Abstract
Dendritic cells (DCs) play a key role in the pathogenesis of HIV infection. HIV interacts with these cells through 2 pathways in 2 temporal phases, initially via endocytosis and then via de novo replication. Here the transcriptional response of human DCs to HIV-1 was studied in these phases and at different stages of the virus replication cycle using purified HIV-1 envelope proteins, and inactivated and viable HIV-1. No differential gene expression was detected in response to envelope. However, more than 100 genes were differentially expressed in response to entry of viable and inactivated HIV-1 in the first phase. A completely different set of genes was differentially expressed in the second phase, predominantly in response to viable HIV-1, including up-regulation of immune regulation genes, whereas genes encoding lysosomal enzymes were down-regulated. Cathepsins B, C, S, and Z RNA and protein decreased, whereas cathepsin L was increased, probably reflecting a concomitant decrease in cystatin C. The net effect was markedly diminished cathepsin activity likely to result in enhanced HIV-1 survival and transfer to contacting T lymphocytes but decreased HIV-1 antigen processing and presentation to these T cells.
Introduction
Human immunodeficiency virus type 1 (HIV-1) infection of activated T lymphocytes is normally rapid and productive and results in cell death. However in macrophages, dendritic cells (DCs), and astrocytes, exposure to HIV-1 results in a less productive infection, which may result in subtle yet complex changes.1-4
Upon contact with sentinel immature DCs in the periphery, foreign glycoprotein antigens bind to C-type lectin receptors (CLRs) and are rapidly endocytosed and processed by the endolysosomal pathway resulting in processing and presentation via MHC class II.5 This process triggers DC maturation and migration to the draining lymph node.6
DCs present in the anal, vaginal, and cervical mucosa as well as the male foreskin are key target cells for sexual transmission of HIV-1.7-9 It is now thought that HIV-1 uses DCs to gain access to large numbers of CD4 T lymphocytes in the submucosa and lymph nodes, and elicits a productive infection.10-13 HIV DC/T-cell interaction is probably also important in many other locations in the body. How DCs take up HIV-1 and transfer it to T cells has been recently demonstrated,11,14-17 but the gene changes underlying the effects of exposure to HIV-1 have not been comprehensively studied and reported. A global analysis of overall gene expression profiles would provide an integrated picture of the complex and subtle changes occurring within these cells after exposure to HIV-1. To date, microarray analysis has been used to examine gene expression in HIV-1–infected T-cell lines,18-20 and HIV-1 and/or monomeric gp120–treated peripheral blood mononuclear cells (PBMCs),20,21 macrophages,4,22 and astrocytes.1,3,22
DCs have unique aspects to their interactions with HIV-1. However, to date, no studies have been carried out on the effects of gp120 on DC gene expression and only one study has investigated the effect of HIV-1 (and its accessory protein Tat) on dendritic cell gene expression.23 This study used low-titer virus stocks and investigated the transcriptional effects between 1 and 14 days after infection. After HIV-1 binding to CLRs on monocyte-derived DCs (MDDCs) the majority (> 95%) of HIV-1 is endocytosed and subject to acid proteolytic digestion over 6 to 12 hours.24 A minority is transferred to CD4/CCR5, resulting in fusion of the virus envelope with the plasma membrane and de novo infection, apparent only at a later phase more than 24 hours after infection. Therefore, after contact between DCs and T cells, HIV-1 is also transferred to them in 2 phases, first from endosomes and then later from the cytosol.14
Due to their high endocytic capacity, DCs are far less susceptible to infection with HIV-1 than other cells such as T cells and macrophages, as 95% of bound HIV-1 is endocytosed and traffics to late endosomes where it is destroyed.14 Treatment of DCs with low-titer HIV-1 stocks results, initially, in very low infection levels (eg, an MOI of 0.1 often used for infection of T cells leads to < 0.5% of DCs infected de novo). This does not allow the investigation of the direct effects of the initial interaction between HIV-1 and DCs or the early stages of infection on gene and protein expression, as any changes would be masked by bystander effects on the vast majority of uninfected cells and asynchronous replication cycles in different cells. We have therefore used microarrays to elucidate gene expression profiles in MDDCs during a single growth cycle after treatment with high titer (MOI 10), highly purified CCR5 using HIV-1BaL, which infects DCs productively as used previously by ourselves and others.11,25 The use of a relatively high-titer inoculum is consistent with the high burst size (> 100-10 000 virions) of HIV-1 or SIV derived from potentially neighboring T cells.26,27 It is also consistent with the higher concentrations expected to be transmitted by cell to cell transmission in the genital tract and with the effects of certain factors in semen, which may increase the efficiency of mucosal infection.28,29
Experiments were carefully designed to determine which genes were differentially expressed in response to 3 different stages of the virus life cycle: binding, entry, and replication. This was achieved by treating DCs with viable virus, virus that was chemically inactivated with aldrothiol-2 (AT-2), or with the HIV-1 viral glycoprotein gp120 in both its monomeric and trimeric forms. The matched concentration of AT-2–inactivated HIV-1 acts as both a specificity control for live HIV-1 and a comparator in the replication cycle as it does not progress to reverse transcription. Gene expression was examined during the 2 phases of the interactions of HIV-1 with DCs at 6, 24, and 48 hours after treatment. The results allowed identification of altered expression of relevant clusters of functionally related genes. Downstream analysis confirmed the significance of such clusters, such as those involved in DC maturation,11 interferon-stimulated genes (A.N.H. and A.L.C., unpublished data, March 2009), and lysosomal enzymes (this study).
Methods
Preparation of MDDCs
CD14+ monocytes derived from peripheral blood mononuclear cells (PBMCs) using CD14 magnetic beads (Miltenyi Biotec) were converted to immature MDDCs (iMDDCs) by culture in granulocyte macrophage colony-stimulating factor (GM-CSF) and interleukin-4 (IL-4) for 6 days as described previously.11 iMDDCs expressed on their surface high MR, DC-SIGN, and HLA-DR and moderate to low CD40, CD80, and CD86, and are negative for CD14 and CD83. At day 6, fresh media and cytokines were added before experiments were performed.
Preparation of naive T lymphocytes
Naive T lymphocytes were isolated from PBMCs using a magnetic bead isolation kit (Miltenyi Biotec).
Preparation of high-titer purified HIV-1 (BaL) virus stocks
Purified high-titer viable HIV-1BaL stocks in the order of 5 × 1010 TCID50 mL−1 were produced using tangential filter concentration and ultracentrifugation as described previously.11,14,30 A matched part of the inocula was inactivated with aldrithriol-2 (AT-2). Virus content was determined by p24 gag enzyme-linked immunosorbent assay (Beckman-Coulter) and as 50% tissue culture infectious dose (TCID50) value generated in TZM-B1 cells (National Institutes of Health [NIH] AIDS Research and Reference Reagent Program, contributed by John Kappes and Xiaoyun Wu) measured by luciferase reporter gene expression after a single round of infection.31 The endotoxin levels of these virus stocks were below the detectable limit of 0.005 U/mL or 0.0005 ng/mL (limulus amebocyte lysate assay; Sigma), and testing for residual interferon α, β, γ, and TNF-α by enzyme-linked immunosorbent assay was negative.
