Abstract
The role of hematopoietic cytokines in lineage commitment remains uncertain. To gain insight into the contribution of cytokine signaling to myeloid lineage specification, we compared granulocyte colony-stimulating factor (G-CSF) and macrophage colony-stimulating factor (M-CSF) signaling in Ba/F3 cells expressing both the G-CSF and M-CSF receptors and in lineage-negative murine marrow cells. G-CSF and M-CSF serve as prototypes for additional cytokines that also influence immature myeloid cells. G-CSF specifically activated signal transducer and activator of transcription 3 and induced Src homology region 2 domain-containing phosphatase 2 (SHP2) phosphorylation, whereas M-CSF preferentially activated phospholipase Cγ2, and thereby extracellular signal-regulated kinase (ERK), to stabilize c-Fos and stimulate CCAAT/enhancer-binding protein (C/EBP)α(S21) phosphorylation. In contrast, activation of Jun kinase or c-Jun was similar in response to either cytokine. Inhibition of ERK prevented induction of c-Fos by M-CSF and reduced C/EBPα phosphorylation and formation of colony-forming unit–monocytes. SHP2 inhibition reduced ERK activation in G-CSF, but not M-CSF, and reduced colony-forming unit–granulocytes, underscoring divergent pathways to ERK activation. Phorbol ester mimicked the effect of M-CSF, activating ERK independent of SHP2. In summary, M-CSF activates ERK more potently than G-CSF, and thereby induces higher levels of c-Fos and phospho-C/EBPα(S21), which may directly interact to favor monopoiesis, whereas G-CSF activates signal transducer and activator of transcription 3 and SHP2, potentially shifting the balance to granulopoiesis via gene induction by C/EBPα homodimers and via effects of SHP2 on regulators besides ERK.
Introduction
Granulocytic and monocytic myeloid cells develop from a common granulocyte-monocyte progenitor (GMP), which in turn develops from a lymphoid-myeloid progenitor or the common myeloid progenitor, and ultimately the hematopoietic stem cell.1 Mechanisms that regulate myeloid lineage choice are relevant to host defense mechanisms and also to leukemic transformation of immature cells. Several transcription factors contribute to myeloid lineage specification.2 Mice lacking PU.1 or CCAAT/enhancer-binding protein (C/EBP)α have greatly reduced monocytes, granulocytes, and GMP.3-5 Elevated PU.1 favors monocyte as opposed to granulocyte lineage commitment, and C/EBPα induction of PU.1 transcription may thereby favor monopoiesis.6,7 The C/EBPα:c-Jun or C/EBPα:c-Fos leucine zipper heterodimers direct monocyte development when introduced into immature murine marrow progenitors.8 The redundant Egr-1 and Egr-2 transcriptional repressors favor monopoiesis while inhibiting granulopoiesis, in competition with the Gfi-1 repressor that inhibits monopoiesis to favor granulopoiesis.9 Gfi-1 also directly interacts with and inhibits PU.1 to limit monopoiesis.10
Whereas much is known regarding signals emanating from the macrophage colony-stimulating factor (M-CSF) receptor (MCSFR) or the granulocyte colony-stimulating factor (G-CSF) receptor (GCSFR), their role in myeloid lineage specification remains uncertain. The GCSFR dimerizes upon interaction with G-CSF, but lacks intrinsic tyrosine kinase activity. The dimeric receptor binds Jak kinases via a membrane-proximal region, and these in turn phosphorylate 4 GCSFR tyrosine residues to attract and in turn activate signal transducer and activator of transcription 1 (STAT1), STAT3, STAT5, Src homology region 2 domain-containing phosphatase 2 (SHP2), and the Ras/Raf/mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) pathway.11 The GCSFR also activates the Lyn and Hck members of the Src kinase family.11 The MCSFR dimerizes upon interaction with M-CSF, activating its intrinsic tyrosine kinase domain to autophosphorylate 7 tyrosines on its cytoplasmic tail. These in turn have been found to activate the Ras/Raf/MEK/ERK pathway, STAT1, phospholipase Cγ (PLCγ), Src kinases, and phosphatidylinositol 3-kinase.12
Whereas these signaling events most likely influence lineage maturation, their precise roles in this process or in earlier commitment events during myelopoiesis remain uncertain and are context dependent. Mice lacking STAT3 retain the ability to develop neutrophils, although granulocyte maturation is impaired during stress hematopoiesis.13,14 Absence of Lyn, Hck, and Fgr increases rather than impairs granulopoiesis.15 Activation of protein kinase C (PKC) by PLCγ or by phorbol esters enables monocytic differentiation of myeloid cell lines.16 Use of the U0126 MEK inhibitor to block ERK activation prevents FDC-P1 cells from differentiating into monocytes in response to M-CSF,17 induces bone marrow cells immortalized with the E2A-Pbx1 oncogene to differentiate into granulocytes instead of monocytes in M-CSF, and increases production of granulocytes from normal marrow mononuclear cells placed in liquid culture.18,19 Conversely, the same MEK inhibitor interfered with both granulocytic differentiation of the 32Dcl3 cell line and monocytic differentiation of M1 cells.20
To help define the role of cytokine receptor signaling in monocyte versus granulocyte lineage commitment, we reasoned that it would be useful to directly compare GCSFR and MCSFR signals in the same cellular context. Moreover, we thought it most relevant to conduct this comparison using immature, normal myeloid cells. Therefore, after performing initial analyses using a Ba/F3 subclone expressing both exogenous MCSFR and the GCSFR, we assessed signaling events using lineage-negative murine marrow cells, comprised predominantly of immature myeloid cells and expressing similar levels of both receptors. Whereas G-CSF specifically activated STAT3 and stimulated SHP2 phosphorylation, M-CSF specifically activated PLCγ2 and preferentially activated ERK in marrow cells. In contrast, both receptors activated c-Jun N-terminal kinase (JNK) and c-Jun similarly. Increased activation of ERK by M-CSF stabilized c-Fos and induced C/EBPα(S21) phosphorylation, whereas SHP2 inhibition reduced activation of ERK by G-CSF, but not M-CSF, suggesting that SHP2 acts independently of ERK to favor granulopoiesis. Moreover, ERK inhibition preferentially reduced colony-forming unit–monocyte (CFU-M) numbers, whereas SHP2 inhibition specifically reduced colony-forming unit–granulocytes (CFU-G). Based on these findings, a model whereby specific G-CSF or M-CSF signaling to transcriptional regulators influences myeloid lineage specification will be discussed.
