Abstract
The ability of CD8+ T cells to engage a diverse range of peptide–major histocompatibility complex (MHC) complexes can also lead to cross-recognition of self and nonself peptide-MHC complexes and thus directly contribute toward allograft rejection or autoimmunity. Here we present a novel form of cross-recognition by herpes virus–specific CD8+ cytotoxic T cells that challenges the current paradigm of self/non-self recognition. Functional characterization of a human leukocyte antigen (HLA) Cw*0602-restricted cytomegalovirus-specific CD8+ T-cell response revealed an unusual dual specificity toward a pp65 epitope and the alloantigen HLA DR4. This cross-recognition of HLA DR4 alloantigen was critically dependent on the coexpression of HLA DM and was preferentially directed toward the B-cell lineage. Furthermore, allostimulation of peripheral blood lymphocytes with HLA DRB*0401-expressing cells rapidly expanded CD8+ T cells, which recognized the pp65 epitope in the context of HLA Cw*0602. T-cell repertoire analysis revealed 2 dominant populations expressing T-cell receptor beta variable (TRBV)4-3 or TRBV13, with cross-reactivity exclusively mediated by the TRBV13+ clonotypes. More importantly, cross-reactive TRBV13+ clonotypes displayed markedly lower T-cell receptor binding affinity and a distinct pattern of peptide recognition, presumably mimicking a structure presented on the HLA DR4 allotype. These results illustrate a novel mechanism whereby virus-specific CD8+ T cells can cross-recognize HLA class II molecules and may contribute toward allograft rejection and/or autoimmunity.
Introduction
T-cell receptors (TCRs) expressed on antigen-specific CD8+ and CD4+ T cells recognize unique sets of peptides bound to major histocompatibility complex (MHC) class I or class II molecules, respectively.1,2 These T cells undergo a stringent thymic selection process so that these effector cells recognize foreign peptide epitopes when associated with self-MHC molecules, whereas autoreactive T cells recognizing self-peptides associated with self-MHC are eliminated.3,4 As the human immune system encounters a wide range of pathogens, the MHC has evolved as the most polymorphic region of the human genome, which allows it to present a wide range of epitopes.5 The interaction of self-MHC with the TCR during thymic selection fundamentally shapes the specificity and size of the host TCR repertoire.6
Thus, during thymic selection, T cells undergo an instruction process that allows them to carefully navigate the distinction between self-tolerance and self-restriction, including tolerance to host HLA molecules.7,8 Extensive studies9,10 have firmly established that thymic selection involves another critical maturation process, whereby the CD8+ and CD4+ T cells are destined to recognize either human leukocyte antigen (HLA) class I or class II molecules. Indeed, the authors of an earlier study11 showed that precise specificity for HLA class I and class II molecules for CD8+ and CD4+ T cells, respectively, can be dictated by distinct docking modes of TCR and HLA molecules. Furthermore, the interaction of CD8 and CD4 coreceptor with respective MHC molecules is critical for the efficient selection of mature CD4+/CD8+ T cells, as evidenced by reduced numbers of mature T cells in animals expressing mutant MHC molecules that are unable to interact with these coreceptors.12
One of the conundrums of HLA restriction is that a large proportion of self-restricted T cells can recognize “non-self” HLA molecules (referred to as allorecognition).13-15 Analyses of multiple cloned T-cell populations have demonstrated that up to 10% to 20% of antigen-specific self-HLA restricted T-cell clones cross-react with alloantigens, and there is significant overlap in the repertoire of alloreactive and self-HLA restricted T cells.13,16 There is now sufficient evidence to support the argument that this allorecognition is a result of cross-recognition by self-HLA restricted T cells rather than a distinct population of T cells that have escaped thymic negative selection.14 More importantly, a large proportion of these self-HLA–restricted T cells are directed toward infectious agents (eg, herpesviruses).17,18 A study19 of the immune response to Epstein-Barr virus (EBV) has shown examples of cross-reactivity with alloantigens by T cells expressing highly focused public TCR expanded in unrelated persons. Interestingly, those expressing both the self-restricting and cross-reactive HLA alleles maintain a strong antiviral immune response by recruiting noncross-reactive TCRs to control the virus.18 A structural analysis of a murine TCR in complex with a self-MHC peptide (pMHC) complex or a cross-recognized allo–pMHC complex has revealed that, despite considerable similarities in the self and nonself HLA surfaces; the cross-reactive T cells adopt 2 different docking strategies in recognizing self and nonself HLA molecules.20,21
Here we describe a novel form of cross-reactivity in which HLA Cw*0602-restricted CD8+ T cells specific for a human cytomegalovirus (HCMV)–encoded epitope cross-recognized the HLA DR4 allotype. This cross-recognition of the HLA DR4 alloantigen is dependent on the coexpression of the HLA DM molecule and is constrained toward the B-cell lineage. Interestingly, stimulation of peripheral blood mononuclear cells (PBMCs) with B cells expressing HLA DR4 alloantigen preferentially expanded CD8+ T cells, which recognized the HCMV-encoded epitope in association with the HLA Cw*0602 molecule. These studies illustrate for the first time that virus-specific CD8+ T cells can recognize HLA class II alloantigens and may therefore have the potential to contribute toward graft-versus-host disease and autoimmunity.
Methods
Establishment and maintenance of cell lines
All cell lines were routinely maintained in RPMI 1640 supplemented with 2 mmol/L l-glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin plus 10% fetal calf serum (referred to as growth medium) unless otherwise stated. EBV-transformed lymphoblastoid cell lines (LCLs) were established from HCMV-seropositive donors by exogenous virus transformation of peripheral B cells by use of the B95.8, BL74, and QIMR-WIL virus isolates as described previously. Autologous LCLs, pulsed with HCMV peptides, were used to stimulate T-cell lines and clones or as stimulator cells in intracellular cytokine assays (see below). In addition, the peptide transporter-negative B × T-hybrid cell line. 174 × CEM.T2 (referred to as T2; HLA A2, B51)22 stably expressing HLA DRB*0401 with or without HLA DM (referred to as T2.DR4 and T2.DR4/DM) also were used in this study (kindly provided by Prof Peter Cresswell, Yale University). This study was approved by the QIMR human research ethics committee, and donor informed consent was obtained in accordance with the Declaration of Helsinki.
B-cell blasts were prepared by culturing PBMCs with γ-irradiated (8000 rad) CD40L transfectants in the presence of 200 U/mL rIL-4 and 500 μg/mL cyclosporin A. Cultures were grown for 14 days with media changes every 3 days and were frozen until used in stimulation assays.
Monocytes were enriched by plastic adherence at 37°C for 2 hours. Nonadherent cells were removed by 2 washes with warm growth medium. The remaining adherent cells were then cultured for 7 days in either growth medium alone or growth medium supplemented with granulocyte monocyte colony stimulating factor (GM-CSF; 800 U/mL) and interferon-γ (IFN-γ) (100 U/mL), with fresh growth medium given every 3 days.
Peptide synthesis
Synthetic HCMV peptide epitopes and alanine analogs were synthesized by use of the Merrifield solid-phase method and purchased from Mimotopes.