Treatment of MDDCs or naive CD4 T cells with HIV-1 virions or gp120
Day-6 MDDCs were seeded at 106 cells/mL and treated with either viable HIV-1BaL at an MOI of 10 (3.5 μg p24 per 106 cells) or the equivalent amount of AT-2–treated HIV-1BaL as determined by serial dilution, Western blot, and densitometry (supplemental Figure 1A, available on the Blood website; see the Supplemental Materials link at the top of the online article); or with recombinant monomeric gp120 at 50 ng/mL to 5 μg/mL (purified from laboratory adapted BaL strain from NIH AIDS Research & Reference Reagent Program); or with 1 of 2 strains of recombinant trimeric gp140 at 0.1 mg/mL (derived from JRFL and UG037 R5 HIV-1 strains); or mock treated with media prepared in a similar fashion to the virus stock preparation. Naive CD4 T cells were seeded at 2 × 106 cells/mL and treated with the recombinant monomeric gp120 and trimeric gp140 strains at 50 ng per 106 cells as described by Cicala et al.22
Biotinylated gp120 binding assay
Determination of HIV-1 infectivity by quantitative PCR
Microarray hybridization
Total RNA derived from MDDCs treated with monomeric gp120, viable and AT-2–inactivated HIV-1BaL, or mock from 4 independent experiments was amplified, reverse transcribed, and prepared for hybridization to Human ResGen 8k (Australian Genome Research Facility) glass microarrays containing 8000 human cDNAs spotted in duplicate as described previously.11 The mRNA derived from MDDCs treated with trimeric gp140s was amplified and labeled with biotin using Illumina total prep RNA Amp kit (Ambion) and hybridized to sentrix human 6 (Version 2) expression chips containing 46 000 transcripts (Illumina). A full list of gene expression data can be found at http://www.wmi.usyd.edu.au/research/virus.htm. The microarray data are hosted at http://www.wmi.usyd.edu.au/ourpeople/profiles/cunningham_tony.htm under the heading Supplementary Data.
Analysis of microarray data
The hybridized cDNA microarrays were scanned using an Axon GenePix 4000B scanner (Axon Instruments) and images were processed using GenePix 5.0 software. Following image quantitation, all analyses was undertaken using the R (V 2.0.0) statistical computing environment and BioConductor (V 1.5). All normalization and data analysis were carried out as described previously.11 A linear model was fitted to the full data set, and the closed loop experimental design (supplemental Figure 1B) allowed estimation of the differential expression (DE) between pairs of samples not hybridized together onto a single array via the use of linear contrasts of the parameter estimates from the fitted model. Since estimates of the log2(DE) between pairs of samples not hybridized together onto a single array were linear combinations of at least 2 parameter estimates of log2(DE) between pairs hybridized together, the associated standard errors were larger than those of the parameter estimates corresponding to pairs hybridized together. We used Bayesian linear modeling methods (limma package) to produce estimates of log2(DE) and of the log (posterior odds of differential expression) denoted by “B value” for ranking genes based on their probability of being differentially expressed.28,29,32,33 Thus, the higher the B value the greater the probability of differential expression. For a given DE, larger standard errors for estimates of log2(DE) tend to be associated with smaller B values.
The hybridized Illumina chips were scanned on a beadarray reader (Illumina) and the images quantified using Illumina BeadScan software. Microarray analysis was carried out using Version 3 of BeadStudio software (Illumina). The raw data were normalized using the cubic splines method and the resulting gene list was filtered such that only genes with a detection P value less than .01 or a differential expression score greater than 13 or less than −13 were included. Functional annotation analysis of differentially expressed gene lists derived from either platform was carried out using the web-based DAVID tool (National Institute of Allergy and Infectious Diseases).34,35
Confirmation of differential gene expression by quantitative PCR
Total unamplified RNA was DNase I treated (Promega) and then reverse transcribed using oligo d(T) and superscript III (Invitrogen). The cDNA was then subject to QPCR using defined primers (Sigma) and SYBR Green (Invitrogen). The relative quantitation method (ΔΔCT)36 was used to evaluate the expression of selected genes with the glyceraldehyde-3-phosphate-dehydrogenase (GAPDH), β-actin, and β-2-microglubulin amplicons as an internal control and the normalizer for all data.37 PCR assays were performed for a total of 5 experiments. Primer sequences for GAPDH, β-actin, β-2-M, CD1a, CD40, CD80, CD83, and CXCR4 were used as previously published.11 In addition primers specific to the following genes were used: APA2, CCL3, CCL5, CCL15, CD48, CDK5, CTSB, CTSC, CTSH, CTSL, CTSS, STSZ, CSTA1, CST3, CST7, DNM1, EPS15, IL10RB, IL13RA1, IRF1, IRF2, GIP2, MX1, NFKBIA, OAS1, RAB5A, RPL31, SOCS3, STAT3, and TARBP. The specific sequences are presented in supplemental Table 1.
Detection of lysosomal enzyme expression by Western blot
Mock- or HIV-1BaL–treated MDDCs were lysed in Nonidet P-40 (pH 7) lysis buffer (50 mM citrate/phosphate, 1 mM EDTA, 0.5% Nonidet P-40) resolved by 12% Bis/Tris NuPage gels (Invitrogen), transferred to poly(vinylidene difluoride) (PVDF) membrane (GE Healthcare), blocked, and probed with appropriate dilutions of the respective primary antibody, followed by a secondary IgG antibody coupled with peroxidase. The enhanced chemiluminescence plus Detection Kit (GE Healthcare) was used to visualize the antibody-reactive proteins. Anti–cathepsins B, S, and H and anti–cystatin A antisera were kindly provided by E. Weber (University of Halle). Anti–cathepsins Z and L antibodies were obtained from Abcam, anti–cathepsin C and anti–cystatin F antibodies from R&D, and anti–cystatin C antibody was from Biomol. Anti–β-actin and anti-GAPDH were commercially obtained from Sigma.
Affinity labeling of active cysteine proteases
NP-40 cell lysate protein (7 μg; pH 5) was incubated with reaction buffer (50 mM citrate/phosphate, pH 5, 1 mM ethylenediamine tetraacetic acid, 30 mM dithiothreitol) in the presence of DCG-0N, an activity-based probe for cysteine cathepsins,38 for 1 hour at room temperature. Reactions were terminated by the addition of SDS-reducing sample buffer and immediate boiling. Samples were resolved by 12.5% SDS/polyacrylamide gel electrophoresis, then blotted on a PVDF membrane (GE Healthcare) and visualized using streptavidin-HRP (Vector Laboratories) and the enhanced chemiluminescence plus detection kit (GE Healthcare).
Fluorogenic substrate measurement of cathepsin activity
Catalytic activities of cathepsins Z(X), B, L, and S were determined fluorometrically by cleavage of the common synthetic substrate Z-Phe-Arg-7-amido-4-methylcoumarin (Bachem). NP-40 cell lysate protein (7 μg; pH 5) was incubated with pH 5 buffer (0.1 M citrate, 4 mM dithiothreitol, 4 mM ethylenediamine tetraacetic acid, 6 μM aprotinin, 0.02% Triton X-100) and 10 μM of the substrate at 37°C and excitated at 360 nm. Emitted fluorescence was measured at 465 nm every minute with a fluorescence reader (Tecan SpectraFluor). E-64 (30μM; Roche) completely inhibited enzyme activities, confirming the detection of cysteine protease activities. Samples were measured in triplicates. Catalytic activity of cathepsin S was measured with the recently published specific substrate Mca-GRWPPMGLPWE-Lys(Dnp)-DArg-NH2, which was kindly provided by N. Lützner.39 Cathepsin C activity was specifically measured by cleavage of the synthetic substrate NH2-Gly-Arg-AMC (0.5 μM) in 50 mM pH 5 sodium acetate buffer.
Results
Establishment of criteria for analysis of gene expression changes
To determine the genes differentially expressed in DCs in response to HIV-1 binding, entry and replication, MDDCs were treated with (1) a highly concentrated and purified viable HIV-1BaL stock at an MOI of 10, (2) the equivalent concentration of the same stock of HIV-1BaL, treated with AT-2, (3) recombinant monomeric HIV-1BaL gp120, or (4) mock treatment. MDDCs treated with viable HIV-1BaL showed 15% to 33% infectivity by 48 hours after treatment (supplemental Figure 1C). To determine cellular genes differentially expressed in response to the full virus replication cycle, labeled cDNA derived from mock-treated MDDCs and MDDCs infected with viable HIV-1BaL were hybridized to microarrays. To dissect which genes from this list were differentially expressed at different stages of the replication cycle, 3 additional comparisons were conducted: mock versus gp120 to assess viral binding to MDDCs; AT-2–inactivated HIV-1 versus gp120, which compares gene expression changes resulting from binding alone to binding and entry; and AT-2–inactivated HIV-1 versus viable HIV-1, to examine changes resulting from the later stages of the replication cycle.