Methods
Cell line culture, transduction, and FACS
Ba/F3 cells21 were maintained in RPMI 1640 with 10% heat-inactivated fetal bovine serum (HI-FBS), 1 ng/mL murine interleukin-3 (IL-3; PeproTech), and penicillin/streptomycin. The murine MCSFR cDNA was introduced into the pBabePuro retroviral vector, and the murine GCSFR cDNA was introduced into pBabeNeo.22 These were packaged by cotransfection of 8 μg retroviral vector with 2 μg pEcokat23 and 16 μL Lipofectamine 2000 (Invitrogen) into 293T cells, cultured in 100-mm dishes with Dulbecco modified Eagle medium and 10% HI-FBS. Two days later, viral supernatants were collected and filtered through 0.45-μm filters. Ba/F3 cells were transduced sequentially with these vectors in the presence of Polybrene (4 μg/mL) on tissue-culture plates coated with Retronectin (Takara Shuzo). Puromycin was used at 2 μg/mL and G418 at 1.2 mg/mL. Subclones were isolated by limiting dilution in 96-well plates and screened for MCSFR and GCSFR expression by fluorescence-activated cell sorting (FACS), using a monoclonal anti-murine c-fms (MCSFR or CD115) phycoerythrin (PE)–conjugated antibody (eBioscience) and 5 μg/mL biotin-GCSF generated using the EZ-link biotinylation kit (Pierce), followed by incubation with streptavidin-allophycocyanin (APC). MCSFR expression was also assessed using similarly generated biotin-MCSF and streptavidin-PE. PE anti-fms, biotin-GCSF, and streptavidin–fluorescein isothiocyanate were also used to detect endogenous receptor expression in marrow cells. For cytokine stimulation, Ba/F3 cells were washed twice with phosphate-buffered saline (PBS) and placed in RPMI 1640 with 10% HI-FBS without IL-3 for 1 hour before stimulation with 10 ng/mL either human G-CSF (Amgen) or murine M-CSF (PeproTech). JNK (SP600125), MEK (U0126 or PD98059), PLCγ (U73122), PLC (edelfosine), or SHP2 (NSC-87877; obtained from Acros) inhibitors or 12-O-tetradecanoylphorbol-13-acetate (TPA; Sigma-Aldrich) were added during the start of the cytokine starvation period.
Marrow culture
Six- to 8-week-old C57BL/6 mice were treated with 5-fluorouracil (5-FU) intraperitoneally (150 mg/kg). Marrow cells isolated 5 days later from the long bones were collected, subjected to red cell lysis with NH4Cl, and cultured in Iscove modified Dulbecco medium (IMDM) with 10% HI-FBS, 10 ng/mL murine IL-3, 10 ng/mL murine IL-6, and 10 ng/mL murine stem cell factor (SCF; PeproTech) for 5 days. The cells were then subjected to lineage depletion using immunomagnetic beads and a mixture of lineage antibodies, B220, CD5, Mac-1, Ter119, Gr-1, and 7-4 (StemCell Technologies), yielding 5% of total mononuclear cells. FACS confirmed absence of lineage-marker expression. Cells were then cultured in IMDM with HI-FBS, IL-3, IL-6, and SCF for 1 hour to allow for recovery, washed twice with PBS, transferred to IMDM with HI-FBS, and cytokine starved for 1 hour before addition of 10 ng/mL G-CSF or 10 ng/mL M-CSF. For CFU assays, mononuclear marrow cells were isolated from mice not exposed to 5-FU and plated directly in methylcellulose with 10% HI-FBS, IL-3, IL-6, SCF, and indicated inhibitors or vehicle control. Myeloid CFUs were enumerated 8 days later. Institutional Animal Care and Use Committee approval for the use of mice was obtained from Johns Hopkins University.
Western blotting
Cells were harvested, washed twice with PBS, and lysed for 20 minutes at 4°C on a rotator in 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), pH 7.5, 300 mM NaCl, 2 mM EDTA (ethylenediaminetetraacetic acid), 2 mM sodium orthovanadate, 0.1% Triton X-100, 20 μg/mL phenylmethylsulfonyl fluoride, and 1:500 dilution of protease inhibitor mixture for mammalian cell culture (Sigma-Aldrich). Lysates were cleared by centrifugation at 15 000g. Cleared lysates corresponding to 5 × 105 cells were placed in Laemmli sample buffer, boiled, and separated on 12.5% polyacrylamide gels. Protein transfer to a polyvinylidene difluoride membrane (Hybond-P; Amersham) was accomplished using a semidry transfer apparatus, followed by probing with primary antibodies. Antibodies used were c-Jun (H-79), P-cJun (Ser 63/73 R), ERK (K-23), JNK (D-2), β-tubulin (D-10), C/EBPα (14AA), C/EBPβ (C-19), PU.1 (T-21), PLCγ2 (Q-20; obtained from Santa Cruz Biotechnology), c-Fos, P-JNK (98F2), P-ERK (197G2), STAT3 (79D7), STAT5 (3H7), P-STAT3 (D3A7), P-STAT5 (C11C5), SHP2, P-SHP2 (Tyr542), P-SHP2 (Tyr580), P-C/EBPα (Ser 21), P-PLCγ2 (Y759; obtained from Cell Signaling Technology), and β-actin (AC-15; obtained from Sigma-Aldrich). After application of horseradish peroxidase–conjugated secondary antibody and subsequent washes, a signal was generated using Supersignal West (Pierce). Signals were detected by autoradiography.
Phosphatase and calcium flux assays
Phosphatase activity of immunoprecipitated SHP2 was assessed, as described (supplemental Figure 3, available on the Blood website; see the Supplemental Materials link at the top of the online article). To assess release of Ca2+ into the cytoplasm, Ba/F3 cells removed from IL-3 were placed in Ca2+/Mg2+-free HEPES-buffered saline with 1% FBS and loaded with 2 μg/mL Indo-1AM (Invitrogen) for 30 minutes at 37°C, washed, warmed, and then monitored by FACS for fluorescence at 395 nm (Ca2+ bound) and 500 nm (Ca2+ unbound) with ultraviolet excitation after addition of cytokine.
Results
G-CSF preferentially induces STAT and SHP2 phosphorylation in Ba/F3 cells
Ba/F3 cells were initially defined as having a pro-B phenotype, although their dependence upon IL-3 for survival and proliferation is more reminiscent of the myeloid lineage. Their cytokine dependence makes them attractive as well for study of signaling events in comparison with leukemic cell lines. A Ba/F3 subclone expressing both the MCSFR and the GCSFR at similar levels was derived by retroviral transduction, and expression of the receptors was confirmed by FACS analysis (Figure 1A). FACS using PE anti-MCSFR and biotin-GCSF/streptavidin-APC indicated simultaneous expression of both receptors on the large majority of the cells. Staining the cells separately with biotin-GCSF or biotin-MCSF, followed by streptavidin-PE, indicated similar expression of both receptors, as the biotinylated cytokines were in excess. Consistent with our prior demonstration that activation of exogenous GCSFR in Ba/F3 cells does not lead to induction of C/EBPα, myeloperoxidase, or lactoferrin,24 exposure of Ba/F3(MR,GR) cells to either M-CSF or G-CSF for up to 8 days did not induce monocytic or granulocytic morphology, Mac1, Gr-1, or C/EBPα (supplemental Figures 1-2A).