Generation of polyclonal and clonal HCMV-specific CTLs
To generate polyclonal cytotoxic T lymphocytes (CTLs), 106 autologous PBMCs were incubated with 0.1 μg/mL peptide for 1 hour at 37°C.23 Peptide-sensitized PBMCs were washed free of unbound peptide and cocultured with 4 × 106 PBMCs from HCMV-seropositive healthy donors for 3 days. On day 3, culture media was supplemented with 10 U/mL rIL-2 and 30% MLA-144 supernatant. After 7 days of culture in supplemented growth medium, the cells were used as polyclonal effectors in a standard intracellular cytokine secretion (ICS) assay. In some experiments PBMCs from HCMV-seropositive persons were cocultured with either γ-irradiated (8000 rad) allogeneic DR*0401 LCL (responder to stimulator ratio of 20:1) or γ-irradiated (8000 rad) T2.DR4 or T2.DR4/DM cells (responder to stimulator ratio of 20:1) for 7 days in growth medium. On day 7, these cells were tested for HCMV-specific reactivity by use of an ICS assay.
To generate HCMV-specific CTL clones, PBMCs were initially stimulated as described previously for polyclonal CTLs. After 3 days of culture in growth medium, CTL clones were generated by seeding in 0.35% agarose containing rIL2 (50 U/mL).24 On day 7 of culture at 37°C, CTL clones were harvested from the agarose into 96-well round-bottom microtiter plates and maintained in growth medium containing rIL2 (50 U/mL) and 30% MLA 144 supernatant. Clones were restimulated twice weekly with peptide-sensitized γ-irradiated (8000 rad) autologous LCLs (1000 cells/well) supplemented once weekly with γ-irradiated (2000 rad) allogeneic PBMCs (5000 cells/well, non-DR*0401+). The CTL clones and polyclonal lines described previously were screened for specific activity by the use of an ICS assay on a panel of autologous or allogeneic target cells that were sensitized with synthetic peptides. To confirm HLA restriction for the T-cell recognition, target cells were pretreated with anti-HLA class I (W6/32) or anti-HLA class II (L243) monoclonal antibodies and then exposed to HCMV-specific T cells in an ICS assay.
pMHC tetramer and TRBV staining
PBMCs or HCMV-specific T cells from healthy virus carriers were incubated for 20 minutes at 4°C with an antigen-presenting cell (APC)–labeled HLA-Cw*0602-TRAT tetramer (kindly provided by the National Institutes of Health tetramer facility). Cells were then incubated with CD8 peridinin chlorophyll protein (PerCP), CD3 phycoerythrin (PE)–Cy7, and CD4 fluorescein isothiocyanate (FITC) washed 3 times in FACS buffer (phosphate-buffered saline [PBS] supplemented with 2% fetal calf serum and 0.1% sodium azide) before fixing in FACSfix (PBS with 1% paraformaldehyde and 10 g/L glucose). In some experiments these T cells were also costained with anti-Vβ3 (T-cell receptor V-β [TRBV]28), 13.6 (TRBV6), 16 (TRBV14), 21.3 (TRBV11)–FITC, 1 (TRBV9), 2 (TRBV20), 4 (TRBV29), 5.1 (TRBV5), 5.2 (TRBV5), 5.3 (TRBV5), 7 (TRBV4), 7.2 (TRBV4-3), 8 (TRBV12), 9 (TRBV3), 11 (TRBV25), 12 (TRBV10), 13.1 (TRBV6), 14 (TRBV27), 17 (TRBV19), and 18 (TRBV18), 20 (TRBV30), 22 (TRBV2), and 23 (TRBV13)–PE (Immunotech) and Vβ6.7 (TRBV7; Pierce Endogen). In brief, these cells were initially stained with anti-Vβ antibodies and then tetramer and cell surface antibodies. Cells were acquired as soon as possible by the use of a FACSCanto with FACSDiva software (BD Biosciences), and postacquisition analysis was performed by the use of FlowJo software (TreeStar).
Tetramer dissociation assay
The tetramer dissociation assay was based upon previously published methods. Cells were incubated with PE-conjugated anti-Vβ antibodies at room temperature for 30 minutes. Cells were then incubated with the APC-labeled HLA-Cw*0602-TRAT tetramer at 4°C for 30 minutes. PBMCs also were incubated with PerCP-conjugated anti-CD4, anti-CD14, and anti-CD19 antibodies (BD Biosciences). Cells were washed, resuspended in FACS buffer, incubated at room temperature, and an aliquot of 20 μL was transferred into 150 μL of 2% paraformaldehyde in PBS at the indicated time points. Cells were then analyzed on a FACSCanto, and the mean fluorescent intensity of tetramer+ cells (for T cells clones) or Tetramer+ CD4/CD14/CD19− cells (for PBMCs) was determined. Fluorescence intensity was then normalized as a percentage of the initial time point.
Intracellular cytokine staining
PBMCs from seropositive persons, HCMV-specific T-cell clones, or polyclonal lines were incubated overnight with uncoated or peptide sensitized (0.1 μg/mL) LCLs, B-cell blasts, or monocytes in growth medium supplemented with 1 μg/mL Brefeldin A (BD PharMingen) for analysis of IFN-γ expression. For avidity assays, TRAT-specific T-cell clones were incubated with 5-fold serial dilutions of TRAT peptide for 4 hours at 37°C. These cells were then washed and incubated with PerCP-conjugated anti-CD8, FITC-conjugated anti-CD4, and APC-conjugated anti-CD3 at 4°C for 20 minutes. Cells were washed, then fixed and permeabilized with cytofix/cytoperm (BD PharMingen) at 4°C for 20 minutes. Cells were washed in perm/wash (BD PharMingen), incubated with PE-conjugated anti–IFN-γ (BD PharMingen) at 4°C for 30 minutes, washed again with perm/wash, resuspended in PBS, and acquired on a FACS Canto. In some experiments Vβ antibody staining was performed, followed by staining for cell-surface markers (CD8 PerCP, CD3 PE-Cy7, CD4 APC-Cy7) and IFN-FITC for ICS.
Molecular analysis of TRBV and TRAV usage
Total RNA was extracted from 5 TRAT-specific clones by the use of TRIzol reagent (Invitrogen). Reverse transcription polymerase chain reaction (RT-PCR) was performed by the use of SuperScript III Reverse Transcriptase (Invitrogen) with CβA primer (5′-GTTGCTCCAGGCCACAGCACTG-3′) and CβA primer (5′-TATCTGGAGTCATTGAGGGCGGGCT-3) according to the manufacturer's guidelines. All PCR cycle conditions were 95°C for 12 minutes, denaturation 95°C for 20 seconds, annealing 55°C for 40 seconds, and extension 72°C for 40 seconds for 40 cycles in a PE Applied Biosystems DNA thermocycler. In brief, 0.4 μL cDNA was added to the 25 μL reaction mixture composed of 4 μL 5xdNTP, 2.5 μL 10× buffer, 1.5 μL MgCl2, 0.25 μL AmpliTaq Gold DNA Polymerase (Applied Biosystems), 13.75 μL water, 0.5 μL Cαprimer (5′-GGTGAATAGGCAGACAGACTTGTCACTGGA-3′) or Cβprimer (5′-TTCTGATGGCTCAAACAC-3′), and 5 μL of a T-cell receptor alpha variable (TRAV) family-specific primer (Vα1-34) or TRBV family-specific primer (Vβ1-24).25 The PCR products were separated and visualized on ethidium bromide-stained 2% agarose gels. PCR products were purified and cloned into the pGEM-T vector system (Promega) and sequenced by use of the ABI PRISM Big Dye termination reaction kit (Applied Biosystems). Sequences were analyzed on the Applied Biosystems 3700 Sequencer (Applied Biosystems) and by use of the IMGTV Quest website (http://imgt.cines.fr/IMGT_vquest/vquest?livret=0&Option=human TcR).