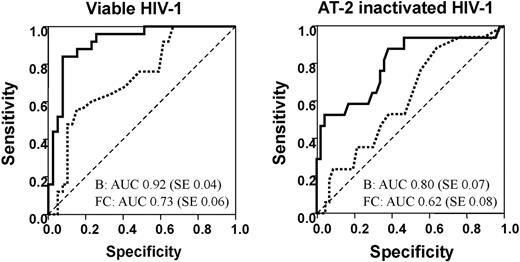
Quantitative PCR (QPCR) was used to validate gene expression changes identified by microarray analysis. Microarray-derived expression levels for a sample of 32 genes were compared with those determined by QPCR. The genes were chosen to cover a variety of treatments and time points, and genes were selected so that a range of B values and fold changes were included. B values are derived from Bayesian linear modeling32,33 and were chosen because they represent a measure of the statistical likelihood that a given gene is differentially expressed. A total of 34 instances in which no differential expression had been detected by the microarray were also included for comparison with QPCR. In all, 128 comparisons were made and a positive QPCR result for all biologic replicates was required as evidence of differential expression. Receiver operating characteristic (ROC) plots were constructed to discriminate between genes identified as being differentially expressed by QPCR and microarray using absolute fold change and B value for both viable and AT-2–treated HIV-1 (Figure 1). The closer the area under the curve (AUC) is to 1, the better the variable is at classifying differentially expressed genes.40 B value was a better classifier than fold change with its ROC curve lying consistently above that for the absolute fold change. Agreement between microarray and QPCR results was greater for live than AT-2–inactivated HIV-1. This was expected, as AT-2–inactivated HIV-1 versus media had not been hybridized on a single array but rather calculated indirectly, therefore losing precision. The sensitivity and specificity of different B value and absolute fold change cutoff points for predicting QPCR results were examined (supplemental Table 2). Acceptable sensitivity (> 85% for live and > 59% for AT-2 HIV-1) and high specificity (> 80% for both) were achieved using a B value higher than −0.5 for live HIV-1, and a B value higher than − 2.6 for AT-2 HIV-1.
Determination of methods for defining the statistical significance of differentially expressed genes by comparing microarrays and quantitative PCR data. Receiver operating characteristic (ROC) curves associated with using B values (unbroken line) or the absolute fold change (dashed line) to discriminate between genes identified as being differentially expressed or not by QPCR. The area under the curve (AUC) and its standard error (SE) are shown for each variable. The closer the AUC is to 1, the better the variable is at classifying differentially expressed genes.41
Determination of methods for defining the statistical significance of differentially expressed genes by comparing microarrays and quantitative PCR data. Receiver operating characteristic (ROC) curves associated with using B values (unbroken line) or the absolute fold change (dashed line) to discriminate between genes identified as being differentially expressed or not by QPCR. The area under the curve (AUC) and its standard error (SE) are shown for each variable. The closer the AUC is to 1, the better the variable is at classifying differentially expressed genes.41
Binding HIV-1 envelope proteins does not influence DC gene expression
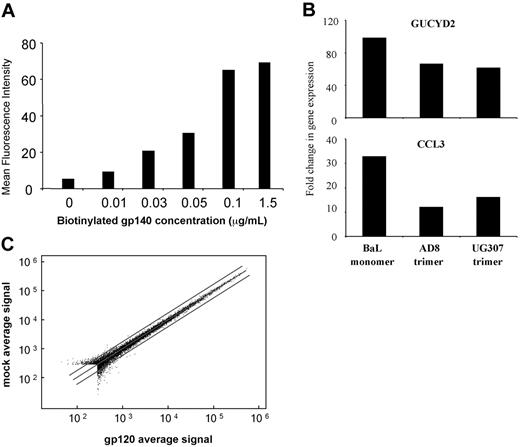
Monomeric gp120 had no effect on host cell gene expression using the 8K cDNA microarrays, which was unexpected. To determine whether gp120 is required to be in its native trimeric state to mediate surface signaling, we obtained 2 independent sources of trimeric gp140 derived from different R5 virus strains (AD8 and UG037). A binding assay was first carried out to ensure that trimeric gp140 was able to efficiently bind to MDDC to saturating levels (Figure 2A). In addition the biologic activity of both trimeric proteins was confirmed by demonstrating their ability to up-regulate GUCYD2 and CCL3 gene expression in naive T cells as previously published22,41 (Figure 2B). MDDCs were treated with saturating concentrations of both gp140 trimers for 2, 6, and 24 hours, and the differential gene expressed was determined using Illumina microarray chips, which assay expression for 44 000 transcripts. No MDDC genes were detected to be changed in their expression in response to treatment with either strain of trimeric gp140 at any time point (Figure 2C).
HIV gp120 binding affects differential expression of T-cell but not DC genes. (A) Binding of HIV-1 envelope proteins to MDDCs. Day-6 immature MDDCs (5 × 106) were incubated with biotinylated monomeric gp120 (BaL strain) and trimeric gp140 (AD8 and UG037 strains) ranging from 0 to 0.15 μg/μL. Biotinylated gp120/140 binding was determined by flow cytometry using PE-conjugated streptavidin. The geometric means of the fluorescent intensity data for the UG037 strain of trimer are displayed in the histogram. (B) Biologic activity of gp120 species. Naive CD4 T cells (5 × 106) were incubated with monomeric gp120 (BaL strain) and trimeric gp140 (AD8 and UG037 strains) at 50 ng per 106 cells for 24 hours. Differential expression of the GUCY2D and CCL3 genes was determined by QPCR. The fold change in gene expression compared with mock-treated cells is displayed in the histogram. (C) Differential MDDC gene expression in response to monomeric and trimeric HIV-1 envelope species. Day-6 immature MDDCs were treated with either recombinant monomeric gp120 (BaL strain) or 1 of 2 trimeric gp140 stocks (AD8 and UG037 strains) for 6, 24, and 48 hours (monomer) or 2, 6, and 24 hours (trimers). Global DC gene expression was determined using Illumina gene expression chips (trimeric gp140s). A representative MA plot is shown with 1.5-fold change lines for trimeric gp140 (UG037 strain) 24 hours after treatment.
HIV gp120 binding affects differential expression of T-cell but not DC genes. (A) Binding of HIV-1 envelope proteins to MDDCs. Day-6 immature MDDCs (5 × 106) were incubated with biotinylated monomeric gp120 (BaL strain) and trimeric gp140 (AD8 and UG037 strains) ranging from 0 to 0.15 μg/μL. Biotinylated gp120/140 binding was determined by flow cytometry using PE-conjugated streptavidin. The geometric means of the fluorescent intensity data for the UG037 strain of trimer are displayed in the histogram. (B) Biologic activity of gp120 species. Naive CD4 T cells (5 × 106) were incubated with monomeric gp120 (BaL strain) and trimeric gp140 (AD8 and UG037 strains) at 50 ng per 106 cells for 24 hours. Differential expression of the GUCY2D and CCL3 genes was determined by QPCR. The fold change in gene expression compared with mock-treated cells is displayed in the histogram. (C) Differential MDDC gene expression in response to monomeric and trimeric HIV-1 envelope species. Day-6 immature MDDCs were treated with either recombinant monomeric gp120 (BaL strain) or 1 of 2 trimeric gp140 stocks (AD8 and UG037 strains) for 6, 24, and 48 hours (monomer) or 2, 6, and 24 hours (trimers). Global DC gene expression was determined using Illumina gene expression chips (trimeric gp140s). A representative MA plot is shown with 1.5-fold change lines for trimeric gp140 (UG037 strain) 24 hours after treatment.
