhibited defects in thymocyte development as well as mature TCR responses.37 In addition, the phosphorylation of serine 376 of SLP-76 induces direct association with 14-3-3 proteins, which play a role in down-modulating T-cell activation.38,39 It is reasonable to believe that more protein-binding motifs of SLP-76 and their respective binding partners will be identified in the future.
In this study, we demonstrated that a SAM domain, which is generally considered a protein interaction motif, plays an important role in SLP-76 function in thymopoiesis. Deletion of the SAM domain in SLP-76 slightly affected the DN3 to DN4 transition mediated by pre-TCR signaling. The more obvious developmental block in the ΔSAM SLP-76 knockin mice occurred at the DP to SP transition, causing defective generation of SP thymocytes and mature T cells. We also used the HY-TCR transgenic system to examine positive and negative thymic selection in SLPm/m mice. Although this system is widely used as a model for negative selection, some studies argue that the lack of DP thymocytes in male HY-TCR transgenic mice is a consequence of the conversion of DN thymocytes into γδ-like cells due to signals delivered by premature TCR-αβ expression on DN thymocytes.40-42 However, other lines of evidence indicate that this argument cannot be the sole explanation, because HY-TCR+ DP thymocytes can indeed be found in the male mice and their deletion can be rescued by CD8β deficiency.43 Therefore, we cautiously concluded that that the disruption of the SAM domain inhibited negative thymic selection.
Our data showed that the SAM domain of SLP-76 is indispensable for optimal mature TCR signaling. The peripheral T cells in SLP-76m/m mice exhibited defective TCR-mediated calcium mobilization and ERK activation, leading to decreased IL-2 production and cell proliferation. Previous studies have shown that SLP-76 interacts with PLC-γ1 in both constitutive and TCR-inducible fashions. The constitutive binding occurs between the SH3 domain of PLC-γ1 and specific sequences within the SLP-76 proline-rich region.13,14,35 Abolishment of such basal association in Jurkat cells greatly reduces TCR-dependent PLC-γ1 phosphorylation, resulting in defective calcium flux and NFAT promoter activity.13,14 In contrast, upon TCR engagement, phosphorylated LAT recruits both PLC-γ1 and Gads, the latter of which constitutively binds SLP-76. In this manner, SLP-76 also associates with PLC-γ1 indirectly. Although SLP-76 is not required for LAT recruitment of PLC-γ1,3 its inducible interaction with LAT and PLC-γ1 appears to stabilize the PLC-γ1–LAT association and is essential for optimal activation of PLC-γ1. Mutation of the Gads-binding site of LAT significantly reduces the binding of PLC-γ1 with LAT, leading to reduced activation of PLC-γ1.44 Similarly, when the Gads-binding site of SLP-76 or Gads itself is deleted, phosphorylation of PLC-γ1 is markedly decreased.20,21,33 These studies suggest that the complicated interactions between LAT, SLP-76, and PLC-γ1 are necessary for the full activation of PLC-γ1, and any disturbance between them could lead to defective TCR-dependent calcium flux. The constitutive and inducible interaction with SLP-76 may help maintain and stabilize PLC-γ1 in a conformation that allows it to be phosphorylated and thus catalytically active.13 Furthermore, recent studies have shown that SLP-76 may additionally contribute to the activation of PLC-γ1 by recruiting Vav and Itk, which have been implicated in direct or indirect phosphorylation of PLC-γ1.45-47 These studies demonstrate that SLP-76 regulates the activity of PLC-γ1 by affecting its phosphorylation. Intriguingly, in our study, the TCR-induced phosphorylation of PLC-γ1 appeared unaffected in SLP-76m/m T cells, despite impaired production of IP3 and defective calcium flux. These data suggested that, apart from regulating the phosphorylation of PLC-γ1, SLP-76 might also directly regulate the catalytic activity of PLC-γ1 through novel mechanisms yet to be determined.
Previous studies using live imaging of Jurkat cells have shown that cross-linking the TCR by immobilized antibodies induces rapid formation of signaling microclusters, which contain tyrosine kinases, LAT, Gads, and SLP-76, at the contact site.34,48-50 The persistence of SLP-76 clustering appeared to be dependent on its multiple domain structures, including the N-terminal phosphotyrosine motif, the Gads-binding motif, the proline-rich P1 domain, and the C-terminal SH2 domain. Cluster formation in cells expressing the aforementioned SLP-76 mutants was rapidly terminated.50 Our data clearly indicated that the deletion of the SAM domain significantly weakened the intensity and stability of the SLP-76 clusters, suggesting that the SAM domain is also critical in the formation of stable and persistent SLP-76 clusters. Whereas the biologic relevance of the microcluster assembly is still not clear, the impaired recruitment of SLP-76 to these structures most likely affects the activity of PLC-γ1 and causes the signaling defects in the SLP-76m/m T cells.
