Abstract
The role of miRNAs in regulating megakaryocyte differentiation was examined using bipotent K562 human leukemia cells. miR-34a is strongly up-regulated during phorbol ester–induced megakaryocyte differentiation, but not during hemin-induced erythrocyte differentiation. Enforced expression of miR-34a in K562 cells inhibits cell proliferation, induces cell-cycle arrest in G1 phase, and promotes megakaryocyte differentiation as measured by CD41 induction. miR-34a expression is also up-regulated during thrombopoietin-induced differentiation of CD34+ hematopoietic precursors, and its enforced expression in these cells significantly increases the number of megakaryocyte colonies. miR-34a directly regulates expression of MYB, facilitating megakaryocyte differentiation, and of CDK4 and CDK6, to inhibit the G1/S transition. However, these miR-34a target genes are down-regulated rapidly after inducing megakaryocyte differentiation before miR-34a is induced. This suggests that miR-34a is not responsible for the initial down-regulation but may contribute to maintaining their suppression later on. Previous studies have implicated miR-34a as a tumor suppressor gene whose transcription is activated by p53. However, in p53-null K562 cells, phorbol esters induce miR-34a expression independently of p53 by activating an alternative phorbol ester-responsive promoter to produce a longer pri-miR-34a transcript.
Introduction
microRNAs (miRNA), small noncoding RNAs that suppress expression of genes bearing partially complementary sequences, participate in regulating hematopoietic cell differentiation and leukemogenesis.1 miRNA expression changes dramatically during cell differentiation. In some cases, alterations in a single miRNA can have a significant impact on cell-lineage commitment and survival. A general requirement for miRNAs during T-cell development was demonstrated by studies in mice selectively deleted of Dicer-1 in the thymus, which show impaired CD8 T-cell development and defects in T-helper cell differentiation and function.2,3 miR-181a regulates T-cell differentiation to effector cells by inhibiting the expression of multiple phosphatases, and miR-155 regulates regulatory T cell–lineage commitment in part by inhibiting SOCS1 expression.4,5 miR-223 regulates granulocytic differentiation through translational inhibition of the transcription factor NFI-A.6 miR-150, whose expression is largely restricted to mature lymphocytes, facilitates B-cell development and megakaryocyte differentiation of megakaryocyte-erythrocyte progenitors (MEPs) by regulating MYB expression.7,8
miRNAs have also been implicated in leukemogenesis and cancer more generally. A high proportion of miRNA genes are encoded in cancer-associated regions associated with loss of heterozygosity, gene amplification, common breakpoint regions, and fragile sites.9 miRNAs can function as either tumor suppressors or oncogenes. Oncogenic miRNAs in hematologic malignancies include miR-155 and the miRNA cluster miR-17-92, which are both amplified in different types of B-cell lymphoma.1 The miR-15a/16-1 cluster, encoded in a region of chromosome 13 that is frequently deleted in B-cell chronic lymphocytic leukemia, is a tumor suppressor miRNA.10,11
miR-34a is another example of a tumor suppressor. miR-34a is encoded within the chromosome region 1p36 whose loss has been associated with glioma, neuroblastoma, pancreatic cancer, and chronic myelogenous leukemia.12-15 miR-34a is part of the p53 regulatory network.16 miR-34a transcription is directly activated by p53, and in turn miR-34a regulates the expression of some p53 target genes.15,17,18 In support of its role as a tumor suppressor, miR-34a has been shown to regulate genes involved in cell-cycle regulation and apoptosis, including CDK4, CDK6, CCND1, E2F3, and SIRT1.14,17,19,20
The human erythroleukemia cell line K562, derived from a chronic myelogenous leukemia patient, resembles a bipotent MEP.21 Phorbol esters, such as 12-O-tetradecanoyl-phorbol-13-acetate (TPA), induce megakaryocyte (MK) differentiation, whereas hemin, sodium butyrate, or Ara-C induces differentiation to erythrocytes.22 TPA-induced MK differentiation of leukemic K562 cells is accompanied by characteristic changes in cell morphology, cell adhesion, cell-cycle arrest, endomitosis, and expression of MK lineage–specific markers, such as platelet-derived growth factor and the αIIbβ3 (CD41/CD61) and α2β1 (CD49b) integrins.22 We used this cellular model to investigate the role of miRNAs in MK differentiation. We found that miR-34a is the most highly up-regulated miRNA during TPA-induced MK differentiation of leukemic K562 cells and is part of a miRNA signature of up-regulated miRNAs specific to the MK lineage. miR-34a contributes to MK differentiation by inhibiting cell proliferation and inducing expression of MK-specific markers, at least in part, through direct regulation of its known targets CDK4 and CDK6 and the newly identified target MYB. miR-34a also enhances MK differentiation of CD34+ hematopoietic progenitor cells. We also show that miR-34a expression in K562 cells is p53 independent. An alternative promoter drives expression of a longer pri-miR-34a transcript in response to phorbol esters.
Methods
Cell culture and reagents
Human erythroleukemia K562 cells (ATCC) were grown in RPMI-1640 medium supplemented with 10% fetal calf serum and penicillin/streptomycin (Invitrogen). For MK differentiation, cells were treated with 10 nM TPA (Sigma-Aldrich). For erythrocyte differentiation, 100 μg/mL hemin (Sigma-Aldrich) was added to the medium for the duration of the experiment. Benzidine staining detected hemoglobin-positive cells. Frozen human umbilical cord blood CD34+ cells were grown in StemSpan H3000 Defined Medium in the presence of the cytokine cocktail CC200 (containing 50 ng/mL recombinant human [rh] thrombopoietin [TPO], 50 ng/mL rh stem cell factor [SCF], and 10 ng/mL rh interleukin-3 [IL-3]; StemCell Technologies).
miRNA and gene microarrays
For miRNA analysis, small RNA libraries were prepared as described previously.23 cRNA was generated and labeled with Cy3-CTP or Cy5-CTP using the Low-Input Linear Amplification Kit (Agilent Technologies). Samples were hybridized on custom oligonucleotide microarrays (Agilent Technologies) containing probes for 286 annotated miRNAs.24 Hybridized microarrays were scanned using the Agilent LP2 DNA Microarray Scanner, and microarray images were analyzed using Feature Extraction Software (Version 7.1.1; Agilent). mRNA microarray analysis was performed using 15 μg total RNA applied to Human Genome U133 Plus 2.0 Arrays (Affymetrix) at the Microarray Core Facility at the Dana-Farber/Harvard Cancer Center core facility. Data were analyzed using Microarray Suite 5.0 software (Affymetrix). Microarray data are available at the public databases ArrayExpress (TPA-induced MK differentiation experiment; accession no. E-MEXP-2213) and Gene Expression Omnibus (miR-34a overexpression experiment; accession no. GSE16674).
miRNA mimics and transfections
K562 cells were transfected by nucleofection (Amaxa) with 5 μg plasmid DNA or 3 μg miRNA mimic or control mimic (Dharmacon) as per the manufacturer's protocol.