Kinetics of gene expression in HIV-1–treated DCs
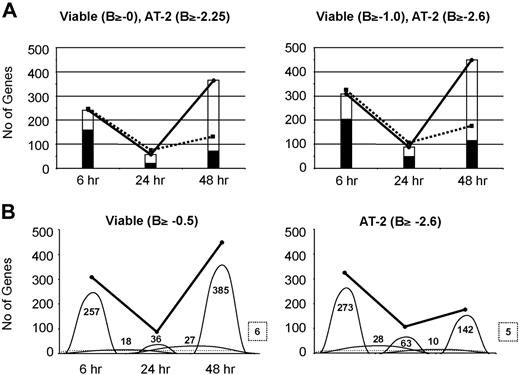
The total numbers of differentially expressed genes were determined in DCs treated with live or inactivated HIV-1BaL for 6, 24, and 48 hours, across the previously determined 2 successive temporal phases of HIV-1 interaction with DCs.14 In addition, the number of genes in common and those that were different between the 2 treatments were quantified (Figure 3A). The pattern of changes was similar whether B values of 0/2.25 or −0.5/2.60 (supplemental Table 2) were used for live or inactivated virus, respectively. The total number of differentially expressed genes decreased significantly from 6 to 24 hours and then rose again at 48 hours, particularly in MDDCs treated with viable virus. The proportion of genes in common was much lower at 48 hours than at 6 or 24 hours, indicating that, as expected, replicating viable HIV-1BaL has a much greater modulatory effect on DC gene expression than the matched inocula of inactivated nonreplicating virus at 48 hours after treatment.
Kinetics of differential DC gene expression induced by HIV-1 uptake and infection. Numbers of differentially expressed genes in response to viable and AT-2–treated HIV-1 are shown. Viable and AT-2–inactivated HIV-1–treated MDDC differentially expressed gene lists were generated after filtering according to B value. (A) Commonly expressed genes in viable and AT-2–inactivated HIV-1–treated MDDCs. The numbers of genes differentially expressed in response to viable and AT-2–inactivated HIV-1 at 6, 24, and 48 hours after treatment are illustrated by the solid and broken lines, respectively. The open histogram shows the numbers of genes differentially expressed in response to viable HIV-1 only, and the shaded area shows the number of genes that were commonly changed in their expression in cells treated with viable virus and those treated with AT-2–inactivated virus. (B) Commonly expressed genes between time points. The numbers of genes differentially expressed in response to viable and AT-2–inactivated HIV-1 at 6, 24, and 48 hours after treatment are illustrated by the solid line. The numbers of specific genes expressed at each time point, or common between 2 time points, are shown by the domes, with the number of genes shown. The dotted line illustrates the numbers of genes differentially expressed at all 3 time points with the number shown in a box with a broken line.
Kinetics of differential DC gene expression induced by HIV-1 uptake and infection. Numbers of differentially expressed genes in response to viable and AT-2–treated HIV-1 are shown. Viable and AT-2–inactivated HIV-1–treated MDDC differentially expressed gene lists were generated after filtering according to B value. (A) Commonly expressed genes in viable and AT-2–inactivated HIV-1–treated MDDCs. The numbers of genes differentially expressed in response to viable and AT-2–inactivated HIV-1 at 6, 24, and 48 hours after treatment are illustrated by the solid and broken lines, respectively. The open histogram shows the numbers of genes differentially expressed in response to viable HIV-1 only, and the shaded area shows the number of genes that were commonly changed in their expression in cells treated with viable virus and those treated with AT-2–inactivated virus. (B) Commonly expressed genes between time points. The numbers of genes differentially expressed in response to viable and AT-2–inactivated HIV-1 at 6, 24, and 48 hours after treatment are illustrated by the solid line. The numbers of specific genes expressed at each time point, or common between 2 time points, are shown by the domes, with the number of genes shown. The dotted line illustrates the numbers of genes differentially expressed at all 3 time points with the number shown in a box with a broken line.
These gene changes were further analyzed according to whether they were persistent across the 2 or 3 time points or identified only at one of the time points (Figure 3B). Most of the gene changes, including those in common (data not shown), were confined to 6, 24, or 48 hours rather than persisting across time points. Concordant functional gene changes between live and inactivated virus followed a similar pattern, indicating a rapid succession of different cellular genes being transcribed corresponding to the 2 successive phases: viral endocytosis and destruction followed by de novo replication.
Gene groups of interest: functional annotation analysis
Using a B value cutoff of −.05 and −2.6 for viable and AT-2–inactivated HIV-1 gene lists, respectively, lists of up- and down-regulated genes were generated for MDDCs treated with viable and AT-2–inactivated virus for 6, 12, and 48 hours, and functional annotation analysis was carried out (Table 1).
Functional annotation of differentially expressed gene groups
| Gene cluster . | 6 h . | 24 h . | 48 h . | |||
|---|---|---|---|---|---|---|
| Viable . | AT2 . | Viable . | AT2 . | Viable . | AT2 . | |
| Up-regulated gene clusters | ||||||
| Endosome | 2.3 | 2.0 | ||||
| GTPase activity | 1.6 | 1.6 | 1.1 | |||
| Immune response | 1.4 | 1.1 | 5.2 | 7.6 | ||
| Induction of apoptosis/apoptosis | 1.0 | 1.0 | 1.6 | 2.3 | 1.4 | |
| NF-kB associated | 1.6 | 1.7 | 1.3 | |||
| TNFR2 signaling | 1.6 | |||||
| TNF/stress signaling | 1.6 | 1.3 | ||||
| MAPK signaling | 1.6 | 1.3 | ||||
| TLR signaling | 1.6 | 1.3 | ||||
| Translation/ribosomal parts | 1.5 | |||||
| Antiapoptosis | 1.5 | 1.4 | ||||
| Response to virus | 3.7 | 4.0 | ||||
| Down-regulated gene clusters | ||||||
| Mitochondria/respiration | 2.0 | 1.6 | 1.1 | 3.2 | 2.5 | |
| Nucleus associated | 1.7 | 1.6 | 1.1 | 2.3 | ||
| RNA splicing | 1.4 | 1.6 | 1.2 | 2.3 | ||
| Translation/ribosomal parts | 3.2 | 2.9 | ||||
| Lysosome | 2.7 | |||||
| Gene cluster . | 6 h . | 24 h . | 48 h . | |||
|---|---|---|---|---|---|---|
| Viable . | AT2 . | Viable . | AT2 . | Viable . | AT2 . | |
| Up-regulated gene clusters | ||||||
| Endosome | 2.3 | 2.0 | ||||
| GTPase activity | 1.6 | 1.6 | 1.1 | |||
| Immune response | 1.4 | 1.1 | 5.2 | 7.6 | ||
| Induction of apoptosis/apoptosis | 1.0 | 1.0 | 1.6 | 2.3 | 1.4 | |
| NF-kB associated | 1.6 | 1.7 | 1.3 | |||
| TNFR2 signaling | 1.6 | |||||
| TNF/stress signaling | 1.6 | 1.3 | ||||
| MAPK signaling | 1.6 | 1.3 | ||||
| TLR signaling | 1.6 | 1.3 | ||||
| Translation/ribosomal parts | 1.5 | |||||
| Antiapoptosis | 1.5 | 1.4 | ||||
| Response to virus | 3.7 | 4.0 | ||||
| Down-regulated gene clusters | ||||||
| Mitochondria/respiration | 2.0 | 1.6 | 1.1 | 3.2 | 2.5 | |
| Nucleus associated | 1.7 | 1.6 | 1.1 | 2.3 | ||
| RNA splicing | 1.4 | 1.6 | 1.2 | 2.3 | ||
| Translation/ribosomal parts | 3.2 | 2.9 | ||||
| Lysosome | 2.7 | |||||
Differentially expressed gene lists were generated for viable HIV-1–treated cells at 6, 24, and 48 hours using a B-value cutoff of −0.5 and for AT-2–inactivated cells using a B-value cutoff of −2.6. Separate lists were made for up and down-regulated genes. The web-based DAVID functional annotation software34 was used to group the genes according to their function, and the degree to which their expression was enriched (overrepresented) against the background of all the genes spotted on the arrays was determined. The numbers refer to the DAVID-generated enrichment factor. Numbers are shown only for those gene groups with an enrichment score greater than 1.