Results
HCMV-specific CD8+ T cells show dual specificity for a viral epitope presented on HLA Cw*0602 and the alloantigen HLA DR4
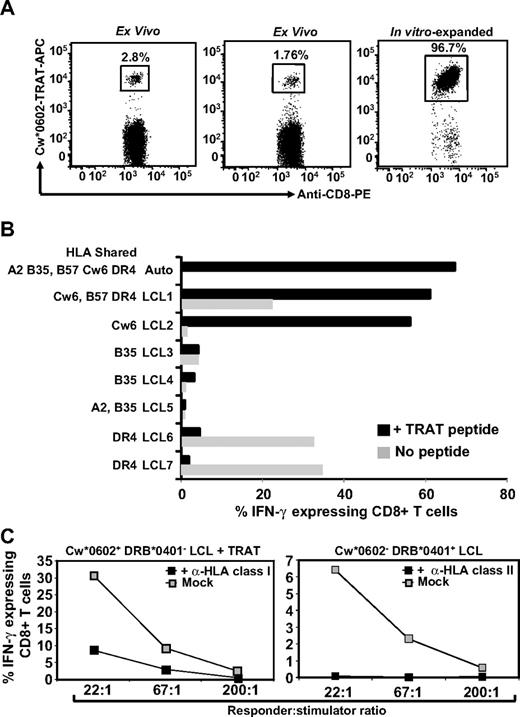
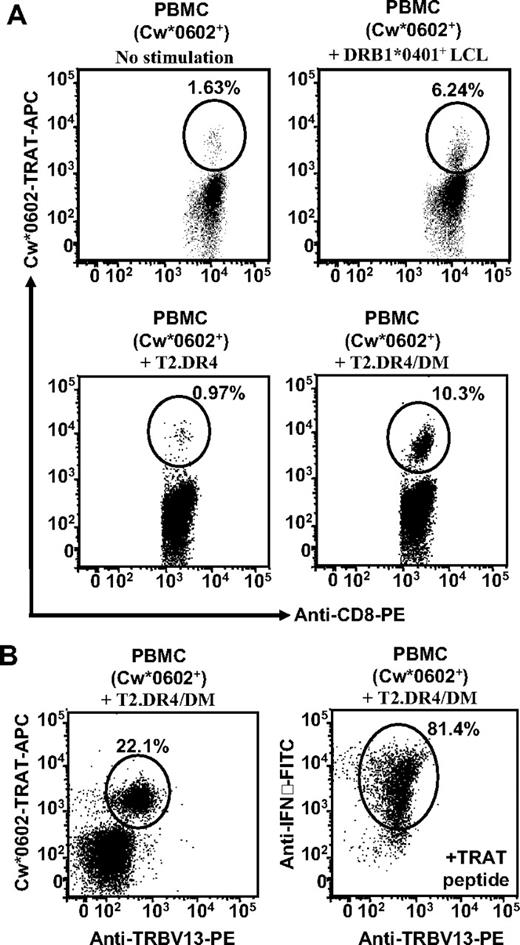
While characterizing HCMV-specific T-cell responses in healthy virus carriers, we mapped a peptide epitope from pp65 (TRATKMQVI; referred to as TRAT). Ex vivo analysis of PBMCs from healthy virus carriers with the use of pMHC tetramers revealed very high precursor frequencies for this epitope (1.2%-3% of CD8+ T cells; Figure 1A). These precursor frequencies were comparable with those observed for HLA A2, B7, and A24-restricted epitopes from HCMV (data not shown). In vitro stimulation of these T cells with synthetic TRAT peptide epitope rapidly expanded these cells, and within 10 days more than 95% of CD8+ T cells from one of the donors were specific for this epitope (Figure 1A). In-depth analysis of these in vitro-expanded polyclonal TRAT-specific CD8+ T cells revealed an unusual pattern of reactivity. Although these T cells were activated by peptide-sensitized autologous and HLA-matched Cw6+ LCLs, unexpectedly a strong reactivity against LCLs expressing the HLA DR4 allotype, without the TRAT peptide was also observed (Figure 1B). This appeared to be MHC class II-specific alloreactivity by CD8+ T cells because although these T cells shared DR4 with these stimulator cells, further analysis revealed that they expressed different subtypes of HLA-DR4. Importantly, HLA-mismatched (HLA Cw*0602 without peptide or HLA DR4-negative) LCLs were not recognized by these T cells (data not shown). Figure 1B illustrates the T-cell reactivity of one such line from donor SB (HLA A2, B35, B57, Cw*0602, DRB*0402). To confirm the class of HLA molecules involved in this T-cell recognition, we incubated the APCs with anti-HLA class I or class II monoclonal antibodies and then exposed them to TRAT-specific T cells. Data presented in Figure 1C show that the T-cell recognition of LCLs coated with TRAT peptide epitope was blocked with anti–class I molecule, whereas the HLA DR4 recognition by same T cells was completely blocked by anti-HLA class II antibodies. It is important to mention here that this inhibition was highly specific because anti–class I and –class II antibodies did not block the recognition of HLA DR4+ LCLs or TRAT peptide-coated HLA Cw0602+ LCLs, respectively (data not shown).
Characterization of HLA Cw*0602-restricted TRATKMQVI-specific T cells. (A) Staining of fresh PBMCs and in vitro expanded T cells with APC-labeled Cw*0602-TRAT tetramer. Representative data from 2 persons (for fresh PBMCs) and one polyclonal T-cell line are shown. These cells were costained with anti-CD3 and anti-CD8 antibodies. (B) HLA restriction analysis of in vitro–expanded TRAT-specific T-cell lines. Autologous LCL and allogeneic LCLs sharing 1 or more HLA class I or class II alleles were used as APCs. These cells were either coated with synthetic TRAT peptide or left uncoated and then exposed to T cells. These T cells were then analyzed for IFN-γ expression by the use of ICS assays. Data presented show the percentage of IFN-γ expressing T cells after exposure to these LCLs. (C) Effect of anti-HLA class I or class II antibody on the T-cell recognition. Peptide sensitized Cw*0602+ and DRB*0401− or unsensitized Cw*0602− and DRB*0401+ LCLs were exposed to TRAT-specific T cells in the presence or absence of anti-HLA class I or anti-HLA class II antibodies. After incubation, these T cells were analyzed for IFN-γ expression using ICS assays.
Characterization of HLA Cw*0602-restricted TRATKMQVI-specific T cells. (A) Staining of fresh PBMCs and in vitro expanded T cells with APC-labeled Cw*0602-TRAT tetramer. Representative data from 2 persons (for fresh PBMCs) and one polyclonal T-cell line are shown. These cells were costained with anti-CD3 and anti-CD8 antibodies. (B) HLA restriction analysis of in vitro–expanded TRAT-specific T-cell lines. Autologous LCL and allogeneic LCLs sharing 1 or more HLA class I or class II alleles were used as APCs. These cells were either coated with synthetic TRAT peptide or left uncoated and then exposed to T cells. These T cells were then analyzed for IFN-γ expression by the use of ICS assays. Data presented show the percentage of IFN-γ expressing T cells after exposure to these LCLs. (C) Effect of anti-HLA class I or class II antibody on the T-cell recognition. Peptide sensitized Cw*0602+ and DRB*0401− or unsensitized Cw*0602− and DRB*0401+ LCLs were exposed to TRAT-specific T cells in the presence or absence of anti-HLA class I or anti-HLA class II antibodies. After incubation, these T cells were analyzed for IFN-γ expression using ICS assays.