Colony assays and lentivirus infections
A total of 5 × 104 human cord blood CD34+ cells (StemCell Technologies) were infected overnight with high-titer miRNA-expressing lentiviruses. Cells were washed the next day with phosphate-buffered saline and plated for colony-forming units–megakaryocyte (CFU-MK) assays using the MegaCult-C kit (StemCell Technologies) following the manufacturer's instructions. After incubation at 37°C for 12 days, MK colonies were scored after CD41 staining.
Quantitative RT-PCR
Analysis of mature miRNA expression relative to U6 snRNA was performed using miRNA-specific quantitative reverse-transcribed polymerase chain reaction (RT-PCR; TaqMan MicroRNA Assays; Applied Biosystems). Quantitative RT-PCR mRNA analysis was performed in triplicate using the SYBR Green master mix (Applied Biosystems) and the Bio-Rad iCycler and normalized to GAPDH. Primers were from PrimerBank25 (primer identification nos. 4885497a2, MYB; 4502735a3, CDK4; 4502741a2, CDK6; and 7669492a3, GAPDH).
RACE PCR analysis
RACE PCR was performed using total RNA extracted from TPA-treated K562 cells and the FirstChoice RLM-RACE Kit (Ambion) according to the manufacturer's instructions using primers: P1, 5′-AGAGCTTCCGAAGTCCTGG-3′ and P2, 5′-TTGCTCACAACAACCAGCTAAGA-3′ for the 5′-RACE PCR; and P3, 5′-ACCGGCCAGCTGTGAGTGTTTCTTT-3′ and P4, 5′-TGGCAGTGTCTTAGCTGGTTGTT-3′ for the 3′-RACE PCR. Reaction products were analyzed by agarose gel electrophoresis, purified using the High Pure PCR Cleanup Micro Kit (Roche Applied Science), cloned (CloneJET PCR Cloning Kit), and sequenced.
Additional methods are provided in the supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Results
miRNA signature of TPA-induced MK differentiated K562 cells
To explore the role of miRNAs during K562 cell MK differentiation, we compared miRNA expression in undifferentiated versus TPA-treated cells by miRNA microarray. Within 2 days of TPA treatment, K562 cells stopped proliferating, became adherent, and expressed the MK marker CD41 (supplemental Figure 1A, data not shown). Total RNA was extracted from untreated K562 cells or from cells cultured for 2 or 4 days after TPA treatment. Small RNA libraries were prepared from each sample by size fractionation, adapter ligation, and PCR amplification.23 cRNA was hybridized to microarrays containing probes for 286 annotated miRNAs. miRNA expression of samples harvested 2 and 4 days after TPA treatment was similar (supplemental Figure 1B). Total miRNA expression increased with differentiation (supplemental Table 1). We focused on a subset of 12 highly up-regulated miRNAs, which included miRNAs that increased by 6-fold or more and whose signal saturated at day 4 (miR-34a, miR-375, miR-139, miR-409-3p, and the miRNA clusters miR-132/212 and miR-221/222) and miRNAs that increased by 50-fold or more (miR-181b, miR-299-5p, and miR-134). We also selected miR-181a, which increased 19-fold, because it is part of the miR-181 cluster (Figure 1A). The up-regulation of each of these miRNAs was validated by Northern blot (Figure 1B). One miRNA that was unchanged (miR-25) and one that was down-regulated (miR-218) after TPA by microarray analysis were probed as controls.
miRNA signature of TPA-treated K562 cells differentiated into megakaryocytes. (A) miRNAs up-regulated at least 6-fold 4 days after TPA treatment by microarray. Background counts were ≤ 500 and the assay saturated at 65 232 counts. Reproducibility of the miRNA microarray in 2 TPA-treated K562 samples is shown in supplemental Figure 1B. (Supplemental Table 1 shows data for all miRNAs analyzed.) (B-C) Change in miRNA expression after TPA or hemin treatment analyzed by Northern blot. U6 snRNA is a loading control. The miRNAs most induced by TPA were not induced by hemin (supplemental Figure 1C). MK differentiation by TPA and erythroid differentiation by hemin were verified (supplemental Figure 1A). miR-24 expression increased after either TPA or hemin treatment.
miRNA signature of TPA-treated K562 cells differentiated into megakaryocytes. (A) miRNAs up-regulated at least 6-fold 4 days after TPA treatment by microarray. Background counts were ≤ 500 and the assay saturated at 65 232 counts. Reproducibility of the miRNA microarray in 2 TPA-treated K562 samples is shown in supplemental Figure 1B. (Supplemental Table 1 shows data for all miRNAs analyzed.) (B-C) Change in miRNA expression after TPA or hemin treatment analyzed by Northern blot. U6 snRNA is a loading control. The miRNAs most induced by TPA were not induced by hemin (supplemental Figure 1C). MK differentiation by TPA and erythroid differentiation by hemin were verified (supplemental Figure 1A). miR-24 expression increased after either TPA or hemin treatment.
K562 cells can be differentiated into the erythroid lineage by hemin.26 To determine whether the subset of highly up-regulated miRNAs in TPA-treated K562 cells was specific to MK differentiation, we analyzed their expression in hemin-differentiated K562 cells by Northern blot. The 12 highly up-regulated miRNAs in TPA-treated K562 cells were specific to MK because none was up-regulated during hemin-induced erythroid differentiation (Figure 1C, supplemental Figure 1C). Furthermore, analysis of global miRNA expression in TPA versus hemin-differentiated K562 cells by miRNA microarray revealed very limited shared changes in miRNA expression between both lineages.27 Of note, up-regulation of the miR-24 clusters (containing miR-24, miR-23, and miR-27, although up-regulation was < 6-fold) and down-regulation of miR-17-5p/18a/20a and miR-106a/19b/92 clusters were common to both differentiation pathways, suggesting that these miRNAs might regulate common processes during terminal differentiation of hematopoietic cells. miR-24 up-regulation during MK and erythroid differentiation of K562 cells was also validated by Northern blot (Figure 1C).