At 6 hours after HIV-1BaL treatment, enriched gene clusters of up- and down-regulated genes were very similar in cells treated with both viable and AT-2–inactivated virus. In the up-regulated enriched gene lists, those associated with the endosome and GTPase activity were the most enriched and included RAB7, RAB11A, RAB35, and RHOG (supplemental Table 3). Others included those associated with the induction of apoptosis and the innate immune response. In the down-regulated gene lists, those associated with mitochondrial respiration and the nucleus (especially RNA splicing) represented the most enriched gene clusters.
At 24 hours after treatment with viable HIV-1, gene groups associated with NF-κB, TNFR2, TNF, MAPK, and toll-like receptor (TLR) signaling pathways, GTPase activity, and apoptosis were strongly enriched. No significantly enriched up-regulated gene groups were detected in MDDCs treated with AT-2–inactivated virus. However, many down-regulated gene groups were enriched. As at 6 hours, these included gene groups associated with mitochondrial respiration, the nucleus (especially RNA splicing), and translation. With viable HIV-1, down-regulated gene groups were less enriched, but similar to AT-2–inactivated HIV-1.
At 48 hours after treatment, many gene groups were strongly enriched in response to both viable and inactivated virus. The most enriched up-regulated gene groups were those associated with the innate immune response (particularly in response to viral infection). Genes associated with NF-κB, TNF, and MAPK signaling pathways were enriched in AT-2–inactivated HIV-1–treated cells but not those treated with viable virus, suggesting that viable HIV-1 shuts down expression of these genes. Genes associated with translation were enriched in cells treated with viable HIV-1 but not AT-2–inactivated HIV-1. In addition, a mixture of both apoptotic and antiapoptotic genes was up-regulated in response to both inactivated and viable HIV-1. No strongly enriched down-regulated gene groups were detected with AT-2 inactivated HIV-1. However viable HIV-1 down-regulated gene groups associated with the lysosome were of particular interest, including enzymes such as hydrolases (ACP5), cathepsins (CTSC), and other lysosomal proteins such as LIPA and LYPLA3 (Table 2). Down-regulated genes associated with mitochondrial respiration and translation were also enriched in MDDCs treated with viable HIV-1.
Down-regulated lysosomal genes in response to viable HIV-1
| Acc no. . | Symbol . | Name . | FC . | B . |
|---|---|---|---|---|
| R08817 | ACP5 | Acid phosphatase 5, tartrate resistant | −3.0 | 4.4 |
| AA644088 | CTSC | Cathepsin C | −1.8 | 0.2 |
| AA457725 | GABARAP | GABA(A) receptor-associated protein | −1.5 | 0.0 |
| AA418683 | HPS1 | Hermansky-Pudlak syndrome 1 | −1.6 | 0.3 |
| AA630104 | LIPA | Lipase A, lysosomal acid, cholesterol esterase | −1.9 | 0.3 |
| W72651 | LYPLA3 | Lysophospholipase 3 (lysosomal phospholipase A2) | −1.8 | 0.3 |
| AW025732 | NCF2 | Neutrophil cytosolic factor 2 | −2.2 | 1.8 |
| AA630449 | NPC2 | Niemann-Pick disease, type C2 | −1.5 | 0.7 |
| AI954240 | SGSH | N-sulfoglucosamine sulfohydrolase (sulfamidase) | −2.0 | 5.4 |
| AA063637 | PPT1 | Palmitoyl-protein thioesterase 1 | −1.4 | −0.3 |
| Acc no. . | Symbol . | Name . | FC . | B . |
|---|---|---|---|---|
| R08817 | ACP5 | Acid phosphatase 5, tartrate resistant | −3.0 | 4.4 |
| AA644088 | CTSC | Cathepsin C | −1.8 | 0.2 |
| AA457725 | GABARAP | GABA(A) receptor-associated protein | −1.5 | 0.0 |
| AA418683 | HPS1 | Hermansky-Pudlak syndrome 1 | −1.6 | 0.3 |
| AA630104 | LIPA | Lipase A, lysosomal acid, cholesterol esterase | −1.9 | 0.3 |
| W72651 | LYPLA3 | Lysophospholipase 3 (lysosomal phospholipase A2) | −1.8 | 0.3 |
| AW025732 | NCF2 | Neutrophil cytosolic factor 2 | −2.2 | 1.8 |
| AA630449 | NPC2 | Niemann-Pick disease, type C2 | −1.5 | 0.7 |
| AI954240 | SGSH | N-sulfoglucosamine sulfohydrolase (sulfamidase) | −2.0 | 5.4 |
| AA063637 | PPT1 | Palmitoyl-protein thioesterase 1 | −1.4 | −0.3 |
The table shows the GenBank43 accession number symbol and name, the fold change increase in its expression (FC), and B value (B), for all lysosomal genes down-regulated in MDDC response to treatment with viable HIV-1 for 48 hours. No lysosomal genes were down-regulated in response to treatment with AT-2–inactivated HIV-1.
Effects of HIV-1 treatment of MDDCs on the expression of cathepsins and cystatins
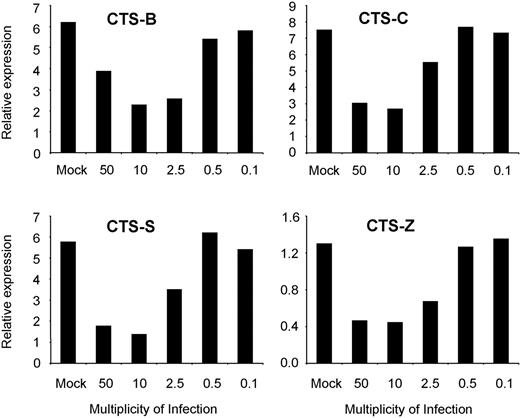
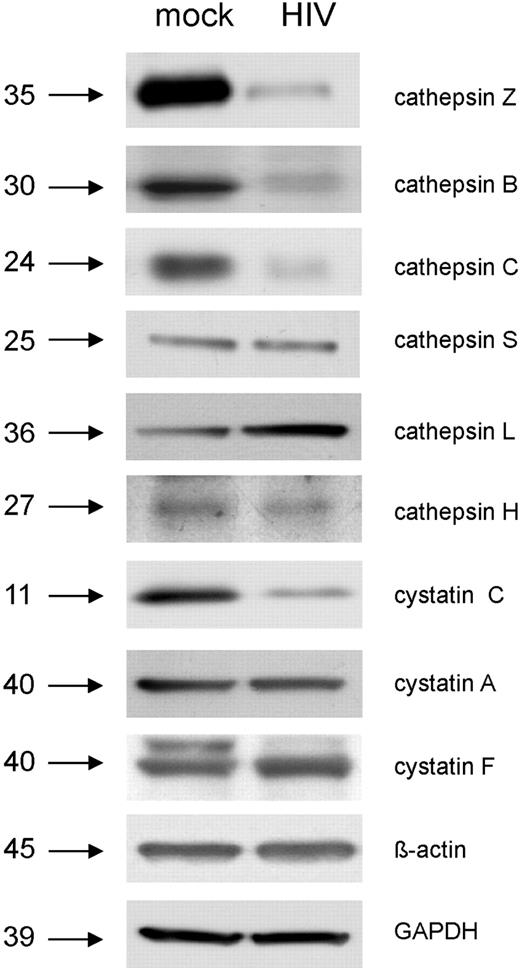
The microarray data presented in Tables 1 and 2 strongly suggest that HIV-1 down-regulates lysosomal activity in MDDCs during the later phase of de novo infection (48 hours after treatment). To further explore this possibility, we determined the expression of genes that encode a range of cathepsins and cystatins in MDDCs treated with viable and AT-2–inactivated HIV-1BaL for 48 hours by QPCR (Table 3). We also determined whether down-regulation of cathepsin gene expression in response to viable HIV-1BaL occurs in a dose-dependent manner (Figure 4). Next, expression of the proteins that these genes encode in MDDCs after treatment with viable HIV-1BaL was examined using Western blotting (Figure 5). Cathepsins B, C, S, and Z RNAs were all markedly decreased in cells treated with viable but not with AT-2–inactivated HIV-1Bal, and this occurred in a dose-dependent manner with no down-regulation seen with an MOI lower than 2. The corresponding proteins were all clearly down-regulated in their expression in response to HIV-1BaL, except for cathepsin S, which appeared unchanged. In contrast, cathepsins H and L RNAs were increased in response to both viable and AT-2–inactivated HIV-1BaL. However, only the cathepsin L protein was up-regulated as determined by Western blot; the cathepsin H protein levels were unchanged. Cystatin C, which negatively regulates cathepsin L,43 was down-regulated at both the gene and protein level in cells treated with HIV-1BaL. Cystatin F, which negatively regulates cathepsin C,44 was up-regulated as RNA and also slightly at the protein level; cystatin A RNA increased but protein expression remained unchanged.