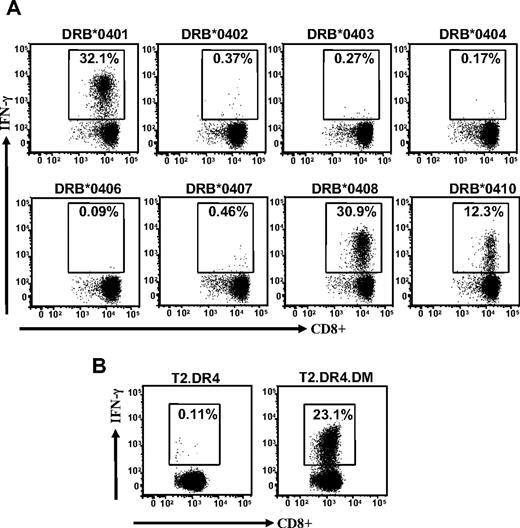
Interestingly, this allorecognition was not constrained to one particular subtype of the HLA-DR4 antigen because LCLs expressing HLA DRB*0401, DRB*0408, and DRB*0410 were also recognized, whereas LCLs expressing DRB*0402, DRB*0403, DRB*0404, DRB*0406, and DRB*0407 were not recognized (Figure 2A). To further confirm this allorecognition, we used T2 cells expressing DR4, with or without HLA DM, as APCs in an ICS assay. Data presented in Figure 2B show that T2 cells with HLA DR4 and HLA DM were efficiently recognized by TRAT-specific CD8+ T cells, whereas T2 cells with HLA DR4 alone were not recognized by these effector cells. Taken together, these experiments clearly demonstrate a cross-recognition of HLA DR4 allotype by virus-specific CD8+ T cells that recognize cognate peptide epitope in association with HLA Cw*0602. More importantly, the allorecognition of the HLA DR4 allotype is critically dependent on the presence of class II peptide editor, HLA DM.
Cross-recognition of HLA DR4 alloantigen by TRAT-specific T cells. (A) EBV-transformed LCLs expressing different HLA DR4 subtypes were used as APCs. After incubation these T cells were then analyzed for IFN-γ expression by the use of ICS assays. (B) TRAT-specific T cells were exposed to T2 cells expressing HLA DRB*0401 with or without peptide editor HLA DM and then assessed for IFN-γ expression by use of the ICS assay.
Cross-recognition of HLA DR4 alloantigen by TRAT-specific T cells. (A) EBV-transformed LCLs expressing different HLA DR4 subtypes were used as APCs. After incubation these T cells were then analyzed for IFN-γ expression by the use of ICS assays. (B) TRAT-specific T cells were exposed to T2 cells expressing HLA DRB*0401 with or without peptide editor HLA DM and then assessed for IFN-γ expression by use of the ICS assay.
Cross-recognition by HCMV-specific CD8+ T cells is preferentially focused toward the B-cell linage
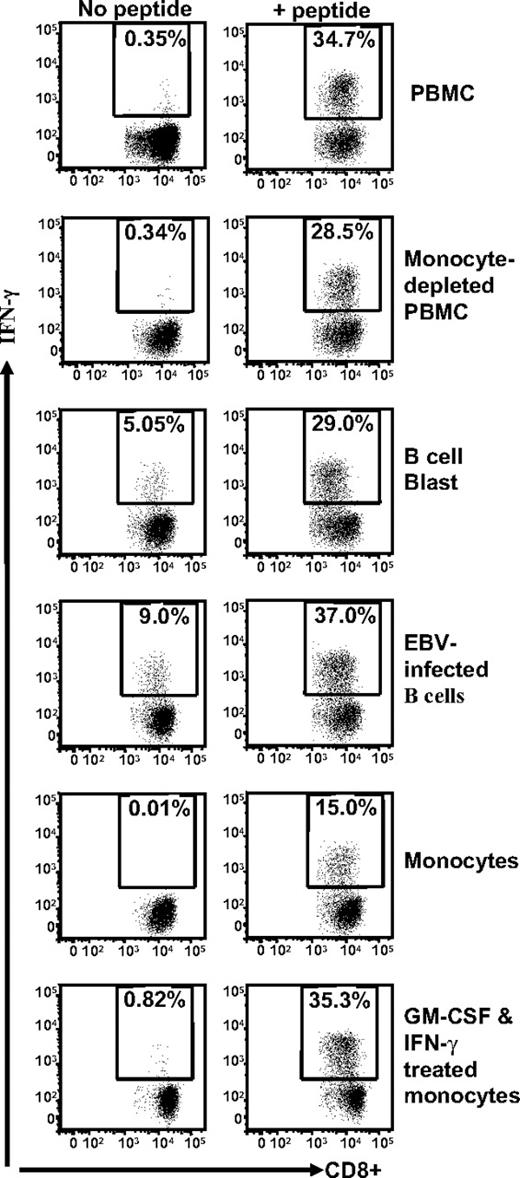
Because the data presented in Figures 1 and 2 are based upon stimulation of TRAT-specific T cells by EBV-transformed LCLs, it was important to determine whether the anti-DR4 allorecognition was dependent on the presence of EBV in the APCs. To address this issue, we tested several cell types, including EBV-negative B-cell blasts, fresh monocytes, and GM-CSF and IFN-γ–treated monocytes as APCs. These cells were taken from a person who was HLA Cw*0602+ and HLA DRB*0401+. Data presented in Figure 3 demonstrate that although all different cell types were able to present the TRAT peptide epitope to HCMV-specific T cells, the allorecogntion by these effector cells was only observed with EBV-negative B-cell blasts and EBV-transformed LCLs. In addition, we also tested HLA DRB*0401+ phytohemagglutinin–stimulated T-cell blasts and melanoma cells, and these cells were not cross-recognized by TRAT-specific T cells (data not shown). These results suggest that a peptide derived from a protein preferentially expressed in B cells is recognized in association with HLA DR4 by TRAT-specific CD8+ T cells.
Stimulation of TRAT-specific T cells with human cells of different lymphoid lineages. HLA Cw*0602+ and DRB*0401+ fresh PBMCs, monocyte-depleted PBMCs, B-cell blast, EBV-infected B-cell line, monocytes, and GM-CSF and IFN-γ–treated monocytes were presensitized with TRAT peptide or left uncoated and then exposed to TRAT-specific T cells. After incubation, these T cells were analyzed for IFN-γ expression by the use of ICS assays.
Stimulation of TRAT-specific T cells with human cells of different lymphoid lineages. HLA Cw*0602+ and DRB*0401+ fresh PBMCs, monocyte-depleted PBMCs, B-cell blast, EBV-infected B-cell line, monocytes, and GM-CSF and IFN-γ–treated monocytes were presensitized with TRAT peptide or left uncoated and then exposed to TRAT-specific T cells. After incubation, these T cells were analyzed for IFN-γ expression by the use of ICS assays.