miR-34a inhibits proliferation, induces G1 arrest, and promotes MK differentiation of K562 cells
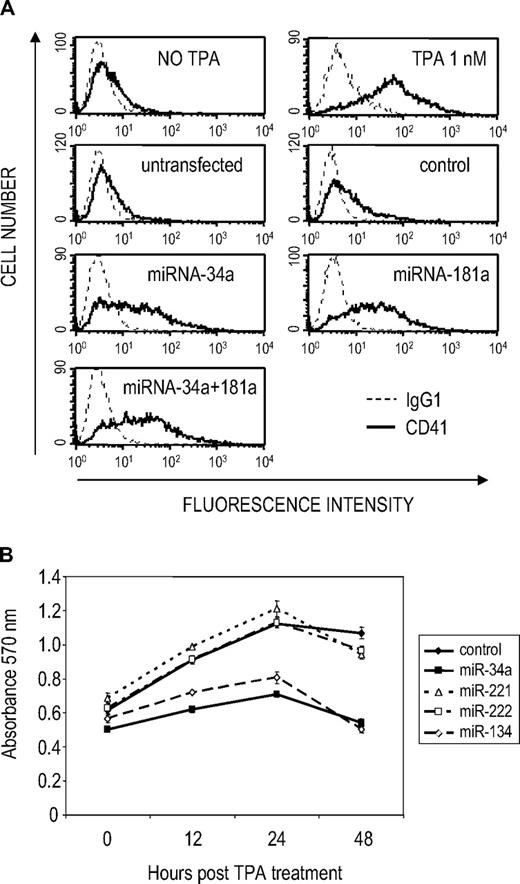
Our next goal was to determine whether any of the miRNAs up-regulated by TPA might play a role in regulating MK differentiation. We first evaluated the effect of miRNA overexpression on MK markers. K562 cells were transfected with miRNA mimics corresponding to each of the up-regulated miRNAs or a control miRNA mimic and treated 48 hours later with a suboptimal amount of TPA (0.1 nM), which does not induce differentiation of untransfected or mimic control-transfected K562 cells (Figure 2A). MK differentiation was assessed by flow cytometry for CD41 and CD61 (Figure 2A, supplemental Figure 2A; data not shown). Overexpression of miR-34a and miR-181a, but not the other up-regulated miRNAs, induced expression of the MK integrins. Of note, CD41 was not up-regulated without suboptimal TPA, even if cells were transfected with miRNA combinations (data not shown).
miR-34a enhances megakaryocytic differentiation and inhibits proliferation in K562 cells. (A) Overexpression of miR-34a or miR-181a in K562 cells induces the MK integrin CD41. K562 cells were transfected with the indicated miRNA mimic and 24 hours later stimulated with a suboptimal amount of TPA (0.1 nM). Efficient uptake of dsRNA oligonucleotides into transfected K562 cells is shown in supplemental Figure 1D. CD41 expression was analyzed by flow cytometry 72 hours after TPA stimulation. As positive control, K562 cells were treated with 1 nM TPA. Levels of expression of the transfected miRNAs at the time of analysis are shown in supplemental Figure 2B. (B) miR-34a and miR-134 inhibit K562 cell proliferation, assessed by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) cell proliferation assay. Cells were transfected with the indicated miRNA and then treated with a suboptimal amount of TPA as indicated. Values are mean ± SD.
miR-34a enhances megakaryocytic differentiation and inhibits proliferation in K562 cells. (A) Overexpression of miR-34a or miR-181a in K562 cells induces the MK integrin CD41. K562 cells were transfected with the indicated miRNA mimic and 24 hours later stimulated with a suboptimal amount of TPA (0.1 nM). Efficient uptake of dsRNA oligonucleotides into transfected K562 cells is shown in supplemental Figure 1D. CD41 expression was analyzed by flow cytometry 72 hours after TPA stimulation. As positive control, K562 cells were treated with 1 nM TPA. Levels of expression of the transfected miRNAs at the time of analysis are shown in supplemental Figure 2B. (B) miR-34a and miR-134 inhibit K562 cell proliferation, assessed by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) cell proliferation assay. Cells were transfected with the indicated miRNA and then treated with a suboptimal amount of TPA as indicated. Values are mean ± SD.
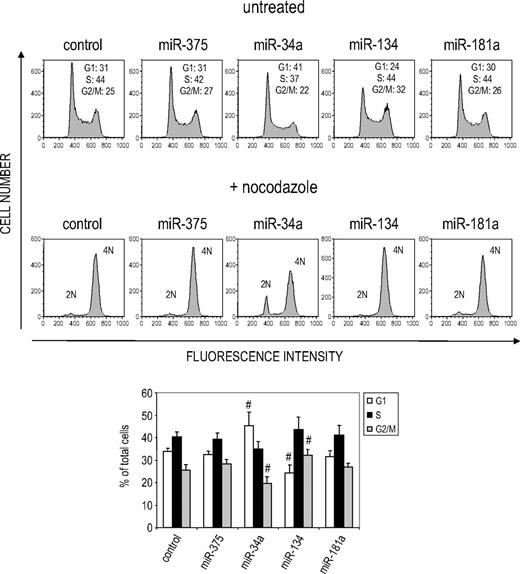
We also evaluated the effect of miRNA overexpression on cell proliferation. Overexpression of miR-34a and miR-134, but not other miRNAs, inhibited cell proliferation in transfected K562 cells treated with suboptimal TPA (0.1 nM, Figure 2B; data not shown). To understand how these miRNAs might regulate cell proliferation, we performed cell-cycle analysis (Figure 3). Both miRNAs perturbed the cell cycle. miR-34a overexpression significantly increased the proportion of G1 phase cells and significantly reduced the G2/M compartment, whereas miR-134 had the opposite effect (reduced G1, increased G2/M). As expected, when miRNA-transfected cells were treated with nocodazole for 16 hours, miR-34a–overexpressing samples showed increased diploid cells (2N). However, no significant alteration of cell- cycle profile was observed in cells transfected with 2 other up-regulated miRNAs, miR-181a or miR-375. Taken together, these results suggest that multiple miRNAs contribute to MK differentiation of K562 cells and identify miR-34a as an important contributor that regulates both cell-cycle progression and MK marker expression.
miR-34a and miR-134 inhibit cell-cycle progression at different phases of the cell cycle. K562 cells, transfected with the indicated miRNA mimic, were analyzed by propidium iodide staining 48 hours later. Replicate samples were either left untreated or treated with nocodazole (100 ng/mL) for 16 hours before propidium iodide staining. The percentage of cells in the G1, S, or G2 phase of the cell cycle is indicated. The bar graph at the bottom represents the mean ± SD of 4 independent experiments. #Significant differences (P < .05) relative to the control sample by Student 2-sided t test.
miR-34a and miR-134 inhibit cell-cycle progression at different phases of the cell cycle. K562 cells, transfected with the indicated miRNA mimic, were analyzed by propidium iodide staining 48 hours later. Replicate samples were either left untreated or treated with nocodazole (100 ng/mL) for 16 hours before propidium iodide staining. The percentage of cells in the G1, S, or G2 phase of the cell cycle is indicated. The bar graph at the bottom represents the mean ± SD of 4 independent experiments. #Significant differences (P < .05) relative to the control sample by Student 2-sided t test.