M-CSF or G-CSF activates the JNK and ERK pathways in Ba/F3 cells. (A) FACS analysis for MCSFR (MR) and GCSFR (GR) in a Ba/F3 subclone transduced with retroviral vectors expressing these receptors. Cells were stained either with anti-MCSFR-PE, biotin-GCSF, and streptavidin-APC (top panel), or separately with biotin-GCSF or biotin-MCSF and streptavidin-PE (bottom panels). (B) Ba/F3(MR,GR) cells were cultured with IL-3 or in its absence (−) for 3 hours (top panel) or 1 hour (bottom panel). Total cellular protein extracts from equal numbers of cells were then subjected to Western blotting for indicated total or phosphorylated proteins (top). (C) Ba/F3(MR,GR) cells removed from IL-3 for 1 hour were cultured for 0 to 120 minutes in M-CSF or G-CSF. Total cellular protein extracts were then subjected to Western blotting for the indicated proteins.
M-CSF or G-CSF activates the JNK and ERK pathways in Ba/F3 cells. (A) FACS analysis for MCSFR (MR) and GCSFR (GR) in a Ba/F3 subclone transduced with retroviral vectors expressing these receptors. Cells were stained either with anti-MCSFR-PE, biotin-GCSF, and streptavidin-APC (top panel), or separately with biotin-GCSF or biotin-MCSF and streptavidin-PE (bottom panels). (B) Ba/F3(MR,GR) cells were cultured with IL-3 or in its absence (−) for 3 hours (top panel) or 1 hour (bottom panel). Total cellular protein extracts from equal numbers of cells were then subjected to Western blotting for indicated total or phosphorylated proteins (top). (C) Ba/F3(MR,GR) cells removed from IL-3 for 1 hour were cultured for 0 to 120 minutes in M-CSF or G-CSF. Total cellular protein extracts were then subjected to Western blotting for the indicated proteins.
Although signaling studies with hematopoietic cells are at times performed after removal of both FBS and cytokines, followed by cytokine addition, we sought to optimize a milder protocol, thereby preventing activation of stress signaling. We confirmed that removal of IL-3 alone from Ba/F3(MR,GR) cells for 3 hours did not increase c-Jun or c-Fos levels, nor did it induce JNK or c-Jun phosphorylation (Figure 1B top panel), demonstrating lack of a stress response. We also found that removal of IL-3 for 1 hour markedly reduced ERK or STAT3 phosphorylation (Figures 1B and 2C bottom panels), and in subsequent experiments with cell lines or marrow cells, cytokines were removed for only 1 hour before stimulation with cytokines. Ba/F3(MR,GR) cells were withdrawn from IL-3 and challenged with M-CSF or G-CSF for 10, 30, 60, or 120 minutes (Figure 1C). In this experiment, we focused on signaling to several transcription factors involved in control of myeloid development. Both cytokines induced phosphorylation of c-Jun, JNK, and ERK and increased c-Fos levels without affecting expression of total c-Jun, JNK, ERK, C/EBPα, C/EBPβ, or PU.1, or phosphorylation of C/EBPα(S21). The doublet in the ERK and P-ERK lanes represents ERK1 and ERK2. The 2 bands in the JNK and P-JNK lanes each contain alternative splice variants of JNK1 and JNK2. These findings were reproducible with independent Ba/F3 extracts, and similar findings were also observed when the human U937 myeloid leukemia cell line, expressing both receptors endogenously, was challenged with M-CSF or G-CSF (data not shown). Activation of ERK by either M-CSF or G-CSF was cytokine dose dependent (supplemental Figure 2B).
To gain insight into communication between signaling pathways leading to c-Jun activation and c-Fos induction, we carried out a similar experiment in the presence of JNK, MEK1, or PLCγ inhibitors compared with a no inhibitor control, preparing extracts 30 minutes after cytokine addition (Figure 2A). The JNK inhibitor suppressed c-Jun and JNK phosphorylation, as expected, but did not affect ERK activation or c-Fos induction. MEK1 inhibition blocked ERK activation and c-Fos induction, but increased JNK and c-Jun phosphorylation. In addition to the major doublet most evident in the last 4 lanes, the P-JNK antibody also detected minor, most likely nonspecific bands. Of note, ERK phosphorylation and stabilization of c-Fos have been seen previously in NIH 3T3 fibroblasts.25 PLCγ inhibition prevented both ERK activation and c-Fos induction, consistent with ERK to c-Fos signaling in these hematopoietic cells, and PLCγ inhibition also led to a striking increase in P-JNK and P-Jun, consistent with PLCγ's ability to inhibit JNK signaling through NF-κB activation. Reduction of ERK activation and c-Fos induction was also seen with a second MEK1 inhibitor or with a general PLC inhibitor (Figure 2B). The results with these 5 inhibitors were seen in a second, independent set of experiments. Together, these data provide evidence for a novel pathway comprising PLCγ, ERK, and c-Fos together with a separate JNK to c-Jun pathway in the Ba/F3 hematopoietic cell line, although they do not uncover MCSFR- or GCSFR-specific signaling.
PLCγ/ERK/c-Fos, JNK/c-Jun, STAT, and SHP2 signaling from the MCSFR or GCSFR in Ba/F3 cells. (A) Ba/F3(GR,MR) cells withdrawn from IL-3 for 1 hour were cultured for an additional 30 minutes in the absence of cytokine (C), with M-CSF (M), or with G-CSF (G), together with no inhibitor or with a JNK (10 μM SP600125), MEK1/2 (10 μM U0126), or PLCγ (2 μM U73122) inhibitor, followed by Western blotting. (B) Ba/F3 cells were evaluated similarly after culture with no inhibitor or with a MEK1 (50 μM PD98059) or a general PLC (10 μM edelfosine) inhibitor. A vertical line has been inserted to indicate repositioned gel lanes. (C) Ba/F3(GR,MR) cells withdrawn from IL-3 for 1 hour were cultured for an additional 30 minutes in the absence of cytokine (C), with M-CSF (M), or with G-CSF (G), together with no inhibitor or with a JNK (10 μM SP600125), MEK1/2 (10 μM U0126), or PLCγ (2 μM U73122) inhibitor, followed by Western blotting (top panel). Ba/F3(GR,MR) cells were analyzed for P-STAT3 or STAT3 expression in IL-3, after removal of IL-3 for 1 hour, or after an additional 30 minutes in the absence of cytokine, with M-CSF, or with G-CSF (bottom panel). (D) Ba/F3 cells withdrawn from IL-3 were cultured for 30 minutes with no cytokine, M-CSF, or G-CSF. Total cellular protein extracts were then subjected to Western blotting for P-SHP2 Y542 and Y580, total SHP2, and tubulin.