Summary of cathepsin and cystatin gene and protein differential expression and enzymatic activity in HIV-1–treated MDDCs
| Name . | Microarray (FC) . | QPCR (FC) . | Western blot . | Function . | ||
|---|---|---|---|---|---|---|
| Viable . | Inactivated . | Viable . | Inactivated . | |||
| Cathepsin B | – | – | −3.5 | No change | ↓↓↓ | ↓↓↓ |
| Cathepsin C | −1.8 | No change | −2.0 | No change | ↓↓↓ | ↓↓ |
| Cathepsin Z | – | – | −2.6 | No change | ↓↓↓ | ↓↓↓ |
| Cathepsin S | −1.3 | No change | −1.8 | No change | ↓↓ | ↓↓ |
| Cathepsin L | +2.1 | +1.6 | +5.0 | +1.5 | No change | ↓↓↓ (BZLS) |
| Cathepsin H | +1.8 | +1.5 | +1.9 | +1.9 | No change | – |
| Cystatin C | −1.9 | No change | −2.5 | −2.1 | ↓↓ | – |
| Cystatin F | +2.3 | +1.5 | +3.0 | +1.5 | ↑ | – |
| Cystatin A | +1.9 | No change | +2.1 | No change | No change | – |
| Name . | Microarray (FC) . | QPCR (FC) . | Western blot . | Function . | ||
|---|---|---|---|---|---|---|
| Viable . | Inactivated . | Viable . | Inactivated . | |||
| Cathepsin B | – | – | −3.5 | No change | ↓↓↓ | ↓↓↓ |
| Cathepsin C | −1.8 | No change | −2.0 | No change | ↓↓↓ | ↓↓ |
| Cathepsin Z | – | – | −2.6 | No change | ↓↓↓ | ↓↓↓ |
| Cathepsin S | −1.3 | No change | −1.8 | No change | ↓↓ | ↓↓ |
| Cathepsin L | +2.1 | +1.6 | +5.0 | +1.5 | No change | ↓↓↓ (BZLS) |
| Cathepsin H | +1.8 | +1.5 | +1.9 | +1.9 | No change | – |
| Cystatin C | −1.9 | No change | −2.5 | −2.1 | ↓↓ | – |
| Cystatin F | +2.3 | +1.5 | +3.0 | +1.5 | ↑ | – |
| Cystatin A | +1.9 | No change | +2.1 | No change | No change | – |
The table summarizes the differential expression of genes encoding cathepsin and cystatin as well as associated protein levels and the enzymatic activity of cathepsins in MDDCs treated with HIV-1BAL (MOI1) for 48 hours. If a gene was not spotted on the microarray or a function assay was not performed, then this is marked with a dash.
Down-regulation of cathepsin gene expression in response to HIV-1 occurs in a dose-dependent manner. Day-6 MDDCs were treated with HIV-1BaL at MOIs ranging from 50 to 0.1, or mock treated. After 48 hours, cathepsins B, C, S, and Z gene expression was determined by QPCR. All data were normalized to GAPDH expression. Representative data from 1 of 2 experiments are shown.
Down-regulation of cathepsin gene expression in response to HIV-1 occurs in a dose-dependent manner. Day-6 MDDCs were treated with HIV-1BaL at MOIs ranging from 50 to 0.1, or mock treated. After 48 hours, cathepsins B, C, S, and Z gene expression was determined by QPCR. All data were normalized to GAPDH expression. Representative data from 1 of 2 experiments are shown.
Determination of lysosomal enzyme levels in response to HIV-1 infection by Western blot. Day-6 immature MDDCs were exposed to HIV-1BaL (MOI 10) or mock treated for 48 hours. The relative quantities of lysosomal enzymes present in HIV-1–treated cells compared with mock were then determined by Western blot. Both GAPDH and beta actin were used as loading controls.
Determination of lysosomal enzyme levels in response to HIV-1 infection by Western blot. Day-6 immature MDDCs were exposed to HIV-1BaL (MOI 10) or mock treated for 48 hours. The relative quantities of lysosomal enzymes present in HIV-1–treated cells compared with mock were then determined by Western blot. Both GAPDH and beta actin were used as loading controls.
Effects of HIV-1 on cathepsin functional activity in MDDCs
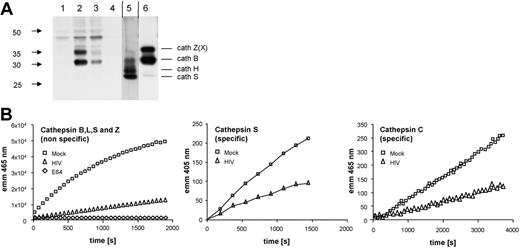
We next carried out functional assays on lysosomal activity in MDDCs infected with HIV-1BaL for 48 hours. First, an affinity labeling assay was performed with the active-site label DCG0N (Figure 6A). This probe irreversibly binds to the active site of cysteine cathepsins and can later be detected by blotting against the biotin moiety of DCG0N. The resulting bands thus directly represent cathepsin activities in a semiquantitative manner that can be compared with the known labeling pattern of the controls (monocyte organelle lysate and macrophage cell lysate).45 Both cathepsins B and Z showed a significant decrease in activity, whereas cathepsins H and S activity was below the detection limit. In addition, we carried out different fluorogenic substrate assays to measure cathepsin enzymatic activity (Figure 6B). The combined activity of cathepsins Z, B, L, and S was significantly reduced in HIV-1BaL–treated MDDCs as well as the specific activities of cathepsins S and C.
Determination of cathepsin enzyme activity in HIV-1–treated MDDCs. Day-6 immature MDDCs were exposed to HIV-1BaL (MOI 10) or mock treated for 48 hours and then lysed in a low pH buffer. (A) Affinity labeling of active cysteine proteases. Cell lysates were incubated with the activity-based probe for cysteine cathepsins DCG-0N. Labeled cathepsins then detected by Western blot. Lanes: (1) Mock MDDCs, leupeptin, (2) mock MDDCs, (3) HIV-1BaL-treated MDDCs, (4) boiled HIV-1BaL–treated MDDCs, (5) monocyte organelles, (6) macrophage cell lysate. Vertical lines denote a section of the Western blot that was included with a lower exposure time. (B) Fluorogenic substrate measurement of cathepsin activity. The combined catalytic activities of cathepsins X, B, L, and S were determined fluorometrically by cleavage of the common synthetic substrate Z-Phe-Arg-7-amido-4-methylcoumarin. Specific catalytic activities of cathepsins S and C were measured using the substrates Mca-GRWPPMGLPWE-Lys(Dnp)-DArg-NH2 and NH2-Gly-Arg-AMC, respectively. Samples were measured in triplicate.