miR-34a regulates MYB
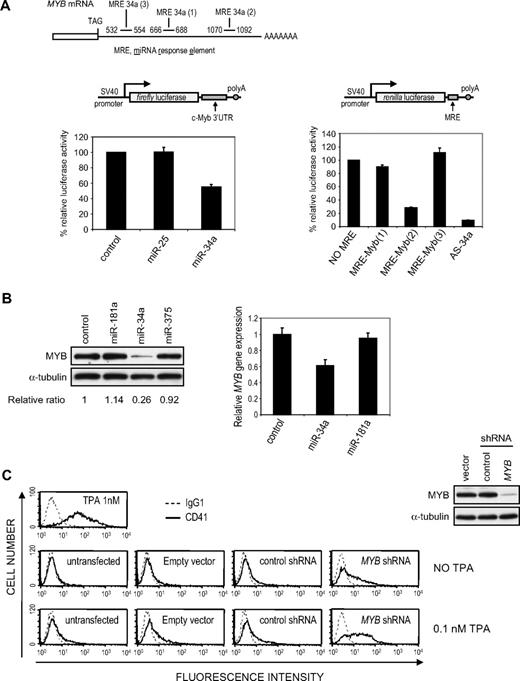
Because miR-34a was the most up-regulated miRNA and exogenous miR-34a induced both MK differentiation markers and cell- cycle arrest, we sought to identify miR-34a target genes in K562 cells. Candidate miR-34a target genes were chosen for experimental validation by combining in silico predictions28,29 and changes in microarray mRNA expression after TPA treatment or enforced expression of miR-34a (supplemental Tables 2-3). MYB, an important transcriptional regulator of hematopoietic cell differentiation, was an attractive candidate target gene. MYB mRNA and protein were significantly down-regulated after TPA treatment of K562 cells by mRNA microarray (4.6-fold decrease; supplemental Table 2), Northern blot, and immunoblot (supplemental Figure 6). After miR-34a overexpression in K562 cells, MYB mRNA also decreased 1.9-fold by microarray analysis (supplemental Table 3). To determine whether MYB is directly regulated by miR-34a, the full-length MYB 3′-untranslated region (3′-UTR) was cloned into a luciferase reporter plasmid, and the effect of miR-34a overexpression on luciferase activity was assessed (Figure 4A left). miR-34a overexpression reduced luciferase activity by 45%. Overexpression of miR-25, which is not predicted to regulate MYB and whose expression does not change during TPA-induced differentiation, had no effect. The 3′-UTR of MYB has 3 rna22-predicted miRNA response element (MREs; Figure 4A, supplemental Figure 3A). MRE2 is also predicted by TargetScan as a high probability, although poorly conserved, MRE with a 7-mer seed match. When each of the predicted MREs was tested in luciferase assays, miR-34a only regulated the reporter encoding MRE2 (Figure 4A right). The 70% reduction in luciferase expression from the MRE2-containing reporter was almost as great as that obtained with a perfectly complementary miR-34a sequence (90% reduction). Deletion of MRE2 or mutation of its seed in the context of the full-length 3′-UTR significantly increased reporter luciferase activity (supplemental Figure 3B) but did not restore it to control levels. This suggests that, although MRE2 is a bona fide binding site for miR-34a, other regions might contribute to miR-34a regulation of MYB.
miR-34a directly regulates MYB expression. (A) miR-34a regulates luciferase activity of a reporter vector containing either the full-length 3′-UTR of MYB (left) or individual predicted MRE (right). Relative luciferase activity was assayed in 293T cells 48 hours after cotransfection with a luciferase reporter plasmid and the indicated miRNA mimic. AS-34a designates a reverse complementary sequence of miR-34a. (B) Overexpression of miR-34a in K562 cells down-regulates MYB protein and mRNA. K562 cells, transfected with the indicated miRNA mimic, were analyzed for MYB protein (left) and mRNA (right) by immunoblot or quantitative PCR, respectively, 48 hours later. Protein expression relative to α-tubulin was quantified by densitometry. mRNA normalized to GAPDH mRNA is shown relative to the ratio in K562 cells transfected with a miRNA mimic control. (C) MYB knockdown (right) up-regulates the MK-specific marker CD41 in cells treated with a suboptimal amount of TPA (0.1 nM). As positive control for CD41 induction, K562 cells were treated with 1 nM TPA.
miR-34a directly regulates MYB expression. (A) miR-34a regulates luciferase activity of a reporter vector containing either the full-length 3′-UTR of MYB (left) or individual predicted MRE (right). Relative luciferase activity was assayed in 293T cells 48 hours after cotransfection with a luciferase reporter plasmid and the indicated miRNA mimic. AS-34a designates a reverse complementary sequence of miR-34a. (B) Overexpression of miR-34a in K562 cells down-regulates MYB protein and mRNA. K562 cells, transfected with the indicated miRNA mimic, were analyzed for MYB protein (left) and mRNA (right) by immunoblot or quantitative PCR, respectively, 48 hours later. Protein expression relative to α-tubulin was quantified by densitometry. mRNA normalized to GAPDH mRNA is shown relative to the ratio in K562 cells transfected with a miRNA mimic control. (C) MYB knockdown (right) up-regulates the MK-specific marker CD41 in cells treated with a suboptimal amount of TPA (0.1 nM). As positive control for CD41 induction, K562 cells were treated with 1 nM TPA.
miR-34a overexpression in undifferentiated K562 cells reduced MYB protein by 74% and mRNA by 40% (Figure 4B). Two other miRNAs up-regulated after TPA treatment, miR-375 and miR-181a, not predicted to target MYB, had no effect. MYB knockdown using a retroviral vector encoding a MYB shRNA also significantly up-regulated CD41 expression in untreated K562 cells, which was further increased by adding a suboptimal amount of TPA (0.1 nM; Figure 4C). The increased CD41 expression after MYB knockdown was comparable with that obtained when miR-34a was overexpressed (Figure 2A). Analogous results were obtained after transfection with 3 different MYB siRNAs (not shown). Cell proliferation also decreased in MYB knockdown samples (not shown), consistent with previous observations.30
The TargetScan algorithm predicts 5 of the other highly up-regulated miRNAs (miR-132/212, miR-221/222, and miR-134) as potential MYB regulators. We therefore analyzed MYB protein in K562 cells overexpressing these miRNAs and tested by luciferase reporter assay their regulation of the full-length MYB 3′-UTR (supplemental Figure 4). miR-221 and miR-134 also directly regulated MYB, although not as strongly as miR-34a. Therefore, miR-34a and other up-regulated miRNAs suppress MYB expression.
miR-34a down-regulates CDK4 and CDK6 in K562 cells
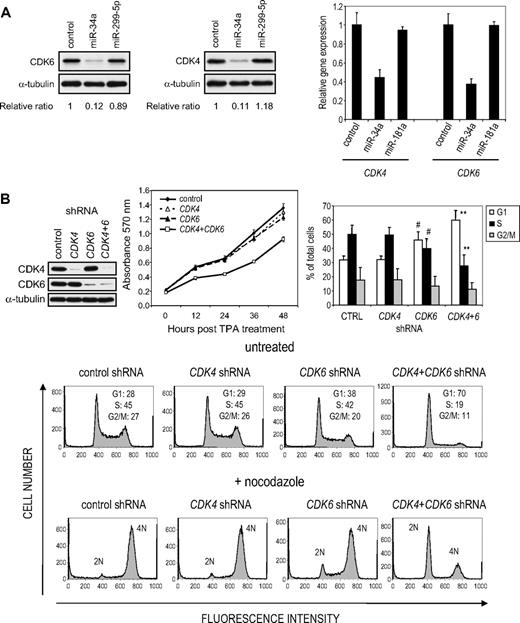
CDK4 and CDK6 have previously been shown to be regulated by miR-34a in colon, lung, and prostate tumor cell lines.17,19,31,32 Because these kinases drive progression from G1 to S phase and overexpressing miR-34a increases the G1 compartment (Figure 3), it is probable that these kinases are also regulated by miR-34a in K562 cells. Both CDK4 and CDK6 mRNAs are also down-regulated in K562 cells after TPA treatment (3.7- and 2.6-fold, respectively; supplemental Table 2) or enforced miR-34a expression (3.2- and 3.0-fold, respectively; supplemental Table 3). Furthermore, CDK4 and CDK6 proteins decreased in TPA-treated cells by immunoblot (supplemental Figure 6). Previous reports identified one CDK4 and 2 CDK6 miR-34a MREs.17,19,32 To verify that miR-34a regulates CDK4 and CDK6 in K562 cells, we analyzed their expression by immunoblot. Overexpressing miR-34a significantly reduced CDK4 and CDK6 protein (89% and 87%, respectively; Figure 5A). Overexpression of miR-299-5p, which is not predicted to target either gene, did not alter either. In addition, miR-34a overexpression in K562 cells also significantly reduced CDK4 and CDK6 mRNAs analyzed by quantitative PCR (Figure 5A). rna22 predicts 8 CDK6 miR-34a MREs, of which MRE3 is also predicted by TargetScan (supplemental Figure 5A). Because these MREs have not been previously tested, we analyzed their activity in luciferase reporter assays (supplemental Figure 5B). Insertion of CDK6 MRE3 and MRE5 strongly reduced luciferase activity (80% and 65%, respectively), whereas MRE2 and MRE8 each had a moderate effect (25% and 40%, respectively).
miR-34a regulates CDK4 and CDK6 in K562 cells. (A) K562 cells, transfected with the indicated miRNA mimic, were analyzed for CDK4 and CDK6 protein and mRNA by immunoblot (left) or quantitative PCR (right), respectively, 48 hours later. Protein expression relative to α-tubulin was quantified by densitometry. mRNA normalized to GAPDH mRNA is shown relative to the ratio in K562 cells transfected with a miRNA mimic control. (B) Simultaneous knockdown (top left) of CDK4 and CDK6, but not either alone, inhibits proliferation and increases the G1 compartment in K562 cells. Cell proliferation was evaluated using the MTT cell proliferation assay (top middle). Cell-cycle analysis (representative flow cytometry histograms, bottom) was performed 72 hours after transfection in nocodazole-treated and untreated cells. The bar graph (top right) represents the mean ± SD of 4 independent experiments performed without nocodazole synchronization. Statistical analysis was performed using the Student 2-sided t test: #P < .05; **P < .01.
miR-34a regulates CDK4 and CDK6 in K562 cells. (A) K562 cells, transfected with the indicated miRNA mimic, were analyzed for CDK4 and CDK6 protein and mRNA by immunoblot (left) or quantitative PCR (right), respectively, 48 hours later. Protein expression relative to α-tubulin was quantified by densitometry. mRNA normalized to GAPDH mRNA is shown relative to the ratio in K562 cells transfected with a miRNA mimic control. (B) Simultaneous knockdown (top left) of CDK4 and CDK6, but not either alone, inhibits proliferation and increases the G1 compartment in K562 cells. Cell proliferation was evaluated using the MTT cell proliferation assay (top middle). Cell-cycle analysis (representative flow cytometry histograms, bottom) was performed 72 hours after transfection in nocodazole-treated and untreated cells. The bar graph (top right) represents the mean ± SD of 4 independent experiments performed without nocodazole synchronization. Statistical analysis was performed using the Student 2-sided t test: #P < .05; **P < .01.
Knockdown of both CDK4 and CDK6 significantly reduced cell proliferation by causing cells to accumulate in G1, but knocking down either by itself had no significant effect on proliferation (Figure 5B). CDK6 knockdown on its own slightly, but significantly, increased the proportion of G1 phase cells (38% vs 28% in control cells). These experiments are consistent with previous work showing that CDK4 and CDK6 have redundant roles in controlling the G1 to S transition33,34 and highlight the potential relevance of miR-34a as a regulator of both kinases.
To address whether miR-34a down-regulates MYB, CDK4, and CDK6 during MK differentiation, we analyzed the change of expression of miR-34a and its targets over time in TPA-differentiated K562 cells (supplemental Figure 6). miR-34 became faintly detectable 12 hours after adding TPA and increased over the next 4 days. However, MYB and CDK4 were down-regulated before miR-34a was up-regulated. CDK6 expression correlated inversely with the induction of miR-34a. Of note, MYB was transiently reexpressed 24 to 48 hours after TPA treatment and became undetectable again only when miR-34a was highly expressed. Similarly, CDK4 protein levels increased at later time points. These data suggest that, although miR-34a might not be responsible for the initial down-regulation of CDK4 or MYB, miR-34a might act to suppress their reexpression to maintain the postmitotic MK state.