PLCγ/ERK/c-Fos, JNK/c-Jun, STAT, and SHP2 signaling from the MCSFR or GCSFR in Ba/F3 cells. (A) Ba/F3(GR,MR) cells withdrawn from IL-3 for 1 hour were cultured for an additional 30 minutes in the absence of cytokine (C), with M-CSF (M), or with G-CSF (G), together with no inhibitor or with a JNK (10 μM SP600125), MEK1/2 (10 μM U0126), or PLCγ (2 μM U73122) inhibitor, followed by Western blotting. (B) Ba/F3 cells were evaluated similarly after culture with no inhibitor or with a MEK1 (50 μM PD98059) or a general PLC (10 μM edelfosine) inhibitor. A vertical line has been inserted to indicate repositioned gel lanes. (C) Ba/F3(GR,MR) cells withdrawn from IL-3 for 1 hour were cultured for an additional 30 minutes in the absence of cytokine (C), with M-CSF (M), or with G-CSF (G), together with no inhibitor or with a JNK (10 μM SP600125), MEK1/2 (10 μM U0126), or PLCγ (2 μM U73122) inhibitor, followed by Western blotting (top panel). Ba/F3(GR,MR) cells were analyzed for P-STAT3 or STAT3 expression in IL-3, after removal of IL-3 for 1 hour, or after an additional 30 minutes in the absence of cytokine, with M-CSF, or with G-CSF (bottom panel). (D) Ba/F3 cells withdrawn from IL-3 were cultured for 30 minutes with no cytokine, M-CSF, or G-CSF. Total cellular protein extracts were then subjected to Western blotting for P-SHP2 Y542 and Y580, total SHP2, and tubulin.
We also compared the effects of M-CSF or G-CSF on the STAT1, STAT3, and STAT5 transcription factors or SHP2, a tyrosine phosphatase that contributes to ERK activation26 (Figure 2C top panel lanes 1-3, and D). G-CSF specifically activated STAT1, STAT3, and STAT5, as was reproduced in independent experiments. Activation of STAT3 by G-CSF exceeded its baseline phosphorylation in IL-3 (Figure 2C bottom panel). Activation of each of the STAT proteins was impaired by PLCγ inhibition, activation of STAT1 or STAT5 was impaired by JNK inhibition, and STAT activation was unimpaired by MEK inhibition (Figure 2C top panel lanes 4-12). Either M-CSF or G-CSF induced phosphorylation of SHP2 Y542 and Y580 in this context; however, the degree of induction by G-CSF was strikingly larger. Combining G-CSF with M-CSF did not alter induction of ERK, SHP2, or STAT3 phosphorylation by G-CSF or M-CSF alone (supplemental Figure 2C). Phosphorylation of SHP2 in G-CSF did not induce SHP2 phosphatase activity toward a peptide substrate (supplemental Figure 3A), although phosphatase activity was reduced by the NSC-87877 SHP2 inhibitor (supplemental Figure 3B). As we will discuss, SHP2 phosphorylation potentially alters SHP2 protein interactions to direct it to specific substrates. Of note, as with ERK and STAT3, removal of IL-3 reduced basal SHP2 phosphorylation to nearly undetectable levels (supplemental Figure 3C). Preferential activation of STAT proteins and modification of SHP2 by G-CSF in this Ba/F3 line are striking given the possibility that supraphysiologic receptor levels might obscure differences.
M-CSF preferentially activates ERK and elevates c-Fos, whereas G-CSF induces STAT3 and SHP2 phosphorylation in marrow progenitors
We next sought to optimize use of lineage-negative murine marrow cells for comparing M-CSF and G-CSF signaling. Upon recovery from isolation by culture with IL-3, IL-6, and SCF for 2 hours, the lin− population demonstrated expression of MCSFR and GCSFR, but lacked the Mac-1 and Gr-1 markers found on mature granulocytes or monocytes (Figure 3A). Cells expressing both receptors as well as a similar number predominantly expressing one or the other receptor are evident. The majority of cells express similar levels of GCSFR, MCSFR, or both, although a small subset expresses increased MCSFR. Although heterogeneous, the lineage-negative marrow population is sufficiently abundant to allow analysis of signal transduction via Western blotting, and events that occur in this marrow subset may reflect events that occur in a bipotent myeloid progenitor. Of note, GMP, isolated as Sca-1−c-kit+FcγRhighCD34+ cells by FACS, generate CFU-granulocytes/monocytes (CFU-GM), but also a significant number of CFU-G and CFU-M myeloid colonies, indicating heterogeneity even among this highly purified progenitor fraction,27 and isolation of GCSFR+MCFR+ cells would be problematic due to low numbers and because bound antibodies would activate the receptors.
M-CSF specifically induces P-ERK and c-Fos, whereas G-CSF specifically induces P-STAT3 and P-SHP2 in marrow progenitors. (A) Murine bone marrow (mBM) cells isolated from mice exposed to 5-FU were lineage depleted; cultured for 2 hours with IL-3, IL-6, and SCF; and subjected to FACS for MCSFR/GCSFR or Mac-1/Gr-1. (B) Lineage-negative mBM cells were cultured in media containing IL-3, IL-6, and SCF for 1 hour, removed from cytokine for 1 hour, and then cultured with no cytokine (C), M-CSF (M), or G-CSF (G) for 30 minutes. Total cellular proteins were then subjected to Western blotting. (C) Lineage-negative mBM cells cultured similarly were analyzed for P-SHP2, total SHP2, and tubulin.
M-CSF specifically induces P-ERK and c-Fos, whereas G-CSF specifically induces P-STAT3 and P-SHP2 in marrow progenitors. (A) Murine bone marrow (mBM) cells isolated from mice exposed to 5-FU were lineage depleted; cultured for 2 hours with IL-3, IL-6, and SCF; and subjected to FACS for MCSFR/GCSFR or Mac-1/Gr-1. (B) Lineage-negative mBM cells were cultured in media containing IL-3, IL-6, and SCF for 1 hour, removed from cytokine for 1 hour, and then cultured with no cytokine (C), M-CSF (M), or G-CSF (G) for 30 minutes. Total cellular proteins were then subjected to Western blotting. (C) Lineage-negative mBM cells cultured similarly were analyzed for P-SHP2, total SHP2, and tubulin.