HLA DR4 allotype recognition by TRAT-specific CD8+ T cells is mediated through TRBV13
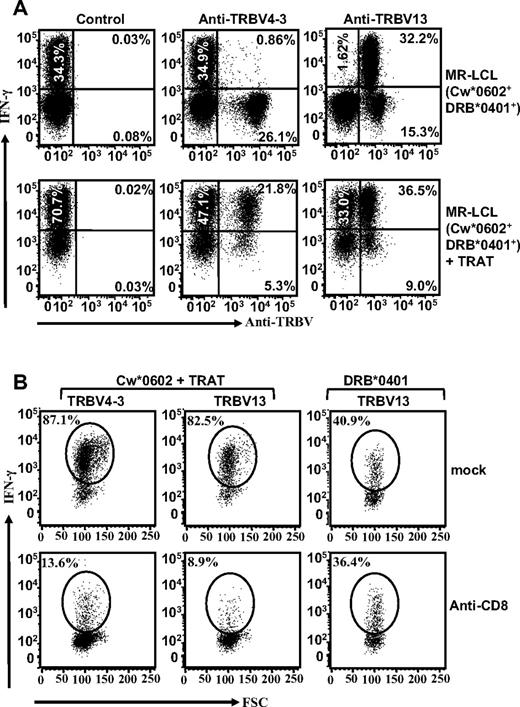
In the next set of experiments, we analyzed fresh PBMCs from HLA Cw*0602+ healthy virus carriers by the use of pMHC- tetramers in combination with a panel of monoclonal TRBV antibodies. Ex vivo analysis of TRAT-specific T cells from multiple donors revealed a highly focused TCR repertoire, with 2 dominant populations of T cells expressing TRBV4-3 or TRBV13 (data not shown). To determine which population was cross-reacting with the HLA DR4 allotype, we expanded these cells in vitro by the use of TRAT peptide. After expansion, these T cells were exposed to HLA Cw*0602+ and HLA DRB*0401+ EBV-transformed LCLs either uncoated or precoated with TRAT peptide epitope and then costained with anti-TRBV and anti-IFN-γ antibodies. Data presented in Figure 4A show that both TRBV4-3+ and TRBV13+ T cells expressed IFN-γ after stimulation with peptide-sensitized LCLs, whereas TRBV13+ T cells also recognized uncoated LCLs.
HLA DR4 allotype recognition by TRAT-specific CD8+ T cells is mediated through TRBV13 and is not dependent on CD8 interaction with TcR. (A) TRAT-specific T cells were exposed to peptide-coated and -uncoated HLA Cw*0602+ and DRB*0401+ LCLs and assessed for IFN-γ expression by the use of ICS assays. These T cells were also stained with a panel of anti-TRBV antibodies. Data from 2 of the anti-TRBV antibodies that showed positive staining are shown in this figure. T cells positive for TRBV13 showed IFN-γ expression after exposure to peptide-coated and -uncoated HLA Cw*0602+ and DRB*0401+ LCLs (right panels) and TRBV4-3+ T cells expressed IFN-γ after exposure to peptide-coated HLA Cw*0602+ and DRB*0401+ LCLs only. (B) TRAT-specific T cells were exposed to peptide-coated and -uncoated HLA Cw*0602+ and DRB*0401+ LCLs in the presence or absence of anti-CD8 antibody and then assessed for IFN-γ expression by the use of ICS assays. These T cells were costained with TRBV-specific antibodies to identify TRBV4-3- and TRBV13-positive populations. Data presented in the left and panel show IFN-γ expression by TRAT-specific T cells after incubation with peptide-coated LCLs. The right panels show IFN-γ expression by TRBV13-positive TRAT-specific T cells in the presence or absence of anti-CD8 antibody.
HLA DR4 allotype recognition by TRAT-specific CD8+ T cells is mediated through TRBV13 and is not dependent on CD8 interaction with TcR. (A) TRAT-specific T cells were exposed to peptide-coated and -uncoated HLA Cw*0602+ and DRB*0401+ LCLs and assessed for IFN-γ expression by the use of ICS assays. These T cells were also stained with a panel of anti-TRBV antibodies. Data from 2 of the anti-TRBV antibodies that showed positive staining are shown in this figure. T cells positive for TRBV13 showed IFN-γ expression after exposure to peptide-coated and -uncoated HLA Cw*0602+ and DRB*0401+ LCLs (right panels) and TRBV4-3+ T cells expressed IFN-γ after exposure to peptide-coated HLA Cw*0602+ and DRB*0401+ LCLs only. (B) TRAT-specific T cells were exposed to peptide-coated and -uncoated HLA Cw*0602+ and DRB*0401+ LCLs in the presence or absence of anti-CD8 antibody and then assessed for IFN-γ expression by the use of ICS assays. These T cells were costained with TRBV-specific antibodies to identify TRBV4-3- and TRBV13-positive populations. Data presented in the left and panel show IFN-γ expression by TRAT-specific T cells after incubation with peptide-coated LCLs. The right panels show IFN-γ expression by TRBV13-positive TRAT-specific T cells in the presence or absence of anti-CD8 antibody.
To test whether the T-cell recognition of HLA Cw*0602+ and HLA DRB*0401+ EBV transformed LCLs either uncoated or precoated with TRAT peptide epitope was dependent on CD8 interaction with TcR, these cells were preincubated with anti-CD8 antibody and then costained with anti-TRBV and anti–IFN-γ antibodies. Data presented in Figure 4B show that although T-cell recognition of HLA Cw*0602+ LCLs coated with TRAT peptide epitope significantly blocked with anti-CD8 antibody, the T-cell recognition of DRB*0401 remained unaffected by this antibody.
Stimulation of PBMCs with LCLs expressing HLA DR4 allotype rapidly expands TRAT-specific TRBV13+ CD8+ T cells
To determine whether the TRAT-specific memory cells could be activated by the use of allostimulation, HLA Cw*0602+ PBMCs were stimulated in vitro with γ-irradiated HLA DR4+ LCLs, T2.DR4/DM, or T2.DR4 cells. The resulting polyclonal T-cell populations were tested for TRAT-specific expansions by the use of pMHC tetramers. Data presented in Figure 5A show stimulation of PBMCs with HLA DR4 LCLs, and T2.DR4/DM cells showed 4- to 6-fold expansion of TRAT-specific T cells, whereas unstimulated or T2.DR4 stimulated cells showed no expansion of TRAT-specific T cells. More importantly, further characterization of the TRAT-specific T cells, expanded after stimulation with T2.DR4/DM, revealed that more than 98% of these cells expressed TRBV13 and that the majority of these cells expressed IFN-γ after exposure to TRAT peptide-coated APCs (Figure 5B). It is important to stress here that HLA DR4 cross-reactive, HCMV-specific T cells are not found in all HLA Cw*0602+ persons. We have expanded TRAT-specific CD8+ T cells from 8 different donors, and of these only 3 (37.5%) showed HLA DR4 cross-reactivity.
Stimulation of PBMCs with LCLs expressing HLA DR4 allotype rapidly expands TRAT-specific TRBV13+ CD8+ T cells. (A) PBMCs from HLA Cw*0602+ persons were stimulated with either HLA DRB1*0401+ LCL, T2.DR4, or T2.DR4/DM cells and then cultured for 7 days. After incubation these cells were assessed for expansion of TRAT-specific T cells (by the use of HLA-peptide tetramers) and IFN-γ expression by the use of ICS assays. PBMCs stimulated with DRB1*0401+ LCL or T2.DR4/DM cells showed strong expansion of TRAT-specific T cells. (B) T cells expanded after stimulation with T2.DR4/DM were costained with HLA-peptide tetramers and anti-TRBV13 antibodies. Representative data from 1 of the 3 unrelated donors are presented.
Stimulation of PBMCs with LCLs expressing HLA DR4 allotype rapidly expands TRAT-specific TRBV13+ CD8+ T cells. (A) PBMCs from HLA Cw*0602+ persons were stimulated with either HLA DRB1*0401+ LCL, T2.DR4, or T2.DR4/DM cells and then cultured for 7 days. After incubation these cells were assessed for expansion of TRAT-specific T cells (by the use of HLA-peptide tetramers) and IFN-γ expression by the use of ICS assays. PBMCs stimulated with DRB1*0401+ LCL or T2.DR4/DM cells showed strong expansion of TRAT-specific T cells. (B) T cells expanded after stimulation with T2.DR4/DM were costained with HLA-peptide tetramers and anti-TRBV13 antibodies. Representative data from 1 of the 3 unrelated donors are presented.
Clonotypic analysis of the TCR repertoire for TRAT-specific CD8+ T cells
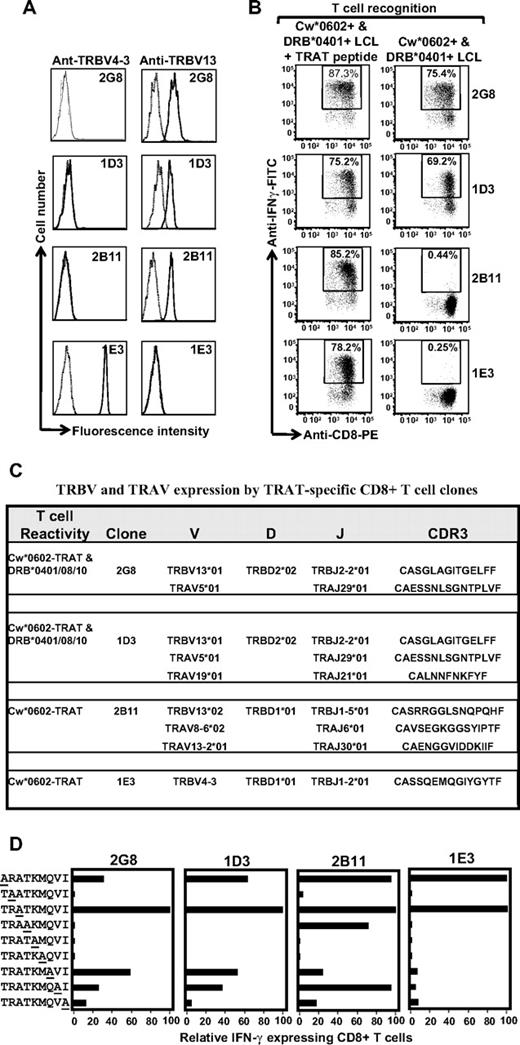
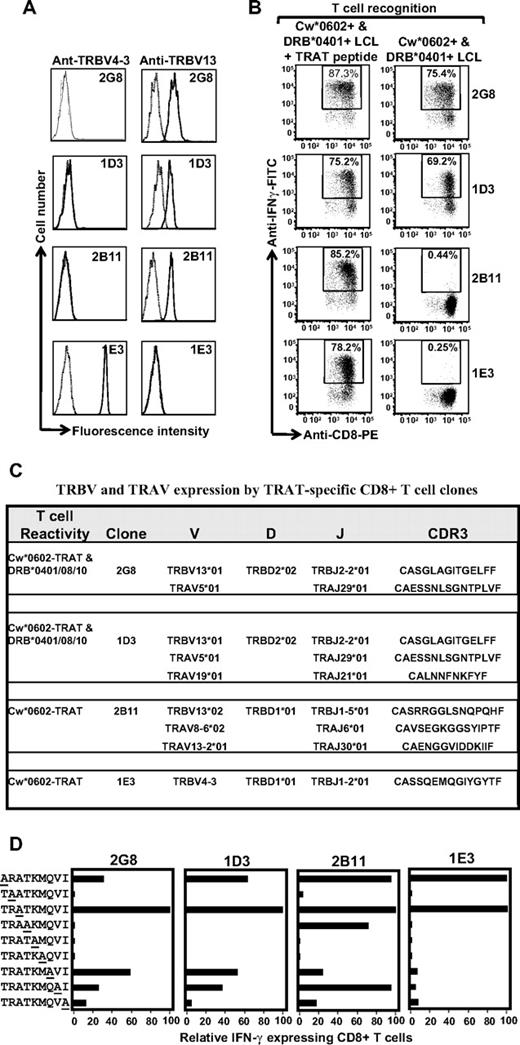
To complement the TCR repertoire analysis based on antibody staining, we next sequenced the CDR3 regions by the use of multiple T-cell clones. A large number of CD8+ T-cell clones specific for the TRAT epitope were isolated and extensively characterized. Representative data from 4 such clones are presented in Figure 6. Analysis of TRBV expression by the use of specific antibodies showed that T-cell clones 2G8, 1D3, and 2B11 expressed TRBV13, whereas IE3 was positive for TRBV4-3. Functional analysis of these clones revealed that although all 4 clones expressed IFN-γ after stimulation with peptide-coated Cw*0602+ LCLs, an interesting pattern of responses was observed when these effector cells were exposed to uncoated HLA DR4+ LCLs. As expected, the TRBV4-3+ 1E3 clone did not respond to the HLA DR4 allotype; however, only 2 of the TRBV13+ T-cell clones (2G8 and 1D3) expressed IFN-γ after exposure to HLA DR4+ LCLs, whereas the 2B11 clone did not show cross-reactivity against these LCLs (Figure 6B). These observations suggested variation in the CDR3 sequence of these clones. Indeed data presented in Figure 6C show that the 2G8 and 1D3 clones used identical TRBV genes and CDR3 sequences, whereas the 2B11 clone expressed an allelic variant of TRB13 and quite a distinct CDR3 sequence compared with other clones. In addition, these clones also expressed different TRAV chains (Figure 6C).
Clonotypic analysis of the TCR repertoire for TRAT-specific CD8+ T cells. (A) HLA Cw*0602-restricted TRAT-specific CD8+ T-cell clones were expanded by use of the agarose cloning method and then assessed for TRBV expression by the use of specific monoclonal antibodies. Representative data from 4 of the 25 clones are presented. (B) These T-cell clones were then assessed for reactivity against peptide-coated and -uncoated HLA Cw*0602+ and DRB*0401+ LCLs by the use of ICS assays. Clones 2G8 and ID3 showed reactivity against peptide-coated and -uncoated LCLs, whereas clones 2B11 and IE3 showed reactivity against peptide-coated LCLs only. (C) Amino acid sequences of CDR3 regions of TRAV and TRBV chains expressed by TRAT-specific clones. (D) T-cell recognition of alanine analogs of the TRAT peptide epitope. HLA-Cw*0602+ LCLs were presensitized with the individual peptides (0.1 μg/mL) and then exposed to TRAT-specific CTL clones. An effector:target ratio of 5:1 was used in the assay. The T-cell reactivity was assessed by the use of standard ICS assays. Data presented in each of the subpanels show relative IFN-γ expression in the presence of alanine analogs compared with the wild-type peptide.
Clonotypic analysis of the TCR repertoire for TRAT-specific CD8+ T cells. (A) HLA Cw*0602-restricted TRAT-specific CD8+ T-cell clones were expanded by use of the agarose cloning method and then assessed for TRBV expression by the use of specific monoclonal antibodies. Representative data from 4 of the 25 clones are presented. (B) These T-cell clones were then assessed for reactivity against peptide-coated and -uncoated HLA Cw*0602+ and DRB*0401+ LCLs by the use of ICS assays. Clones 2G8 and ID3 showed reactivity against peptide-coated and -uncoated LCLs, whereas clones 2B11 and IE3 showed reactivity against peptide-coated LCLs only. (C) Amino acid sequences of CDR3 regions of TRAV and TRBV chains expressed by TRAT-specific clones. (D) T-cell recognition of alanine analogs of the TRAT peptide epitope. HLA-Cw*0602+ LCLs were presensitized with the individual peptides (0.1 μg/mL) and then exposed to TRAT-specific CTL clones. An effector:target ratio of 5:1 was used in the assay. The T-cell reactivity was assessed by the use of standard ICS assays. Data presented in each of the subpanels show relative IFN-γ expression in the presence of alanine analogs compared with the wild-type peptide.
Alanine replacement peptides for the TRATKMQVI epitope were used to further understand the possible mechanism of cross recognition and delineate the interaction of the TCRs with the HLA-peptide complex. Data presented in Figure 6D indicate that TRAT-specific CTL clones 2G8 and ID3 interact specifically with amino acids across the length of the peptide, whereas the 2B11 clone could tolerate more peptide amino acid changes. The 1E3 clone was highly peptide specific, tolerating amino acid substitution at only 2 peptide positions, a feature that could perhaps explain its lack of cross-reactivity with HLA-DR4. Taken together these studies delineate a novel form of cross-reactivity and demonstrate how a distinct pattern in TRBV usage and CDR3 sequences can dictate allorecognition.
TRBV4-3+ and TRBV13+ TRAT-specific T cells display differential pMHC avidity
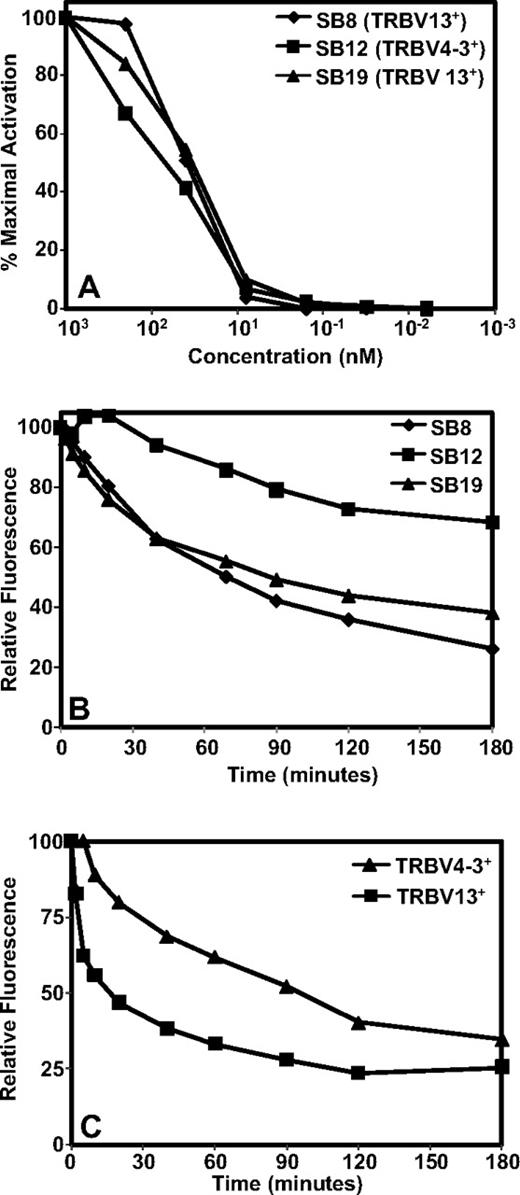
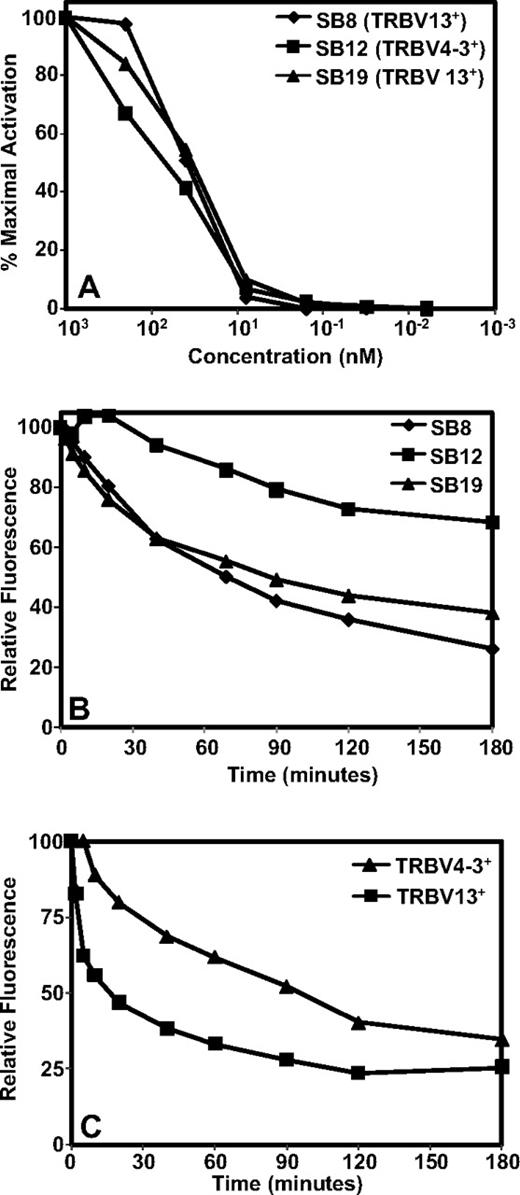
In the next set of experiments we conducted functional characterization of TRBV4-3+ and TRBV13+ T-cell clones by using different avidity indicators to detect activation threshold or TcR binding to pMHC complex. First, we used T-cell responsiveness to synthetic TRAT peptide epitope as a measure of T-cell avidity because it provides the optimal epitope threshold to activate CD8+ T-cell effector function. Three different clones; SB8 (TRBV13+ and HLA DR4 cross-reactive), SB12 (TRBV4-3+, non-HLA DR4 cross-reactive), and SB19 (TRBV13+ and non-HLA DR4 cross-reactive) were compared for their ability to produce IFN-γ under conditions of decreasing peptide stimulation (Figure 7A). All 3 different T-cell clones showed minimal difference in their response to the decreasing concentration of TRAT peptide.
TRBV4-3+ and TRBV13+ TRAT-specific T-cell clones display differential pMHC avidity. (A) TRAT-specific T-cell clones were exposed to varying concentrations of synthetic TRAT peptide and then assessed for IFN-γ expression by the use of ICS assays. (B-C) Tetramer dissociation kinetics of TRAT-specific T-cell clones and TRAT-specific T cells in fresh PBMCs. T cells or PBMCs were initially incubated with PE-labeled anti-TRBV antibodies and then incubated with the APC-labeled HLA-Cw*0602-TRAT tetramer at 4°C for 30 minutes. These cells were washed, resuspended in FACS, and then incubated at room temperature for the indicated time points. T cells were then analyzed on a FACSCanto. Data in panel B are based on T-cell clones, whereas panel C shows data from fresh PBMCs.
TRBV4-3+ and TRBV13+ TRAT-specific T-cell clones display differential pMHC avidity. (A) TRAT-specific T-cell clones were exposed to varying concentrations of synthetic TRAT peptide and then assessed for IFN-γ expression by the use of ICS assays. (B-C) Tetramer dissociation kinetics of TRAT-specific T-cell clones and TRAT-specific T cells in fresh PBMCs. T cells or PBMCs were initially incubated with PE-labeled anti-TRBV antibodies and then incubated with the APC-labeled HLA-Cw*0602-TRAT tetramer at 4°C for 30 minutes. These cells were washed, resuspended in FACS, and then incubated at room temperature for the indicated time points. T cells were then analyzed on a FACSCanto. Data in panel B are based on T-cell clones, whereas panel C shows data from fresh PBMCs.
Recent studies have shown that the dissociation rate of pMHC complex from the TcR can be used as most accurate measures of T-cell function/avidity because it determines the duration of pMHC binding and its potential impact on T-cell signaling. Data presented in Figure 7B show that TRBV4-3+ TRAT-specific T-cell clones showed a much slower tetramer dissociation rate compared with the TRBV13+ TRAT-specific clones. These observations were also observed when tetramer dissociation rates were compared on T cells from fresh PBMCs (Figure 7C).
Discussion
It is now firmly established that cross-recognition of non-self HLA molecules by CD8+ T cells with primary specificity for a self-MHC bound foreign epitope contribute significantly to alloreactivity.14,21 This report extends evidence in support of this view with a unique form of cross-recognition by virus-specific CD8+ T cells, which may have important implications for our current understanding of self/nonself recognition. While characterizing the HCMV-specific CD8+ T cells recognizing an HLA Cw*0602-restricted epitope from pp65 protein, we noticed an unusual cross-recognition of target cells expressing HLA DR4 alloantigen. More importantly, T cells that recognize an HCMV epitope could be expanded in vitro from HCMV-seropositive persons by the use of allostimulation with HLA DR4-expressing APCs. One of the intriguing features of this allorecognition was that recognition of HLA DR4 alloantigen was critically dependent on the expression of the MHC class II peptide editor HLA DM and constrained to the B-cell lineage. These observations suggested that the mature class II molecules bound to endogenously processed B cell–specific peptides were recognized by these virus-specific CD8+ T cells.
Of the major subtypes of HLA DR4 tested, DRB*0401 and DRB*0408 were strongly recognized, whereas a lower level of recognition of DRB*0410 was also observed. In contrast, APCs expressing DRB*0402, DRB*0403, DRB*0404, DRB*0406, and DRB*0407 were not recognized. It is interesting to note that DRB*0401 and DRB*0408 show high levels of homology within the peptide binding pocket with a single highly conserved change from lysine to arganine at position 71.26-28 However, DRB*0402, DRB*0403, DRB*0404, DRB*0406, and DRB*0407 and DRB*0410 alleles show a series of changes at residues 37 (Y→S), 57 (D→S), 67 (L→I), 70 (Q→D), 71 (K→E), 74 (A→E), and 86 (G→V).29,30 Of particular interest are the residues 57 (aspartic acid) and 86 (glycine), which are conserved in DRB*0401 and DRB*0408 alleles, and any change in these residues partially or completely abrogated the cross-recognition by HCMV-specific CD8+ T cells. Although the side chains of amino acids at positions 57 and 86 point into the peptide-binding groove, we cannot rule out the possibility that they may be partially accessible to direct contact with the TCR. Indeed, it would be interesting to resolve the structure of the HLA DR4-peptide complex bound to TRAT-specific TRBV13 TcR to delineate the precise interaction of the relevant residues that play a crucial role in the cross-recognition.
Although it was originally thought that an alloresponse would constitute a highly diverse repertoire that would display reactivity toward multiple antigenic peptides presented on HLA molecules,14 subsequent studies17,31,32 have shown that this assumption may not be correct for all alloresponses. Indeed, studies from our group17,33 have shown that, in some persons, a highly restricted T-cell repertoire can be activated against a single alloantigen. The strength of this alloresponse is not determined by multiple allogeneic determinants but by a highly dominant T-cell response to a viral epitope, which unexpectedly cross-reacts with the alloantigen.17,33
The data presented in this study further extend this observation and demonstrate that this allorecognition by virus-specific CD8+ T cells is not only restricted to HLA class I molecules but also HLA class II molecules. Extensive analysis of the T-cell repertoire revealed 2 dominant clonotypes expressing TRBV4-3 or TRBV13, with cross-reactivity exclusively mediated by the TRBV13+ clonotypes. Interestingly, TRBV13+ T cells included 2 distinct subpopulations with unique CDR3 sequence and TRAV usage. Fine specificity analysis using alanine mutants of the TRAT epitope revealed that non–cross-reactive CD8+ T-cell clones showed a distinctive pattern of recognition compared with HLA DR4–cross-reactive TRBV13+ clones, presumably evading an area of the HCMV epitope that mimics a structure presented on the HLA DR4 allotype. Of particular interest was the recognition pattern of TRBV13+ HLA DR4 cross-reactive and non–cross-reactive clones. Although IFN-γ expression for both clonotypes was severely compromised when amino acids at P2 (peptide anchor residue), P4, P5, P6, and P9 (peptide anchor residue) was replaced with alanine residues; a clear difference was observed for residue P7, where replacement of these residues with alanine had a more pronounced effect on non–cross-reactive clones.
Further comparison of TRBV4-3 or TRBV13 clonotypes also revealed the importance of P4 residue. Thus, a binding footprint of TRBV13 focused on P4 and P7 residues may play a crucial role in the HLA DR4 cross-reactivity. Interestingly, functional characterization of T-cell clones using different avidity indicators showed that TRBV13+ clonotypes displayed much lower avidity compared with the TRBV4-3+ T cells. These observations suggested that low-affinity interaction of TRBV13+ TcR with the pMHC complex may enhance the potential cross-reactivity. Indeed, this contention is also supported by a recent study,34 which have indicated that these low-affinity interactions may be more common contributor in the initiation of autoimmune diseases where infectious pathogens have been implicated.
Taken together, these observations have major implications for both transplantation and autoimmune diseases. The present study provides a new paradigm where we have now shown that self HLA class I–restricted T cells can recognize nonself HLA class II molecules. Of particular interest is the association of HLA DR4 with numerous autoimmune diseases.35-37 It is tempting to speculate that HLA Cw*0602-restricted HCMV-specific T cells may override the self-tolerance and thus contribute toward the initiation/progression of autoimmune diseases. HLA Cw*0602 is one of the commonest class I allele in different ethnic groups, with a frequency ranging from 14% to 30%. This finding is especially relevant to the pathogenesis of psoriatic arthritis, which is strongly associated with HLA Cw*0602.38-40 Another potential implication of this observation relates to the possibility of using these cross-reactive HCMV-specific T cells as an immunotherapeutic tools for the treatment of B-cell lymphomas. It would be important to follow these observations in different clinical settings to explore the potential role of HLA class I–restricted T cells in transplant organ rejection or self-reactivity through MHC class II recognition.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
R.K. and S.R.B. are supported by Research Fellowships from National Health and Medical Research Council (NH&MRC). C.S. is supported by a Fellowship from the Leukemia Foundation. This study was supported by funding from NH&MRC.
Authorship
Contribution: M.R., C.S., and R.K. designed this study; M.R., C.S., M.J.B., and S.R.B. conducted various experimental studies; and R.K. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Assoc Prof Rajiv Khanna, Tumour Immunology Laboratory, Division of Infectious Diseases, Queensland Institute Medical Research, 300 Herston Rd, Herston (Qld) 4006 Australia; e-mail: rajiv.khanna@qimr.edu.au.