Determination of cathepsin enzyme activity in HIV-1–treated MDDCs. Day-6 immature MDDCs were exposed to HIV-1BaL (MOI 10) or mock treated for 48 hours and then lysed in a low pH buffer. (A) Affinity labeling of active cysteine proteases. Cell lysates were incubated with the activity-based probe for cysteine cathepsins DCG-0N. Labeled cathepsins then detected by Western blot. Lanes: (1) Mock MDDCs, leupeptin, (2) mock MDDCs, (3) HIV-1BaL-treated MDDCs, (4) boiled HIV-1BaL–treated MDDCs, (5) monocyte organelles, (6) macrophage cell lysate. Vertical lines denote a section of the Western blot that was included with a lower exposure time. (B) Fluorogenic substrate measurement of cathepsin activity. The combined catalytic activities of cathepsins X, B, L, and S were determined fluorometrically by cleavage of the common synthetic substrate Z-Phe-Arg-7-amido-4-methylcoumarin. Specific catalytic activities of cathepsins S and C were measured using the substrates Mca-GRWPPMGLPWE-Lys(Dnp)-DArg-NH2 and NH2-Gly-Arg-AMC, respectively. Samples were measured in triplicate.
Discussion
Viruses shape their intracellular environment through alterations in host cell gene transcription, protein translation, and posttranslational modification, often initiating these changes by signaling through cell surface receptors46-48 or at subsequent stages in their replication cycle. The binding of HIV-1 gp120 to the chemokine receptor CCR5 and CXCR4 in both macrophages and T cells provides an example of such signaling.49,50 Although genomic screens have been used to examine the range of host cell gene expression and protein changes in HIV-1–infected cells,1,21,22 the effect of individual recombinant proteins or simulating antibodies has been easier to verify because of their relative purity compared with an HIV-1 inoculum.22,51 In some cell types, the extent of gene transcriptional changes is less than others (eg, macrophages versus T lymphocytes for HIV-1)4,19 and the effect of HIV-1 on the cellular transcriptome is often less dramatic than that of other organisms (eg, HIV-1 versus Mycobacterium tuberculosis).52
Special circumstances in HIV-1 infection of DCs, where more than 95% of the HIV-1 inoculum is endocytosed and destroyed within the first 12 hours, markedly reduce the proportion available for de novo infection.14,53 There are 2 potential phases of HIV-1 effects on the cellular transcriptome, the first through HIV-1 binding, entry, and possibly effects from within the endosome during the first 12 hours, and the second during the phase of de novo HIV-1 replication, particularly beyond 24 hours after infection.14
Because the HIV-1 MOI is essentially lower for DC infection compared with other cell types, thus resulting in smaller changes in gene transcription, and because of low signal to noise distinction by microarrays, we used a highly rigorous analysis of the microarray data. A large sample of genes was compared with QPCR-derived gene expression (Figure 1). This facilitated the establishment of thresholds that were an appropriate compromise between sensitivity and specificity and allowed selection of a manageable number of functionally altered genes for further downstream analysis. The use of B values for reproducibility of gene transcriptional changes was more significant than measures of the magnitude of the change.
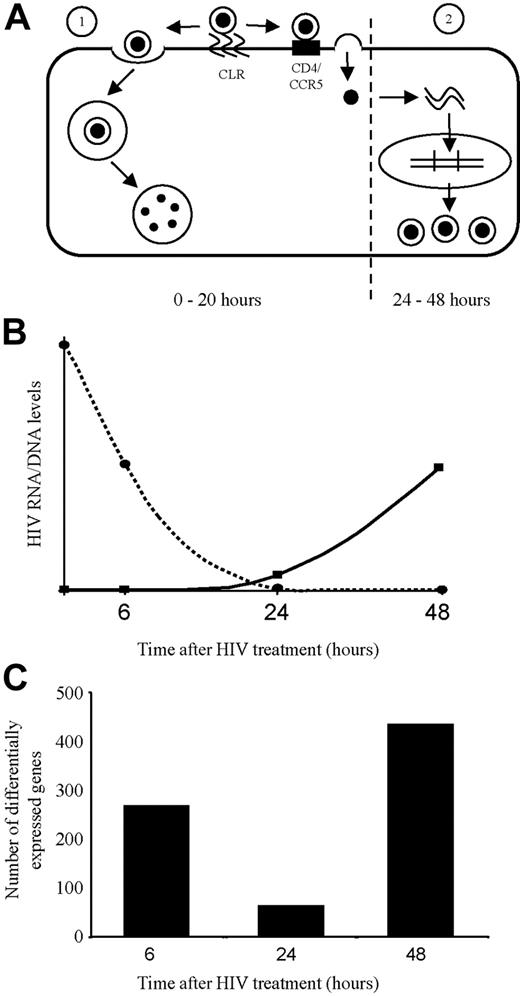
As illustrated in Figures 3 and 7, the overall transcriptional changes were biphasic and correlated well with our previously documented kinetics of the biphasic fate of HIV-1 in DCs; endocytic destruction, or de novo replication.14 Furthermore, the second-phase changes beyond 24 hours after infection were mostly seen in the DCs infected with live virus, demonstrated as discordant results between live and AT-2–treated HIV-1. This second phase of transcriptional changes increased from 24 to 48 hours after infection, correlating with the kinetics of de novo HIV-1 replication in a single cycle. In contrast, the first phase changes were apparent at 6 hours and declined by 24 hours. In this phase, there was a good correlation between the number and nature of the gene transcriptional changes induced by live HIV-1 and AT-2–inactivated HIV-1. It was interesting to note that most of the gene transcriptional changes were transient and only a minority persisted across the 2 phases (ie, at 2 or 3 time points). These transient changes are also consistent with a model of 2 different phases of induction: binding/entry/signaling from the endosome versus steps in the de novo replication cycle.
Kinetics of differential gene expression corresponds with the 2 phases of HIV-1 processing by dendritic cells (DCs). (A) Routes of entry of HIV-1 into DCs. Following initial binding of HIV-1 to C-type lectin receptors (CLRs) on the cell surface, HIV-1 can enter the DC by 1 of 2 routes. (1) More than 95% of the virus is rapidly endocytosed resulting in processing by the endolysosomal pathway and degradation by acid proteolysis in late endosomes. (2) The remaining less than 5% of attached virus is transferred to cell surface CD4/CCR5 resulting in fusion of the viral envelope with the DC plasma membrane and entry of the capsid into cytoplasm, resulting in reverse transcription, HIV-1 cDNA generation (first detected < 24 hours after infection = second phase of de novo replication), and then the later stages of viral replication and assembly. (B) Corresponding HIV-1 nucleic acid levels in DCs after HIV-1 treatment. The graph shows a representation of the HIV-1 nucleic acid levels after treatment of DCs. As shown by the dotted line, at time 0 the high levels of HIV-1 nucleic acid represents input virions. The nucleic acid levels then rapidly decrease to undetectable levels by 20 hours after infection as the vast majority of HIV-1 enters and is destroyed by late endosomes. As shown by the solid line, 20 hours after infection viral nucleic acid levels begin to slowly rise, representing progeny virions resulting from productive infection by the small amount of virus that was able to fuse with the plasma membrane and initiate a productive infection. (C) Temporal differential gene expression corresponds to the kinetics of the 2 phases of HIV-1 processing by DCs. At 6 hours, a burst of gene expression was seen as a consequence of viral entry into the DC (predominantly via endocytosis). By 24 hours, most of the virus that entered via endocytosis has been degraded and virions that entered by fusion with the plasma membrane have yet to undergo replication. Correspondingly very few genes are differentially expressed at this time point. At 48 hours, a fresh burst of gene expression is detected, representing genes differentially expressed as a result of HIV-1 replication in DCs.