Lineage-negative murine marrow cells cultured with FBS, IL-3, IL-6, and SCF for 1 hour after isolation were removed from all 3 cytokines for an additional hour and then challenged with no cytokine, M-CSF, or G-CSF for 30 minutes, followed by Western blot analysis (Figure 3B). M-CSF induced ERK phosphorylation and c-Fos expression more effectively than G-CSF, by 2- to 3-fold, whereas activation of JNK or c-Jun was similar in either cytokine. STAT3 activation was detected only in G-CSF, little STAT5 activation was evident, and STAT1 activation was not detected (data not shown). As in Ba/F3(MR,GR) cells, SHP2 phosphorylation on Y542 and Y580 was stimulated to a much greater extent by G-CSF compared with M-CSF in lineage-negative marrow cells (Figure 3C). Each of these findings was evident in 2 separate experiments or in 3 experiments for M-CSF induction of P-ERK and c-Fos. Thus, G-CSF specifically induced STAT3 and SHP2 phosphorylation in Ba/F3 cells and in lineage-negative marrow cells, whereas M-CSF preferentially induced ERK phosphorylation and elevates c-Fos in marrow cells, although both cytokines induced these events in Ba/F3(MR,GR) cells.
M-CSF and G-CSF use separate pathways to activate ERK and modify C/EBPα
PLCγ inhibition proved excessively toxic to marrow cells, as seen previously,19 and was not pursued further. In contrast, inhibition of MEK with U0126 reproducibly prevented ERK phosphorylation and consequent c-Fos stabilization in lineage-negative marrow cells cultured in M-CSF (Figure 4A), as was seen also in Ba/F3 cells. C/EBPα(S21) phosphorylation was more prominent in M-CSF compared with G-CSF, in this experiment and with independent samples presented in Figure 5B, most likely reflecting increased ERK activity in M-CSF. MEK inhibition reduced C/EBPα(S21) phosphorylation induced by M-CSF in lineage-negative marrow cells (compare lanes 2 and 5), as had been previously seen in cell lines.28
M-CSF and G-CSF signaling with MEK or SHP2 inhibition in marrow progenitors. (A-B) Lineage-negative murine bone marrow (BM) cells were cultured with IL-3, IL-6, and SCF for 1 hour; removed from cytokine for 1 hour; and then cultured with no cytokine, M-CSF, or G-CSF with no inhibitor, 5 μM U0126, or 50 μM NSC-87877 for 30 minutes. Total cellular proteins were then subjected to Western blotting for indicated proteins.
M-CSF and G-CSF signaling with MEK or SHP2 inhibition in marrow progenitors. (A-B) Lineage-negative murine bone marrow (BM) cells were cultured with IL-3, IL-6, and SCF for 1 hour; removed from cytokine for 1 hour; and then cultured with no cytokine, M-CSF, or G-CSF with no inhibitor, 5 μM U0126, or 50 μM NSC-87877 for 30 minutes. Total cellular proteins were then subjected to Western blotting for indicated proteins.
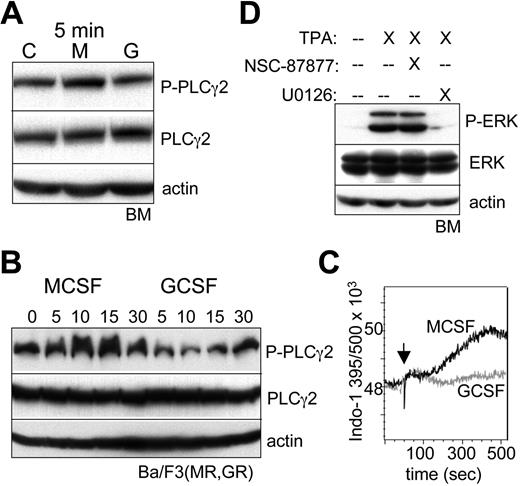
M-CSF preferentially activates PLCγ2, and phorbol ester induces ERK independent of SHP2. (A) Lineage-negative bone marrow (BM) cells were cultured with IL-3, IL-6, and SCF for 1 hour; removed from cytokine for 1 hour; and then cultured with no cytokine, M-CSF, or G-CSF for 5 minutes. Total cellular proteins were then subjected to Western blotting. (B) Ba/F3(MR,GR) cells removed from IL-3 for 1 hour were cultured with M-CSF or G-CSF for 0, 5, 15, or 30 minutes, followed by Western blotting. (C) Ba/F3(MR,GR) cells removed from IL-3 for 1 hour were loaded with Indo-1AM and then cultured with M-CSF or G-CSF, and the Indo-1 fluorescence at 395 nm (Ca2+ bound) and 500 nm (Ca2+ unbound) was assessed by FACS over a 7-minute period. An arrow indicates the time of cytokine addition. (D) Lineage-negative BM cells cultured with IL-3, IL-6, and SCF for 1 hour were removed from cytokine for 1 hour and then cultured with no cytokine in the absence or presence of 100 nM phorbol ester (TPA). U0126 (5 μM) or NSC-87877 (50 μM) was also included, where indicated.
M-CSF preferentially activates PLCγ2, and phorbol ester induces ERK independent of SHP2. (A) Lineage-negative bone marrow (BM) cells were cultured with IL-3, IL-6, and SCF for 1 hour; removed from cytokine for 1 hour; and then cultured with no cytokine, M-CSF, or G-CSF for 5 minutes. Total cellular proteins were then subjected to Western blotting. (B) Ba/F3(MR,GR) cells removed from IL-3 for 1 hour were cultured with M-CSF or G-CSF for 0, 5, 15, or 30 minutes, followed by Western blotting. (C) Ba/F3(MR,GR) cells removed from IL-3 for 1 hour were loaded with Indo-1AM and then cultured with M-CSF or G-CSF, and the Indo-1 fluorescence at 395 nm (Ca2+ bound) and 500 nm (Ca2+ unbound) was assessed by FACS over a 7-minute period. An arrow indicates the time of cytokine addition. (D) Lineage-negative BM cells cultured with IL-3, IL-6, and SCF for 1 hour were removed from cytokine for 1 hour and then cultured with no cytokine in the absence or presence of 100 nM phorbol ester (TPA). U0126 (5 μM) or NSC-87877 (50 μM) was also included, where indicated.