The defects in thymocyte development in Gads−/− mice, in which the LAT-dependent membrane recruitment of SLP-76 is abolished, are much less severe than those in SLP-76−/− mice. The residual signaling seen in Gads−/− mice might result from the membrane recruitment of SLP-76 through Grb2–SLP-76 interaction, PLC-γ1–SLP-76 interaction, or LAT-independent mechanisms, one of which is through the SAM domain. Our data showed that, by further disrupting the SLP-76 SAM domain in the absence of Gads, such residual signaling was markedly decreased. These data suggested that the SAM domain and Gads play synergistic roles in SLP-76 function. How the SLP-76 SAM domain functions in TCR-mediated signaling remains to be determined. It is very possible that this domain interacts with other proteins to stabilize the LAT–Gads–SLP-76–PLC-γ1 signaling complex. Our data presented in this study clearly demonstrate that this domain is critical for SLP-76 function during thymocyte development and T-cell activation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Duke University Cancer Center Flow Cytometry, DNA Sequencing, and Transgenic Mouse facilities for their excellent services.
This work was supported by National Institutes of Heath (Bethesda, MD) grants AI048674 and AI056156. J.L. is supported by a National Science Scholarship from A*STAR, Singapore. W.Z. is a scholar of the Leukemia & Lymphoma Society (White Plains, NY).
National Institutes of Health
Authorship
Contribution: S.S. designed and performed experiments, analyzed and interpreted data, performed statistical analysis, and wrote the paper; J.L., M.Z., J.Z., and D.F. performed research and analyzed data; Q.-j.L. assisted imaging experiments and analyzed and interpreted data from imaging studies; and W.Z. designed research, analyzed and interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Weiguo Zhang, Box 3010, Dept of Immunology, Duke University Medical Center, 1 Research Dr, Durham, NC 27710; e-mail: zhang033@mc.duke.edu.

![Figure 5. Defective T-cell activation in ΔSAM–SLP-76 knockin mice. (A) Splenocytes from SLP-76m/+ and SLP-76m/m littermates were either left untreated (gray area), stimulated with 3 μg/mL plate-bound anti-CD3 (solid line), or stimulated with 20 ng/mL PMA plus 0.5 μg/mL ionomycin (dotted line) for 6 hours and subsequently analyzed by flow cytometry. Surface expression of CD25 and CD69 on CD4+ gated cells is shown. (B) Purified SLP-76m/+ and SLP-76m/m CD4+CD44low T cells were either stimulated with various concentrations of plate-bound anti-CD3 plus 1 μg/mL soluble anti-CD28 (left), or with PMA plus ionomycin (right). After 36 hours, cells were pulsed with [3H]thymidine for an additional 6 hours before being harvested for scintillation counting. Cell proliferation is represented by the counted radioactivity (CPM). (C) Purified SLP-76m/+ and SLP-76m/m CD4+ T cells were stimulated the same as in panel B. Supernatants were harvested after 8 hours, and the IL-2 concentrations were determined by ELISA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/1/10.1182_blood-2008-09-177832/4/m_zh89990938180005.jpeg?Expires=1763849739&Signature=hUjIQ8oTg52h3iy8czn3zufP5gVy~pWdQhM8CtKW2ktrDvkcL0IDdc9VpAEoV3c6D7QBzGzXMPFZdaOwwp9zsnlOQp25OpgIJdI4kIcJhskYZK2doqRqpZ897Jql1brT8PLlDYAdPBqnzhkoTR2AfIJhO~rx6gOjyhzX-xbQp175BVhaJtvOy0lRPO-2Ln-DJNLs9p39PYATXDenKHVafChoSfF3y2dSJifJ26CIU8adYfG8hLhR484Lv9uMs2TgrGmp-0ArAFTwqX8agMPruOm-ipU0G1xykI4jKZ0Wq0jFc1spdqFSEB2WUS66UX375~SRPYCly7vi-ARWccOBoA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)