Enforced miR-34a expression enhances thrombopoietin-induced MK differentiation of primary human CD34+ cells
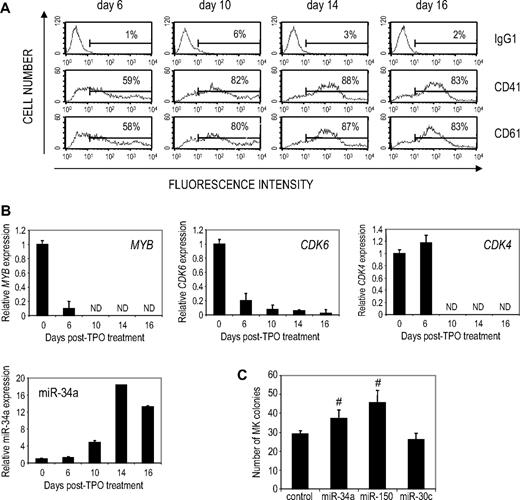
Because results in hematopoietic cell lines may not reflect normal hematopoiesis, our next goal was to determine whether miR-34a also contributes to MK differentiation of primary human hematopoietic stem cells (HSCs) treated with TPO, IL-3, and SCF.35 MK differentiation was assessed by flow cytometry for CD41 and CD61 staining (Figure 6A). CD41 and CD61 were detected by day 6 and increased over 2 weeks, when most cells expressed these markers. miR-34a expression analyzed by quantitative PCR increased with MK differentiation, first becoming significantly increased at day 10 (Figure 6B). MYB, CDK4, and CDK6 mRNA also decreased with MK differentiation. As with K562 cells, the kinetics of miR-34a induction and down-regulation of its targets did not correlate. In particular, MYB and CDK6 were down-regulated before miR-34 expression became appreciable. These findings suggest that miR-34-independent mechanisms control the initial down-regulation of these genes.
Enforced miR-34a expression enhances MK differentiation of human CD34+hematopoietic precursors. (A) CD34+ hematopoietic cells, differentiated to MK by culture in medium containing TPO, SCF, and IL-3, express MK integrins. The proportion of CD41/CD61-positive cells is indicated. (B) Changes in miR-34a and its MYB, CDK4, and CDK6 target mRNAs were evaluated by quantitative PCR. miR-34a expression is normalized to U6 and mRNA is normalized to GAPDH. Gene expression is plotted as expression relative to day 0. ND indicates not detected. (C) CFU-MK assay of miRNA-overexpressing CD34+ HSCs. Primary human CD34+ cord blood cells were infected with lentiviruses encoding the indicated miRNAs, and the effect of miRNA overexpression on CFU-MK numbers was evaluated 12 days later. The bar graph represents the mean ± SD of quadruplicate samples. A representative experiment of 3 independent experiments is shown. #P < .05 relative to control lentivirus-infected cells. Exogenous miR-34a and miR-150 both enhance MK colony numbers.
Enforced miR-34a expression enhances MK differentiation of human CD34+hematopoietic precursors. (A) CD34+ hematopoietic cells, differentiated to MK by culture in medium containing TPO, SCF, and IL-3, express MK integrins. The proportion of CD41/CD61-positive cells is indicated. (B) Changes in miR-34a and its MYB, CDK4, and CDK6 target mRNAs were evaluated by quantitative PCR. miR-34a expression is normalized to U6 and mRNA is normalized to GAPDH. Gene expression is plotted as expression relative to day 0. ND indicates not detected. (C) CFU-MK assay of miRNA-overexpressing CD34+ HSCs. Primary human CD34+ cord blood cells were infected with lentiviruses encoding the indicated miRNAs, and the effect of miRNA overexpression on CFU-MK numbers was evaluated 12 days later. The bar graph represents the mean ± SD of quadruplicate samples. A representative experiment of 3 independent experiments is shown. #P < .05 relative to control lentivirus-infected cells. Exogenous miR-34a and miR-150 both enhance MK colony numbers.
To determine whether miR-34a contributes to MK differentiation of HSCs, CD34+ cells infected with lentiviruses encoding miR-34a or a control sequence were evaluated in CFU-MK assays (Figure 6C). miR-150, a known regulator of the MK lineage,7 and miR-30c, whose expression did not change in TPA-induced megakaryopoiesis (supplemental Table 1), were used as additional controls. miR-34a significantly increased MK colony number by approximately 30%, whereas the miR-30c–expressing virus had no significant effect. As expected, miR-150 also increased MK colonies (∼ 60%). Therefore, miR-34a is able to enhance MK differentiation of primary HSCs, although to a lesser extent than miR-150.
miR-34a up-regulation in TPA-differentiated K562 cells is p53-independent
miR-34a participates in the p53 gene regulatory network.16 However, K562 cells are p53-null.36 K562 cells contain a single cytosine insertion in exon 5, between codons 135 and 136, of TP53, which generates a frameshift mutation leading to an N-terminal truncated protein of 147 amino acids. Although a shorter transcript can be detected by RT-PCR, neither the truncated nor full-length protein is detected by immunoblot (Figure 7B).36 miR-34a is contained within the second exon of an expressed sequence tag (accession no. DB286351) encoded by the negative strand of human chromosome 1.15,17,18 Exon 1 is separated from exon 2 by a 30-kb intron and contains a conserved p53 binding site responsible for miR-34a transactivation. The p53-regulated pri-miR-34a promoter region has been identified 1.5 kb upstream and 0.5 kb downstream of the pri-miR-34a transcription start site (TSS), which includes the conserved p53 binding site.15,17,18 To evaluate whether the identified promoter region drives expression of pri-miR-34a during TPA-induced differentiation of K562 cells, we cloned the putative promoter of expressed sequence tag DB286351 into the promoterless luciferase reporter vector pGL3-basic. Luciferase activity driven by the putative miR-34a promoter was not enhanced by TPA treatment in K562 cells (Figure 7A). TPA treatment increased luciferase activity approximately 400-fold from a control reporter gene in which the luciferase gene was driven by the TPA-responsive CD41 promoter. The putative miR-34a promoter reporter vector was strongly activated (100-fold relative to the promoterless reporter) when a p53 expression plasmid was cotransfected (Figure 7B). These results confirm that K562 cells are p53-null and demonstrate that miR-34a up-regulation during TPA-induced K562 cell differentiation is p53-independent. Therefore, DNA regulatory sequences other than those in the previously described miR-34a promoter drive miR-34a expression.
miR-34a up-regulation after TPA treatment of K562 cells is p53-independent. (A) The p53-dependent miR-34a promoter does not activate luciferase expression after TPA treatment of K562 cells. Cells were transfected with a firefly luciferase reporter vector in which luciferase expression was driven by the DB286351 miR-34a promoter (PM-34a) or the CD41 promoter (PM-CD41) or with the promoterless luciferase vector (pGL3-Basic). Samples were cotransfected with a Renilla luciferase reporter vector for normalization. Luciferase activity was measured 48 hours after TPA treatment. (B) Exogenous expression of p53 in K562 cells activates the DB286351 miR-34a promoter. K562 cells were transfected with promoterless firefly luciferase reporter vector (pGL3) or a reporter containing the miR-34a promoter (PM-34a), a Renilla luciferase reporter vector, and increasing amounts (2, 4, and 8 μg) of pCMV-p53 vector. The total amount of transfected DNA was kept constant using an empty pCMV plasmid. Luciferase activity was measured 48 hours after transfection and was normalized as described. The fold increase in luciferase activity relative to the control sample transfected with the promoterless vector is shown. The bottom panel represents the level of p53 protein in the samples analyzed herein by immunoblot. The membrane was stripped and reprobed for α-tubulin as a loading control. (C) 5′- and 3′-end RACE PCR analysis of pri-miR-34a transcripts in TPA-treated K562 cells. Agarose gel images show the 5′- and 3′-RACE PCR products obtained in K562 cells treated with TPA for 4 days. (D) Two pri-miR-34a transcripts identified by sequencing the 5′-end RACE PCR products from TPA-treated K562 cells and their location in the genome. E indicates exon; the numbers below the exons indicate their length in base pairs. Also indicated is the length of the intronic regions. For comparison, the previously described pri-miR-34a transcript, DB286351, is also shown (top). (E) The genomic region adjacent to the alternate TSS serves as the miR-34a promoter in TPA-treated K562 cells. A DNA genomic fragment comprising 1.5 kb upstream to 0.5 kb downstream of the identified alternative TSS (PM-34a-K1/2) was cloned into pGL3-basic, and the effect of TPA treatment on luciferase activity was evaluated as described.
miR-34a up-regulation after TPA treatment of K562 cells is p53-independent. (A) The p53-dependent miR-34a promoter does not activate luciferase expression after TPA treatment of K562 cells. Cells were transfected with a firefly luciferase reporter vector in which luciferase expression was driven by the DB286351 miR-34a promoter (PM-34a) or the CD41 promoter (PM-CD41) or with the promoterless luciferase vector (pGL3-Basic). Samples were cotransfected with a Renilla luciferase reporter vector for normalization. Luciferase activity was measured 48 hours after TPA treatment. (B) Exogenous expression of p53 in K562 cells activates the DB286351 miR-34a promoter. K562 cells were transfected with promoterless firefly luciferase reporter vector (pGL3) or a reporter containing the miR-34a promoter (PM-34a), a Renilla luciferase reporter vector, and increasing amounts (2, 4, and 8 μg) of pCMV-p53 vector. The total amount of transfected DNA was kept constant using an empty pCMV plasmid. Luciferase activity was measured 48 hours after transfection and was normalized as described. The fold increase in luciferase activity relative to the control sample transfected with the promoterless vector is shown. The bottom panel represents the level of p53 protein in the samples analyzed herein by immunoblot. The membrane was stripped and reprobed for α-tubulin as a loading control. (C) 5′- and 3′-end RACE PCR analysis of pri-miR-34a transcripts in TPA-treated K562 cells. Agarose gel images show the 5′- and 3′-RACE PCR products obtained in K562 cells treated with TPA for 4 days. (D) Two pri-miR-34a transcripts identified by sequencing the 5′-end RACE PCR products from TPA-treated K562 cells and their location in the genome. E indicates exon; the numbers below the exons indicate their length in base pairs. Also indicated is the length of the intronic regions. For comparison, the previously described pri-miR-34a transcript, DB286351, is also shown (top). (E) The genomic region adjacent to the alternate TSS serves as the miR-34a promoter in TPA-treated K562 cells. A DNA genomic fragment comprising 1.5 kb upstream to 0.5 kb downstream of the identified alternative TSS (PM-34a-K1/2) was cloned into pGL3-basic, and the effect of TPA treatment on luciferase activity was evaluated as described.
To identify the promoter of the pri-miR-34a transcript in K562 cells, we performed 5′-RACE PCR analysis using primers that annealed just downstream of the miR-34a sequence (Figure 7C, supplemental Figure 7A). The 5′-RACE PCR product revealed an alternate TSS approximately 20 kb upstream of the previously described TSS (Figure 7D, supplemental Figure 7B). Two different pri-miR-34a transcripts were sequenced that contained 2 exons in addition to exon 1 and exon 2 in pri-miR-34a DB286351 (Figure 7D, supplemental Figure 8). These pri-miR-34 transcripts shared the same TSS and exon E1′ but differed in exon E2′. Of 14 clones sequenced, 13 corresponded to pri-miR-34a-K1, whereas only 1 corresponded to pri-miR-34a-K2, suggesting that pri-miR-34a-K1 was the dominant transcript in TPA-treated K562 cells. To evaluate whether regions adjacent to the TSS could be responsible for TPA-induced up-regulation of miR-34a, a 2-kb genomic DNA fragment (1.5 kb upstream to 0.5 kb downstream of the alternate TSS) was cloned into pGL3-basic. TPA treatment increased luciferase activity approximately 40-fold, whereas no significant increase was observed for the construct driven by the previously described miR-34a promoter (Figure 7E). Therefore, a p53-independent TPA-inducible promoter regulates expression of distinct pri-miR-34a transcripts in p53-null K562 cells. Further work is required to define the transcription factors that regulate the p53-independent phorbol ester–responsive transcripts.
Discussion
Twelve miRNAs (miR-34a, miR-134, miR-139, miR-299-5p, miR-375, miR-409-3p and the miRNA clusters miR-132/212, miR-181a/181b, and miR-221/222) were strongly up-regulated (6- to 100-fold) in TPA-differentiated K562 cells. This miRNA signature was specific to MK differentiation because none of these miRNAs was up-regulated in hemin-differentiated cells. Our results are consistent with a report that down-regulation of miR-221/222, which derepresses c-kit expression, is required for erythroid differentiation of CD34+ hematopoietic precursors.37 Therefore, we can speculate that up-regulation of miR-221/222, and perhaps other miRNAs, might be required to suppress genetic programs that otherwise would favor erythroid differentiation.
To gain insight into the functional relevance of the miRNAs up-regulated during MK differentiation, we analyzed the effect of enforced expression of these miRNAs on MK differentiation of K562 cells. None of the miRNAs alone or in combination induced MK differentiation unless the cells were also exposed to suboptimal TPA. This suggests that either the up-regulated miRNAs on their own cannot regulate differentiation or that comanipulation of other miRNAs that were less profoundly modulated (in either direction) might be required. However, when cells were treated with suboptimal TPA concentrations that did not induce MK differentiation on their own, both miR-34a and miR-181a induced the MK marker CD41. In addition, miR-34a and miR-134 inhibited cell proliferation, altering the cell cycle at different points. miR-34a overexpression induced a G1 phase arrest, whereas miR-134 caused an arrest in G2/S. miR-34a was not only the most up-regulated miRNA during K562 cell MK differentiation, but enforced expression of miR-34a facilitated 2 important features of MK differentiation: cell-cycle arrest and induction of MK marker genes. Therefore, our results suggest that miR-34a might play an especially important role in MK differentiation.
miR-34a directly inhibits expression of the transcription factor MYB, an important regulator of cell differentiation at multiple decision forks in hematopoiesis.38 Previous studies in mice provided compelling evidence supporting a role for MYB as a potent negative regulator of megakaryopoiesis. Mice bearing mutations that compromised MYB function had elevated blood platelet and bone marrow MK counts.38 In addition, evidence supporting a crucial role for MYB as a regulator of the E-MK lineage bifurcation was provided by 2 studies showing that reduced MYB expression in MK/erythrocyte lineage–restricted progenitors increased MKs and reduced erythroid progenitor cell numbers.39,40
In addition to regulation by miR-34a as demonstrated here, MYB has also been shown to be regulated by miR-15a during erythroid differentiation of human CD34+ cells and by miR-150 both in B-cell development and to control MEP differentiation in mice.7,41 However, it is doubtful that these 2 miRNAs regulate MYB expression during MK differentiation of K562 cells because miR-15a is not significantly changed during TPA-induced MK differentiation of K562 cells by microarray (supplemental Table 1), and miR-150, whose signal increases in TPA-treated K562 cells by microarray, could not be detected by Northern blot in either untreated or TPA-treated K562 cells (supplemental Figure 1E). The MYB 3′-UTR encodes for evolutionarily conserved, high likelihood TargetScan-predicted MREs capable of being regulated by miR-150 (3 conserved, 1 nonconserved site) and miR-15/16 (2 conserved sites) as well as for the hematopoietic cell-expressed regulator miR-155 (2 conserved sites) and for miR-200 (1 conserved, 1 nonconserved site). Of note, the miR-34a MRE in the 3′-UTR of MYB is not conserved in mammals. Five other strongly up-regulated miRNAs in our system (miR-134, miR-221/222, miR-132/212) also have nonconserved MYB MREs.28 Of these, miR-221 and miR-134 also appear to regulate MYB in K562 cells. Because down-regulation of MYB is critical for hematopoietic cell differentiation, it is probable that MYB expression is regulated by multiple miRNAs in different hematopoietic contexts. In addition, miRNA-independent mechanisms to down-regulate MYB, including transcriptional regulation, RNA splicing, and proteolytic degradation via ubiquitylation, are also important.42-44 In particular, our kinetics data suggest that miR-34a–independent regulation of MYB expression is needed for initially suppressing MYB expression.
miR-34a is not substantially up-regulated until a day after TPA stimulation of K562 cells. This finding suggests that miR-34a does not participate in the earliest differentiating events but acts to suppress expression of its target genes at a later stage, perhaps to preserve the postmitotic block and maintain the MK lineage. Consistent with this hypothesis is the temporal association of miR-34a expression with late suppression of reinitiated MYB and CDK4 expression. Careful kinetic studies will be required to sort out whether other MYB-regulating miRNAs, such as miR-150, miR-221, or miR-134, might play a role in the initial down-regulation of these targets. Importantly, exogenous expression of miR-34a in primary CD34+ HSCs enhanced MK colony formation in a more physiologic setting. However, further studies using normal hematopoietic precursor cells are needed to determine whether miR-34a function is mediated by regulation of additional targets other than the ones identified in the present study.
The postulated role of miR-34a as a tumor suppressor is supported by our findings that enforced expression of miR-34a arrests K562 cells in the G1 phase of the cell cycle. In this study, we confirmed that miR-34a suppresses the expression of the CDKs that control the G1 to S transition (CDK4 and CDK6) and identified additional miR-34a MREs in the 3′-UTR of CDK6. Knockdown of CDK4 and CDK6, but not of either by itself, mimicked the cell- cycle inhibitory effect of overexpressing miR-34a, suggesting that these CDKs are important miR-34a target genes.
Down-regulation of miRNAs could also be important for megakaryopoiesis. One study suggested that down-regulation of miR-146a via transcriptional inhibition by PLZF and consequent derepression of the miR-146a target gene CXCR4 facilitates megakaryopoiesis.45 Another suggested that miR-155 down-regulation during megakaryopoiesis facilitates MK differentiation by derepressing expression of Ets-1 and Meis1.46 However, these miRNAs were not significantly altered by microarray after TPA treatment of K562 cells. Our seemingly discrepant findings could be the result of differences in timing or to idiosyncratic properties of the cell line, which might not reflect normal cell differentiation.
Several groups have shown that p53 transcriptionally activates miR-34a, which in turn regulates the expression of p53-regulated genes.16 However, K562 cells are p53-null.36 Therefore, miR-34a expression is not driven by p53 in these cells. In this study, we identified alternate pri-miR-34a transcripts induced by TPA. Furthermore, the genomic region adjacent to the alternate TSS used by these transcripts drives their TPA-dependent expression. The tumor suppressor function of miR-34a could therefore be induced by p53 and/or by a phorbol ester-responsive promoter in p53-null tumor cells. miR-34a may serve to modulate the tumor-promoting effects of phorbol esters. The biologic effect of phorbol esters is mediated largely by protein kinase C, which activates key transcription factors, especially AP-1 (fos/jun) and nuclear factor-κB.47-49 PKC-activated transcription factors, such as these, probably bind to the promoter region surrounding the TSS of the TPA-dependent pri-miR-34a transcripts. It will be interesting to analyze whether miR-34a can be expressed in a p53-independent manner in p53-sufficient or p53-null tumor cells of nonhematologic origin and in normal cells. Activation of miR-34a by itself may offer a novel useful approach to tumor therapy by interfering with cell proliferation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Note added in proof:
While this manuscript was in press, Kato et al reported that miR-34 induction postirradiation in Caenorhabditis elegans does not require the p53 homolog cep-1.50
Acknowledgments
The authors thank Gary Gilliland, Oliver Hoffman, and the laboratory scientists (in the laboratory of J.L.) for useful discussions.
Authorship
Contribution: F.N. designed the research, performed the experiments, analyzed the data, and wrote the manuscript; D.G. performed the experiments; E.M. performed the miRNA microarray analysis; M.C. assisted in gene microarray data analysis; I.R. assisted in miRNA target identification; Z.B. performed the miRNA microarray analysis; and J.L. designed the research, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Judy Lieberman, Immune Disease Institute, Harvard Medical School, Warren Alpert Bldg Rm 255, 200 Longwood Ave, Boston, MA 02115; e-mail: lieberman@idi.harvard.edu.