To assess the role of SHP2 activity on ERK activation, we used NSC-87877, which inhibits SHP1 and SHP2, but not several other protein tyrosine phosphatases.29 Lineage-negative marrow cells were cultured with M-CSF or G-CSF for 30 minutes in the absence or presence of NSC-87877 (Figure 4B). In the absence of the inhibitor, M-CSF again preferentially induced ERK phosphorylation. In the presence of the SHP inhibitor, ERK phosphorylation was reduced in response to G-CSF (compare lanes 3 and 6), but, remarkably, not in response to M-CSF (compare lanes 2 and 5). This finding was reproduced also in an independent experiment. SHP2 inhibition did not affect c-Jun or STAT3 phosphorylation, but reduced C/EBPα(S21) phosphorylation specifically in G-CSF, reflecting SHP2 activation of ERK in response to this cytokine and further supporting the concept that SHP2 is activated to a much greater extent by G-CSF than M-CSF.
M-CSF preferentially activates PLCγ2 to activate ERK
As the MCSFR was shown previously to activate PLCγ,12 we compared its activation by M-CSF versus G-CSF in lin− marrow progenitors. Modest activation of PLCγ2 by M-CSF, but not G-CSF, was evident at 5 minutes (Figure 5A), although no difference was apparent at 10 or 30 minutes after cytokine addition, potentially reflecting negative feedback, and activation of PLCγ1 was not detected (data not shown). Similar specific activation of PLCγ2 by M-CSF at 5 minutes was also evident in a second experiment with marrow progenitors (data not shown). Activation of PLCγ2 by M-CSF versus G-CSF was also compared in Ba/F3(MR,GR) cells (Figure 5B). In this setting, M-CSF potently induced PLCγ2 phosphorylation at 5, 10, or 15 minutes, whereas G-CSF was essentially inactive. One predicted consequence of PLCγ2 activation is elevation of inositol-triphosphate, and thereby, cytoplasmic Ca2+ levels. We confirmed rapid elevation of cytoplasmic Ca2+ in Ba/F3(MR,GR) exposed to M-CSF, but not G-CSF, as reflected by an increase in the 395 nm/500 nm fluorescence, beginning 2 minutes after M-CSF addition and reaching a maximum by 7 minutes (Figure 5C).
Rapid activation of PLCγ2 is also expected to generate diacylglycerol, which in turn activates novel PKC isoforms, an activity mimicked by phorbol ester (TPA), and PKC has the capacity to activate Ras/Raf/MEK/ERK signaling. A novel PKC isoform, PKCδ, mediates monocytic maturation of FDC-P1 or 32D cells in response to TPA.16,30 As TPA induces monocytic differentiation of myeloid leukemic cells and normal marrow progenitors,31,32 we evaluated its ability to activate ERK in lin− marrow cells (Figure 5D). Potent activation was seen 30 minutes after TPA addition, and this induction was prevented by the U0126 MEK inhibitor in 2 separate experiments, but not by the NSC-87877 SHP2 inhibitor.
ERK inhibition reduces CFU-M, whereas SHP2 inhibition reduces CFU-G
To assess the functional consequences of ERK inhibition on myeloid CFU formation, mononuclear marrow cells were cultured in IL-3, IL-6, and SCF with either no inhibitor or the U0126 or PD98059 MEK inhibitors, and CFUs were enumerated 8 days later (Figure 6A). Total colonies observed in 3 experiments were 87% plus or minus 6% of control using U0126, and 87% plus or minus 8% of control using PD98059, and colonies were of normal size, indicating that MEK inhibition only minimally affects CFU proliferation, as seen previously.33 However, inhibition of MEK by either inhibitor significantly reduced the number and percentage of CFU-M, in comparison with control cultures. CFU-G numbers were mildly increased with either MEK inhibitor, but not to a significant extent. These findings indicate that ERK activation favors monocyte over granulocyte lineage specification of immature myeloid progenitors. Consistent with the importance of the PLCγ2 to PKC to ERK signaling pathway in monocyte lineage commitment, TPA induction of CFU-M and suppression of CFU-G were partially and significantly reversed by U0126 (Figure 6B). The effect of SHP2 inhibition on the formation of myeloid colonies from cultured murine marrow cells was also assessed (Figure 6C). In concordance with the preferential induction of SHP2 phosphorylation by G-CSF, inhibition of SHP2 led to preferential loss of CFU-G, compared with CFU-M or CFU-GM, without reducing overall CFU yield.
ERK inhibition reduces CFU-M, whereas SHP2 inhibition reduces CFU-G. (A) Marrow mononuclear cells obtained from mice not exposed to 5-FU were cultured in methylcellulose with IL-3, IL-6, and SCF at 10 000 cells/mL, with either no inhibitor, 5 μM U0126, or 5 μM PD98059. CFUs were enumerated 8 days later. Control cultures yielded on average 67 CFU-G, 67 CFU-M, and 55 CFU-GM per 30 000 cells cultured. The percentage of each CFU obtained with the 2 MEK inhibitors relative to the control colony numbers is shown (mean and SE from 3 experiments). *P < .001 comparing CFU obtained with versus without inhibitor (Student t test). (B) Marrow mononuclear cells were cultured similarly in methylcellulose with either no chemicals, TPA alone, or TPA with U0126. Control cultures yielded on average 151 CFU-G, 129 CFU-M, and 132 CFU-GM per 30 000 cells cultured. The percentage of each CFU obtained with 100 nM TPA or TPA combined with the ERK inhibitor relative to the control is shown (mean and SE from 3 experiments). *P < .001 comparing CFU obtained with versus without TPA; #P < .01 comparing CFU obtained with TPA with versus without U0126. (C) Marrow mononuclear cells were cultured with no inhibitor or 50 μM NSC-87877. Control cultures yielded on average 67 CFU-G, 67 CFU-M, and 55 CFU-GM per 30 000 cells cultured. The percentage of each CFU obtained with the SHP2 inhibitor relative to the control is shown (mean and SE from 3 experiments). *P < .01 comparing CFU obtained with versus without inhibitor.
ERK inhibition reduces CFU-M, whereas SHP2 inhibition reduces CFU-G. (A) Marrow mononuclear cells obtained from mice not exposed to 5-FU were cultured in methylcellulose with IL-3, IL-6, and SCF at 10 000 cells/mL, with either no inhibitor, 5 μM U0126, or 5 μM PD98059. CFUs were enumerated 8 days later. Control cultures yielded on average 67 CFU-G, 67 CFU-M, and 55 CFU-GM per 30 000 cells cultured. The percentage of each CFU obtained with the 2 MEK inhibitors relative to the control colony numbers is shown (mean and SE from 3 experiments). *P < .001 comparing CFU obtained with versus without inhibitor (Student t test). (B) Marrow mononuclear cells were cultured similarly in methylcellulose with either no chemicals, TPA alone, or TPA with U0126. Control cultures yielded on average 151 CFU-G, 129 CFU-M, and 132 CFU-GM per 30 000 cells cultured. The percentage of each CFU obtained with 100 nM TPA or TPA combined with the ERK inhibitor relative to the control is shown (mean and SE from 3 experiments). *P < .001 comparing CFU obtained with versus without TPA; #P < .01 comparing CFU obtained with TPA with versus without U0126. (C) Marrow mononuclear cells were cultured with no inhibitor or 50 μM NSC-87877. Control cultures yielded on average 67 CFU-G, 67 CFU-M, and 55 CFU-GM per 30 000 cells cultured. The percentage of each CFU obtained with the SHP2 inhibitor relative to the control is shown (mean and SE from 3 experiments). *P < .01 comparing CFU obtained with versus without inhibitor.