Kinetics of differential gene expression corresponds with the 2 phases of HIV-1 processing by dendritic cells (DCs). (A) Routes of entry of HIV-1 into DCs. Following initial binding of HIV-1 to C-type lectin receptors (CLRs) on the cell surface, HIV-1 can enter the DC by 1 of 2 routes. (1) More than 95% of the virus is rapidly endocytosed resulting in processing by the endolysosomal pathway and degradation by acid proteolysis in late endosomes. (2) The remaining less than 5% of attached virus is transferred to cell surface CD4/CCR5 resulting in fusion of the viral envelope with the DC plasma membrane and entry of the capsid into cytoplasm, resulting in reverse transcription, HIV-1 cDNA generation (first detected < 24 hours after infection = second phase of de novo replication), and then the later stages of viral replication and assembly. (B) Corresponding HIV-1 nucleic acid levels in DCs after HIV-1 treatment. The graph shows a representation of the HIV-1 nucleic acid levels after treatment of DCs. As shown by the dotted line, at time 0 the high levels of HIV-1 nucleic acid represents input virions. The nucleic acid levels then rapidly decrease to undetectable levels by 20 hours after infection as the vast majority of HIV-1 enters and is destroyed by late endosomes. As shown by the solid line, 20 hours after infection viral nucleic acid levels begin to slowly rise, representing progeny virions resulting from productive infection by the small amount of virus that was able to fuse with the plasma membrane and initiate a productive infection. (C) Temporal differential gene expression corresponds to the kinetics of the 2 phases of HIV-1 processing by DCs. At 6 hours, a burst of gene expression was seen as a consequence of viral entry into the DC (predominantly via endocytosis). By 24 hours, most of the virus that entered via endocytosis has been degraded and virions that entered by fusion with the plasma membrane have yet to undergo replication. Correspondingly very few genes are differentially expressed at this time point. At 48 hours, a fresh burst of gene expression is detected, representing genes differentially expressed as a result of HIV-1 replication in DCs.
The lack of significant changes in the transcriptome after treatment of DCs with physiological concentrations of soluble gp120 monomer, or saturating concentrations of soluble gp140 trimer (even using the expanded range of genes available on the Illumina arrays), was unexpected. This was not a strain-specific effect as trimeric HIV-1 envelope proteins from 2 different R5 strains produced similar results. Furthermore the HIV-1 envelope proteins used in this study were functionally active, as they induced the previously reported up-regulation of the transcription of GUCYD222 and CCL341 genes in T lymphocytes (Figure 3B). These results are also consistent with our previous failure to demonstrate gp120-induced changes in DC maturation genes, in contrast to whole virus.11 The results were unexpected as HIV-1 binding to DCs has been shown to signal through DC-SIGN51,54 and in macrophages through CCR5, which should be simulated by gp120 binding. However such signaling resulted in protein phosphorylation55 and may not necessarily be accompanied by transcriptional changes. Furthermore, we have previously shown that HIV-1 monomeric gp120 traffics to different compartments of MDDCs; gp120, to LAMP-positive lysosomes; and HIV-1, only to late endosomes.14 Therefore, the marked first phase changes induced by AT-2–treated HIV-1 are probably due to signaling after viral entry.
Different classes of genes are affected in the 2 phases of HIV-DC interaction. In the first phase, associated with endocytosis, GTPase-mediated signal transduction and immune recognition were up-regulated, consistent with the fate of the virus during this phase. In particular, some were associated with recycling endosomes (eg, Rab35 and Rab11A) that may carry HIV-1 to the surface and into the viral synapse when DCs come into contact with T cells. Second-phase changes included the up-regulation of expression of genes associated with NF-κB and components of the TNFR2, TNF, MAPK, and TLR signaling pathways, as well as those associated with GTPase activity, and a mixture of apoptotic and antiapoptotic genes. In cells treated with viable HIV-1, but not inactivated HIV-1, genes encoding the lysosomal enzymes, such as cathepsins and their regulators, cystatins, were selectively down-regulated. Almost all were confirmed by downstream QPCR, Western blot, and functional assays. Thus, live but not inactivated HIV-1 significantly decreased expression of cathepsins B, C, H, and Z RNA and protein and of S RNA (but not L) at 48 to 96 hours after HIV-1 treatment, and only at MOIs of 2.5 or higher, sufficient for de novo infection of a significant proportion of the DC culture. Increased expression of cathepsin L corresponded with decreased expression of its inhibitor cystatin C. There was a corresponding decrease in substrate binding to the active sites of cathepsins B and Z. The net functional effect was markedly diminished functional activity of all cathepsins tested (Z, B, L, and S together and C and S separately).
This reduced cathepsin activity should markedly diminish late endosomal degradation of HIV-1 and therefore increase survival of newly replicated virus that is released by DCs but re-endocytosed,16,56 and thus would enhance transfer to contacting CD4 lymphocytes across viral synapses. Furthermore, these cathepsins play a key role in a late endosomal processing of viral antigens, and their expression on MHCII. Thus, reduced function would be expected to reduce HIV-1–specific activation of the same contacting CD4 lymphocytes.
Although many of these changes in expression of functional gene clusters clearly favor viral replication and others are intrinsic host cell defensive responses, the role of many others requires further dissection. It is clear that HIV-1 can alter the sequence and nature of transcription of host defensive genes to assist its replication and persistence. This is observed especially in its effects on the expression and function of cathepsins and also on a subset of interferon-stimulated genes and is now being studied intensively. Studies of this nature will allow a better understanding of the pathogenesis of HIV-1 infection and allow comparisons with other cell types to determine which changes in cellular gene expression facilitate HIV-1 infection in most cell types or are specific for infection of DCs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jim Arthos and Rob Center for providing the UG037 and AD8 strains of trimeric gp140, respectively. Many thanks to Najla Nasr, Joey Lai, and Valerie Marsden for help in preparation of virus stocks and Judith Edmonds for assistance with virus stock quantification. Thanks also to Ekkehard Weber who provided antisera against cathepsins S, B, and H, and cystatin A and to Nicolas Lützner who provided the cathepsin S–specific substrate.
This work was supported by a National Health and Medical Research Council (NHMRC) Program Grant ID number 358399 and National Institutes of Health (NIH) RO1 DE015512-01. S.G.T was supported by an NHMRC C. J. Martin Fellowship.
National Institutes of Health
Authorship
Contribution: A.N.H. was chiefly involved in carrying out this work, jointly prepared the paper, infected all MDDCs with virus stocks or gp120, and conducted all cDNA microarray and QPCR experiments with technical assistance from S.K.M.; M.K. conducted all cathepsin and cystatin Western blots as well as affinity labeling and fluorogenic substrate assays; C.R.B. conducted all microarray analysis; K.B. conducted all statistics involved in the establishment of criteria for analysis of gene expression changes; S.G.T. prepared viral stocks and edited the paper; N.N. prepared all the virus stocks; O.T. conducted the Illumina microarray studies using trimeric gp140; S.K.M. provided technical assistance in running QPCR assays; J.L.S. provided academic input into the initial design of the study; B.S. provided academic input into the initial design of the study and assisted with paper preparation; C.D. provided academic input into the design of all cathepsin and cystatin Western blots as well as affinity labeling and fluorogenic substrate assays; and A.L.C. conceived and supervised the study and jointly prepared the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anthony L. Cunningham, Centre for Virus Research, Westmead Millennium Institute, PO Box 412, Darcy Rd, Westmead, NSW 2145, Australia; e-mail: tony_cunningham@wmi.usyd.edu.au.