Discussion
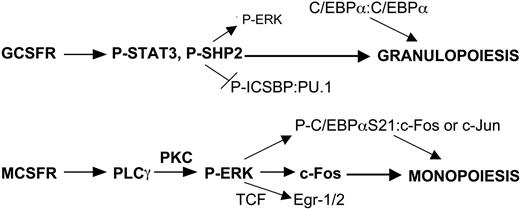
Lineage-specific transcription factor expression provides cells with intrinsic developmental programs. These cell-autonomous events are modified by external signals during development and when the adult organism responds to a changing external environment. Although multiple cytokines regulate the proliferation and survival of hematopoietic stem and progenitor cells, their role in modulating lineage commitment events remains uncertain.34 In this study, we focus on identifying signaling events that potentially influence myeloid progenitors to develop along the granulocyte or monocyte lineages. Although multiple cytokines impact on myeloid cell fate in vivo, G-CSF and M-CSF serve as paradigms for those that favor granulopoiesis or monopoiesis, respectively. To identify signaling pathways relevant to each lineage, we for the first time directly compare GCSFR and MCSFR signaling, focusing our efforts on normal, immature myeloid cells and progenitors from murine marrow. Most previous characterization of GCSFR or MCSFR signaling had been accomplished using cell line models or mature myeloid cells. Several key findings emerge from these studies, which, when combined with prior results discussed below, lead to a model for the role of cytokine signaling in myeloid lineage determination in cooperation with transcriptional regulators (Figure 7). Our results indicate that M-CSF specifically activates PLCγ and ERK in murine marrow cells to elevate c-Fos, whereas G-CSF specifically activates STAT3 and induces SHP2 tyrosine phosphorylation. Notably, SHP2 inhibition reduces ERK activation by G-CSF, but not by M-CSF, suggesting that M-CSF activates ERK independent of SHP2, potentially via specific activation of PLCγ2 and so PKCδ. Both cytokines activated JNK and c-Jun similarly, whereas C/EBPα(S21) phosphorylation occurs more potently in response to M-CSF, of potential functional significance and reflective of increased ERK activity. Finally, ERK inhibition reduced CFU-M while sparing CFU-G, whereas SHP2 inhibition specifically reduced CFU-G, consistent with a role for ERK versus SHP2 activation in monocyte versus granulocyte lineage determination.
Model for induction of specific signals by G-CSF versus M-CSF that contribute to myeloid lineage specification. GCSFR signals specifically activate STAT3 and SHP2. The latter has been shown to dephosphorylate and so inhibit ICSBP cooperation with PU.1, favoring granulopoiesis. MCSFR signals specifically activate PLCγ2 and thereby PKC, leading to more potent ERK activation than occurs via GCSFR-mediated SHP2 activation. As a result, M-CSF signaling more readily stabilizes c-Fos, phosphorylates C/EBPα(S21), and stimulates terniary complex factor (TCF)–mediated Egr-1/2 transcription, each favoring monopoiesis. Reduced phosphorylation of C/EBPα(S21) via G-CSF signaling strengthens its ability to direct granulopoiesis as a homodimer, whereas P-C/EBPα(S21) retains the capacity to induce monopoiesis as a heterodimer with c-Jun or c-Fos.
Model for induction of specific signals by G-CSF versus M-CSF that contribute to myeloid lineage specification. GCSFR signals specifically activate STAT3 and SHP2. The latter has been shown to dephosphorylate and so inhibit ICSBP cooperation with PU.1, favoring granulopoiesis. MCSFR signals specifically activate PLCγ2 and thereby PKC, leading to more potent ERK activation than occurs via GCSFR-mediated SHP2 activation. As a result, M-CSF signaling more readily stabilizes c-Fos, phosphorylates C/EBPα(S21), and stimulates terniary complex factor (TCF)–mediated Egr-1/2 transcription, each favoring monopoiesis. Reduced phosphorylation of C/EBPα(S21) via G-CSF signaling strengthens its ability to direct granulopoiesis as a homodimer, whereas P-C/EBPα(S21) retains the capacity to induce monopoiesis as a heterodimer with c-Jun or c-Fos.
Mutation of MCSFR(Y721) prevents M-CSF-mediated PLCγ2 receptor binding, PLCγ2 activation, and monocytic differentiation of FDC-P1 cells.18 Activation of Src family kinases by the MCSFR also leads to PLCγ2 activation, and Src inhibition prevents FDC-P1 differentiation in response to M-CSF.19 Consistent with a specific role during monopoieisis, we find that M-CSF, but not G-CSF, activates PLCγ2. PLCγ generates diacylglycerol to activate PKCδ, a known mediator of monocytic maturation of hematopoietic cell lines. Of note, activation of Src kinases, as assessed by SrcY416P phosphorylation, was similar upon M-CSF or G-CSF stimulation of lin− marrow cells (data not shown).
What then is the downstream effector of the MCSFR to PLCγ2 to PKC signaling pathway that induces monopoiesis? PLCγ inhibition reduces ERK activation in hematopoietic cells, as demonstrated using the Ba/F3(MR,GR) line in this study and previously in the FDC-P1 monocytic cell line.19 Importantly, we now find that MCSFR signaling activates ERK more potently than GCSFR signaling in normal immature myeloid cells. Similar activation in Ba/F3(MR,GR) cells may reflect excess expression of exogenous receptors. Of note, a recent study implicates ERK signaling also in a still earlier decision during hematopoiesis, commitment of the lymphoid-myeloid progenitor to GMP rather than to the common lymphoid progenitor.33
What then is the consequence of M-CSF–mediated ERK activation? Stabilization of c-Fos by ERK-mediated phosphorylation has been demonstrated previously in nonhematopoietic cells,25 with c-Fos half-life increasing from 30 minutes to 2.5 hours in pulse-chase experiments upon ERK activation and steady-state protein level increasing upon mutation of S362 and S374 to the phosphomimetic aspartic acid, and we now find that M-CSF specifically elevates c-Fos in immature myeloid cells, compared with G-CSF, dependent upon ERK activation. Fos induces monocyte-lineage specification of murine marrow myeloid progenitors via heterodimerization with C/EBPα, but not with c-Jun, suggesting a mechanism whereby MCSFR induction of c-Fos via ERK may stimulate monopoiesis.8 Lack of c-Jun induction or activation by M-CSF suggests further that C/EBPα:c-Fos may be more important that C/EBPα:c-Jun heterodimers for directing monopoiesis. PLCγ inhibition induced JNK and c-Jun phosphorylation in Ba/F3(MR,GR) cells, but was too toxic to evaluate in lin− marrow cells, raising the possibility that M-CSF activation of PLCγ inhibits c-Jun activation, further favoring c-Fos.
Besides c-Fos protein stabilization, preferential ERK activation by M-CSF most likely has additional consequences relevant to myeloid lineage specification. First, ERK phosphorylates Ets factors that bind the serum response factor homodimer to form the ternary transcription complexes that activate the FOS, EGR1, and EGR2 promoters as part of the classic immediate-early growth factor gene response pathway,35,36 and Egr-1 and Egr-2 induce and stabilize monocyte lineage commitment.9,37 Second, ERK phosphorylates CEBPα(S21), as demonstrated using MEK inhibition and by in vitro phosphorylation of GST-C/EBPα by ERK2,28 and as we now demonstrate occurs in normal immature myeloid cells in response to either M-CSF or G-CSF. C/EBPα(S21) phosphorylation prevents C/EBPα from inducing K562 cell granulocytic differentiation,28 suggesting that modification near its N-terminal trans activation domain weakens C/EBPα-mediated gene activation. Similarly, reduced C/EBPα(S21) modification in marrow progenitors in G-CSF, as a consequence of reduced ERK activation by this cytokine, may enable granulopoiesis to mediate by transcriptionally active C/EBPα:C/EBPα homodimers. In contrast, increased phospho-C/EBPα(S21) and c-Fos resulting from strong M-CSF signaling to ERK would favor formation phospho-C/EBPα(S21):c-Fos leucine zipper heterodimers to direct monopoiesis. C/EBPα(S21) phosphorylation varying with level of activated ERK had previously been observed upon exposure of cells expressing the Flt3ITD-activated Flt3 receptor tyrosine kinase to a Flt3 inhibitor.38
As SHP2 mediates ERK activation, we evaluated its phosphorylation on Y542 and Y580 in response to M-CSF or G-CSF. Whereas both cytokines induce phosphorylation of these tyrosines, G-CSF did so with much greater potency in Ba/F3 cells and in normal, immature myeloid cells. Phosphorylation of SHP2 Y542 or Y580 is not necessary for SHP2 activation, although several-fold increase activity has been observed if a stable phosphotyrosine analog is introduced at Y542 or Y580.39 SHP2 phosphorylation is required for increased ERK activation downstream of some growth factors, such as platelet-derived growth factor, but not others, such as epidermal growth factor.40 SHP2 inhibition reduced ERK activation by G-CSF, but not by M-CSF or TPA, indicating a SHP2-independent pathway to ERK, for example, via PLCγ2. We did not detect increased SHP2 activity in response to G-CSF using an artificial substrate; however, SHP2 tyrosine phosphorylation potentially affects its interaction with other proteins to direct target selection. What protein does phosphorylated SHP2 modify or contact to contribute to granulopoiesis? Interferon consensus sequence-binding protein (ICSBP)/interferon regulatory factor-8 is a SHP2 substrate relevant to myeloid lineage specification. Tyrosine phosphorylation of ICSBP allows it to interact with PU.1,41 and SHP2 acts on ICSBP to prevent it from cooperating with PU.1 and IRF2 to activate the NF-1 gene.42 As both ICSBP and higher levels of PU.1 favor monopoiesis,6,43-47 SHP2 inhibition of ICSBP:PU.1 complex formation potentially favors granulopoiesis. The correlation between G-CSF induction of SHP2 phosphorylation and the specific effect of SHP2 inhibition on ERK activation by G-CSF and on CFU-G is striking. Nevertheless, as NSC-87877 also inhibits SHP1, future studies involving specific, regulated SHP1 or SHP2 knockdown mimicking the effect of acute, drug-mediated inhibition will be required to further confirm the specific role of SHP2. Of note, activating SHP2 mutations are found in juvenile myelomonocytic leukemia, a disease associated with monocytosis. Constitutively activated SHP2 activates ERK more potently than does wild-type SHP2,48 mimicking the action of M-CSF we observe and most likely accounting for monocytic as opposed to granulocytic differentiation.
We also demonstrate specific STAT3 activation by G-CSF compared with M-CSF in normal progenitors, although the role of STAT3 in granulopoiesis remains uncertain. Mice lacking STAT3 develop neutrophils at normal and even increased numbers.13 In contrast, neutrophil production in response to G-CSF, modeling emergency granulopoiesis, is impaired in the absence of STAT3.14 In Ba/F3 cells, JNK inhibition reduced STAT1 and STAT5 activation by G-CSF, a previously unrecognized phenomenon. Neither STAT1 nor STAT5 induction was prominent in lineage-negative marrow cells, and STAT5 may play a role in mature granulocytic survival rather than early differentiation decisions.49
Overall, we conclude that M-CSF activates a PLCγ2 to PKC to ERK pathway, whereas G-CSF activates a SHP2 to activate ERK pathway, with M-CSF activating ERK more potently than G-CSF. ERK in turn stabilizes c-Fos, which has the capacity to heterodimerize with C/EBPα or phospho-C/EBPα(S21) to direct monopoiesis. Induction of c-Fos, Egr-1, and Egr-2 transcription and weakening of C/EBPα activity as a result of enhanced ERK signaling may also contribute to monocytic compared with granulocytic development. In addition, G-CSF signaling more potently induces SHP2 phosphorylation, which we find facilitates granulopoiesis independent of its ability to activate ERK. This study represents the first effort to directly compare GCSFR and MCSFR signaling, both in cell lines and in normal, immature myeloid cells, and has uncovered lineage-specific signaling and links between these pathways and transcription factors that mediate lineage specification. Future efforts will require study of GCSFR- versus MCSFR-expressing subsets. Nevertheless, our results support an instructive role for cytokine signaling in regulating myelopoiesis and provide a framework for future functional studies to assess the consequences of perturbing these pathways.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health (grant HL089176) and by the Children's Cancer Foundation (A.D.F.).
National Institutes of Health
Authorship
Contribution: G.D.J. and L.Z. performed experiments; G.D.J., L.Z., and A.D.F. designed the study, analyzed the data, and wrote the paper; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alan D. Friedman, Johns Hopkins University, CRB I Rm 253, 1650 Orleans St, Baltimore, MD 21231; e-mail: afriedm2@jhmi.edu.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal