Abstract
To evaluate internal tandem duplication (ITD) insertion sites and length as well as their clinical impact in younger adult patients with FLT3-ITD–positive acute myeloid leukemia (AML), sequencing after DNA-based amplification was performed in diagnostic samples from 241 FLT3-ITD–mutated patients. All patients were treated on 3 German-Austrian AML Study Group protocols. Thirty-four of the 241 patients had more than 1 ITD, leading to a total of 282 ITDs; the median ITD length was 48 nucleotides (range, 15-180 nucleotides). ITD integration sites were categorized according to functional regions of the FLT3 receptor: juxtamembrane domain (JMD), n = 148; JMD hinge region, n = 48; beta1-sheet of the tyrosine kinase domain-1 (TKD1), n = 73; remaining TKD1 region, n = 13. ITD length was strongly correlated with functional regions (P < .001). In multivariable analyses, ITD integration site in the beta1-sheet was identified as an unfavorable prognostic factor for achievement of a complete remission (odds ratio, 0.22; P = .01), relapse-free survival (hazard ratio, 1.86; P < .001), and overall survival (hazard ratio, 1.59; P = .008). ITD insertion site in the beta1-sheet appears to be an important unfavorable prognostic factor in young adult patients with FLT3-ITD–positive AML. The clinical trials described herein have been registered as follows: AML HD93 (already published in 2003), AML HD98A (NCT00146120; http://www.ClinicalTrials.gov), and AMLSG 07-04 (NCT00151242; http://www.ClinicalTrials.gov).
Introduction
Activating FLT3 mutations occur in approximately 40% of cytogenetically normal acute myeloid leukemia (AML) patients, thus being one of the most frequently affected genes in AML. Three clusters of activating mutations have been identified: FLT3 internal tandem duplications (FLT3-ITDs) in approximately 30%,1-3 point mutations within the activation loop of the second tyrosine kinase domain (FLT3-TKD) in 7% to 10%,4,5 and, in a very low frequency (∼ 2%), point mutations in the juxtamembrane domain (JMD) as well as in the extracellular domain.6,7 The most frequent activating mutations are FLT3-ITDs in exons 14 and 15 ranging in size from 3 to more than hundreds of nucleotides.1-3 Up to 5 different clones of various sizes, insertion sites, and mutant to wild-type ratios can be observed in a proportion of both pediatric and adult AML patients.8-11 The FLT3 receptor consists of an extracellular domain composed of 5 immunoglobulin-like domains, a transmembrane region, a cytoplasmic JMD, and 2 cytoplasmic TKDs (TKD1 and TKD2) interrupted by a short kinase insert.12 The JMD can be furthermore subdivided into 3 distinct parts: the JM-binding (JM-B) motif, which is implicated in activation/inactivation and in stabilizing the inactive kinase conformation; the switch motif (JM-S), which consists of 2 phosphorylation sites and contains the STAT5-binding motif; and the linker/zipper peptide segment (JM-Z), which can undergo large amplitude rotations by pivoting about its attachment point.12 FLT3-ITDs located in the JMD are supposed to disrupt the autoinhibitory conformation of the FLT3 receptor thus promoting constitutive activation of downstream effectors including the RAS/RAF/MEK/ERK kinases, STAT5, and PI3-kinase.13,14 FLT3-ITDs per se have been associated with a worse prognosis. Furthermore, previous studies suggested a prognostic role for the mutant to wild-type allelic ratio and for the size of the ITD.8,9,15-17 Whereas data on mutant to wild-type allelic ratios consistently show an association of high allelic burden with worse prognosis, the prognostic impact of the ITD size has been discussed controversially. Both longer and shorter FLT3-ITDs have been associated with inferior outcome, or have been shown to have no prognostic impact.15-17
Recently, Breitenbuecher et al demonstrated by systematic sequence analysis of FLT3-ITD insertion sites in 753 unselected FLT3-ITD–positive AML patients, who were enrolled in trials of the German AML Cooperative Group, that 28.7% of ITDs integrate in the TKD1 and not, as presumed, in the JMD of the receptor.18 In their study, a FLT3-ITD mutation with insertion site in the beta2-sheet of the TKD1 (FLT3-ITD627E) was shown to mediate constitutive phosphorylation of FLT3 and of STAT5, suggesting that non-JMD ITDs confer constitutive activation of the receptor. In addition, FLT3-ITD627E induced transformation of hematopoietic 32D cells and led to a lethal myeloproliferative disease in a syngeneic mouse model.18 The intramolecular mechanisms of non-JMD ITDs leading to constitutive activation of the FLT3 receptor are still unknown, as are their functional and clinical implications. One might speculate that non-JMD ITDs may induce changes in the structural conformation of the TKD1 or may generate a neo-binding motif, thereby resulting in differential signaling of the FLT3 receptor. Preliminary evidence for this hypothesis comes from a recent study highlighting rewired signaling and differential responsiveness to kinase inhibitors of a particular non-JMD ITD (FLT3-ITD627E).19 Therefore, it might be important to specifically characterize the functional regions affected by the insertion site. In the present study, we aimed to validate the integration of non-JMD ITDs in an independent cohort of younger adult patients with FLT3-ITD–positive AML. In particular, we determined ITD insertion sites according to functional regions, length of ITDs, as well as number of affected clones per patient and correlated the molecular findings with patient characteristics and clinical outcome.
Methods
Patients and treatment
Diagnostic bone marrow or peripheral blood samples were analyzed from 241 younger adult patients (18-60 years) with FLT3-ITD–positive AML (excluding cases with acute promyelocytic leukemia). Patients were treated on 3 consecutive multicenter AMLSG treatment trials: AML HD93 (n = 29),20 AML HD98A (n = 101; NCT00146120),21 and AMLSG 07-04 (n = 111; NCT00151242). The diagnosis of AML was based on French-American-British Cooperative Group criteria,22 and after 2004 on WHO criteria.23 For the present study, the only criterion used to include patients was the presence of an FLT3-ITD. Within the first 2 treatment trials (AML HD93 and AML HD98A), we focused on patients with normal karyotype, whereas in the AMLSG 07-04 trial we also included patients with abnormal cytogenetics. In total, 241 patients were enrolled using either bone marrow (n = 156) or peripheral blood (n = 85) samples from diagnosis for gene sequencing analysis. The 3 clinical studies (AML HD93, AML HD98A, and AMLSG 07-04) were approved by the ethics board of the University Hospital of Heidelberg (AML HD93) and the University Hospital of Ulm (AML HD98A and AMLSG 07-04) and all patients gave informed consent for both treatment and cryopreservation of leukemia samples according to the Declaration of Helsinki.
For all patients, intensive double-induction and consolidation therapy were intended. Patients with a matched related donor (MRD) were allocated to allogeneic stem cell transplantation (SCT); patients who failed to achieve a complete remission (CR), and all patients (with FLT3-ITD–positive AML) who entered the trial after April 2006 were scheduled to receive allogeneic stem cell (SC) transplant from either MRD or matched unrelated donor (MUD).
Detection of FLT3-ITD
Mononucleated cells were isolated by standard Ficoll-Hypaque density gradient centrifugation. Genomic DNA was extracted from 107 cells according to the manufacturer's protocols (QIAGEN AllPrep DNA/RNA Mini Kit [50]; QIAGEN). For FLT3-ITD mutational screening, polymerase chain reaction (PCR) amplification of genomic DNA was carried out as has been described previously.3 PCR products were resolved on a 4% agarose gel stained with ethidium bromide and visualized under an ultraviolet light.
Genescan analysis
For Genescan analysis the ABI3130XL automated sequencer was used. PCR setup and PCR conditions were identical to standard PCR3 with 2 modifications: PCR primer 12R3 was labeled with 6-FAM (Eurofins MWG Operon) and the reaction volume contained 5 ng DNA. For analysis with POP7, a mixture of 1 μL PCR template, 0.3 μL Genescan ROX500, and 20 μL formamide was denatured at 95°C for 2 minutes. The areas under the curves were quantified for FLT3-ITD and the wild-type allele, respectively, by use of the Genemapper Version 4.0 software (Applied Biosystems). The ratio of FLT3-ITD to wild type was expressed as a percentage of the area under the curve for FLT3-ITD divided by the area under the curve for wild-type FLT3.
DHPLC analysis and sequencing reaction
First, PCR amplification was performed on genomic DNA.3 The total reaction volume of 50 μL contained 100 ng DNA, 10 pmol of each primer, deoxynucleoside triphosphates (25 mM each), 3 μL MgSO4, 0.5 U Optimase polymerase, and 5 μL supplied buffer (Transgenomic). The PCR consisted of an initial incubation step at 95°C for 10 minutes followed by 35 cycles at 95°C for 60 seconds, 56°C for 60 seconds, 72°C for 120 seconds, and a final elongation step at 72°C for 10 minutes. PCR products were analyzed on standard 3% agarose gels. DNA amplification was followed by a sensitive denaturating high performance liquid chromatography (DHPLC)–based assay (Wave 3500HT System; MD Transgenomic Inc) under nondenaturating conditions using 10 μL unpurified PCR product. Single mutated fragments were collected in a second run using a fragment collector. In those samples with more than 1 FLT3-ITD, each fragment was collected. Products were reamplified, purified, and directly sequenced using the ABI Ready Reaction Dye Terminator Cycle Sequencing Kit (Applied Biosystems). Sequences were compared with the wild-type sequence. The sensitivity for the detection of an FLT3-ITD with the Wave System, tested with dilution series of the 2 cell lines MV4-11 (homozygous mutation) and HL60 (wild type), was 2.5%.
Categorization of FLT3-ITD insertion sites according to functional regions
Figure 1 shows the schematic structure of the FLT3 receptor according to Griffith et al.12 Based on the functional regions,12 we categorized the FLT3-ITD insertion sites into different groups: insertion site in the JMD, in the hinge region of the JMD, in the beta1-sheet of the TKD1, in the nucleotide binding loop, in the beta2-sheet of the TKD1, and 3′ of the beta2-sheet.
Schematic structure of the FLT3 receptor consisting of 5 immunoglobulin-like folds that make up the ligand-binding extracellular domain (ED), a single transmembrane domain (TD), and a cytoplasmic domain containing the juxtamembrane domain (JMD) as well as the kinase domain divided into 2 parts by a short kinase insert (KI). Of 282 ITDs analyzed in our series, 69.5% (n = 196) of the ITDs had the insertion site in the JMD, whereas 30.5% (n = 86) of the ITDs were localized in the first tyrosine kinase domain (TKD1) of FLT3. ITD indicates internal tandem duplication; TKD1, tyrosine kinase domain-1; and TKD2, tyrosine kinase domain-2.
Schematic structure of the FLT3 receptor consisting of 5 immunoglobulin-like folds that make up the ligand-binding extracellular domain (ED), a single transmembrane domain (TD), and a cytoplasmic domain containing the juxtamembrane domain (JMD) as well as the kinase domain divided into 2 parts by a short kinase insert (KI). Of 282 ITDs analyzed in our series, 69.5% (n = 196) of the ITDs had the insertion site in the JMD, whereas 30.5% (n = 86) of the ITDs were localized in the first tyrosine kinase domain (TKD1) of FLT3. ITD indicates internal tandem duplication; TKD1, tyrosine kinase domain-1; and TKD2, tyrosine kinase domain-2.
Molecular analysis of other gene mutations
AML samples were also analyzed for mutations in the FLT3-TKD (at codons D835 and I836, n = 236), NPM1 (n = 239), CEBPA (n = 203), MLL (partial tandem duplication [PTD]; n = 231), and NRAS (n = 207) genes.24 Furthermore, to improve the accuracy of cytogenetic diagnosis, all cases were analyzed by fluorescence in situ hybridization25 or by polymerase chain reaction (PCR) for the presence of the recurring gene fusions RUNX1-RUNX1T1, CBFB-MYH11, MLL-MLLT3, and PML-RARA.24
Statistical analyses
Pairwise comparisons between patient characteristics (covariates) were performed by Mann-Whitney test for continuous variables and by Fisher exact test for categoric variables. Clinical end points of the study were CR after induction therapy, relapse-free survival (RFS), and overall survival (OS). To evaluate RFS and OS, we used relapse or death during CR, and death, respectively. All end points were based on recommended criteria.26 The median follow up for survival was calculated according to the method of Korn.27 A logistic regression model was used to analyze associations between baseline as well as molecular characteristics and the achievement of CR. Cox regression models with stratification to account for allogeneic SCT in first CR were used to identify prognostic variables for RFS and OS. We estimated missing data for covariates using 50 multiple imputations in chained equations incorporating predictive mean matching.28 All statistical analyses were performed with the use of the Design R package (Version 2.0-12, R Development Core Team) of the R statistical software platform (Version 2.4.1, R Development Core Team).29 P values less than .05 were considered to indicate statistical significance.
Results
Patient characteristics at diagnosis
A total of 1667 patients were screened for FLT3-ITDs within 3 consecutive AMLSG treatment trials. In 385 (23.1%) patients, one or more FLT3-ITDs were identified by conventional qualitative PCR or Genescan analysis; 248 of these patients exhibited a normal karyotype.
In 241 of the 385 patients, sequence analysis of the FLT3-ITD was performed. The only selection criterion was the availability of material. Because in the AML HD93 and the AML HD98A trials only normal karyotype AML patients were screened for the presence of FLT3 mutations, the proportion of patients with normal cytogenetics was very high in our study cohort (86.3%); with regard to normal karyotype AML cases selection was very low, because 208 of the 248 patients could be analyzed. Presenting clinical and cytogenetic data of the patients are given in Table 1.
Molecular characterization of FLT3-ITDs
Two-hundred seven (85.9%) of 241 FLT3-ITD–positive AML patients had one ITD, and 34 (14.1%) revealed more than one ITD (2 ITDs, n = 29 [12.0%]; 3 ITDs, n = 3 [1.2%]; 4 ITDs, n = 2 [0.8%]). In total, 282 ITDs were identified and analyzed. All FLT3-ITD mutations involved exon 14 of the gene and all of them were in-frame fragments with a direct head-to-tail orientation. Thirty-six ITDs also contained intron 14. The median size of the 282 ITDs was 48 nucleotides (range, 15-180 nucleotides). In 208 (73.8%) of the 282 ITDs, integration resulted in a change of the expected amino acid sequence at this site. In 95 ITDs, insertion of additional material between 1 to 18 base pairs preceded the duplicated fragment, whereas in 187 ITDs the duplications were identical to the former sequences of the FLT3 gene. In 80 of the 95 ITDs, the additional nucleotides matched FLT3 sequence, whereas in the 15 remaining ITDs the preceding extra nucleotides were not related to the FLT3 sequence. The size of these extra nucleotides ranged between 5 and 17 base pairs. Of note, the insertion of additional material was always present when the duplicated fragment was not in frame (n = 71), and resulted in an in-frame mutation. In the remaining cases with preceding additional material (n = 24), the duplicated fragment, per se, was in frame and the size of additional material was dividable by 3. Table 2 shows ITD integration sites and ITD sizes of the 282 FLT3-ITD mutations according to the functional regions of the receptor. Molecular characterization of the ITDs revealed that the insertion site was localized in the JMD between amino acids 572 and 609 in the majority of the cases (69.5%), whereas in the remaining 30.5% of the cases ITDs were inserted in 3′ direction of the JMD, predominantly in the beta1-sheet of TKD1. Of note, FLT3-ITD size was strongly correlated with the insertion site, in that the more C-terminal the location of the insertion site, the longer the size of the inserted fragment (P < .001; Table 2).
Analysis of the duplication of specific amino acid motives
Since a recent study highlighted the particular role of the specific tyrosine-rich amino acid stretch Y591 to Y599 (YVDFREYEY) for intracellular signaling,30 we specifically determined how often this specific amino acid motif was involved in an ITD. Duplication of at least one residue in this specific stretch was seen in 271 (96.1%) of the 282 ITDs. The most frequent affected amino acids were at position Y597 with 78.4% (n = 221) followed by R595 with 75.5% (n = 213). The amino acids upstream and downstream of the motif were less frequently involved with Y591 and Y599 in 55.0% (n = 155) and 64.9% (n = 183), respectively. Eleven ITDs did not involve any amino acid of the Y591-Y599 motif, 4 of them showed an insertion in the JMD, and 7 in the TKD1.
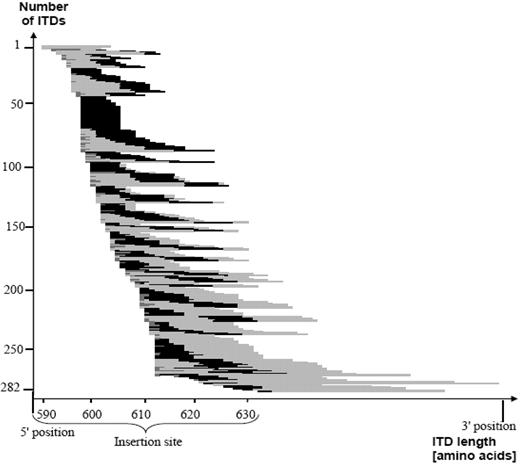
When patients were taken as denominator, in 234 (97.1%) of 241 patients at least one ITD showed the involvement of at least one amino acid of the specific motif. In Figure 2, length of the ITD, insertion site, and relative position of the Y591-Y599 motif within the ITDs are illustrated.
This figure shows the correlation between ITD insertion site, length, as well as the relative position of the Y591-Y599 motif of the 282 ITDs, sorted by insertion site. The light gray color symbolizes the duplicated fragment and the black color demonstrates the involvement of the amino acid motif Y591-Y599. The presence of additional material is depicted in darker gray. The majority of the 282 ITDs included at least a part of the specific amino acid stretch Y591-599. ITD indicates internal tandem duplication.
This figure shows the correlation between ITD insertion site, length, as well as the relative position of the Y591-Y599 motif of the 282 ITDs, sorted by insertion site. The light gray color symbolizes the duplicated fragment and the black color demonstrates the involvement of the amino acid motif Y591-Y599. The presence of additional material is depicted in darker gray. The majority of the 282 ITDs included at least a part of the specific amino acid stretch Y591-599. ITD indicates internal tandem duplication.
Because Y589 and Y591 have been identified as 2 possible candidate STAT5 SH2 docking phosphorylation sites in the JMD, the disruption of the juxtamembrane structure by ITDs might expose these residues and enable activation of STAT5.31 Consistently, the mutant FLT3-ITD-Y589/591F did not activate STAT5, which supports the role of these residues in STAT5 activation.31 Therefore, we determined the combined duplication of the tyrosine residues Y589 and Y591, which could be found in 119 ITDs (42.2%).
Characterization of patients with more than one FLT3-ITD
Thirty-four (14.1%) of the 241 patients had more than one ITD. Table 3 shows the FLT3-ITD insertion sites of these 34 patients. In 14 of these patients, all ITDs occurred exclusively in the JMD, including the hinge region of the JMD. In 12 patients, the ITD insertions were located in the JMD and beta1-sheet of the TKD1, whereas only one single patient had 2 ITDs exclusively in the beta1-sheet of the TKD1; in all other patients at least 1 ITD inserted in the JMD.
Mutant to wild-type FLT3-ITD ratio
In 217 patients genomic DNA for a quantitative Genescan analysis was available. The FLT3-ITD mutant to wild-type ratio ranged from 0.008 to 14.2, with a median of 0.57. There was no difference in mutant to wild-type ratio according to the functionally categorized FLT3-ITD groups (Table 2).
Correlation of FLT3-ITD insertion site with cooperating mutations and presenting clinical characteristics
NPM1 gene mutations, MLL-PTD, CEBPA mutations, FLT3-TKD mutations, and NRAS mutations were detected in 137 (57.3%) of 239, 21 (9.1%) of 231, 12 (5.9%) of 203, 10 (4.2%) of 236, and 9 (4.3%) of 207, of the patients, respectively. The frequencies of these concurrent mutations were equally distributed among the functionally categorized FLT3-ITD groups (Table 4); patients with more than 1 ITD were excluded from the analysis. Therefore, the numbers in the text and the numbers in Table 4 are differing.
The only clinical variable that differed among the 4 functional groups of FLT3-ITD was the WBC count, with lowest counts found in patients with ITDs in the hinge region (AA 604-609; median WBC, 20 × 109/L), compared with those with ITDs inserting C-terminal of amino acid 615 (median WBC, 64 × 109/L; P = .03).
Clinical outcome
Induction therapy.
Clinical data on induction therapy were available in 237 of the 241 patients. One-hundred sixty-six (70.0%) of them achieved a CR, 18 (7.6%) patients died, and 53 (22.4%) patients had refractory AML. To evaluate the impact of the ITD insertion site on response to induction therapy, 2 logistic regression models, one including patients exhibiting a normal karyotype and one comprising all patients, were used including the following variables: ITD insertion site grouped according to the 4 functional regions, ITD mutant to wild-type allelic ratio dichotomized at the median level, number of ITD mutations per patient, insertion of additional nucleotides preceding some of the ITDs, logarithm of WBC counts, age, and the molecular markers NPM1 mutation, MLL-PTD, and CEBPA mutation. Due to the very high correlation of ITD length with ITD insertion site, ITD length was not included in the model to avoid statistical interference.
In both models, FLT3-ITD insertion site in the beta1-sheet of the TKD1 (odds ratio [OR], 0.22; 95% confidence interval [CI], 0.06 to 0.80; and OR, 0.22; 95% CI, 0.07-0.69), insertion site C-terminal of amino acid 615 (OR, 0.15; 95% CI, 0.03-0.66; and OR, 0.13; 95% CI, 0.03-0.54), and logarithm of WBC counts (OR, 0.31; 95% CI, 0.15-0.66; and OR, 0.38; 95% CI, 0.20-0.73) were significantly associated with an inferior CR rate for patients exhibiting a normal karyotype as well as for all patients. The presence of a NPM1 mutation (odds ratio, 2.15; 95% CI, 1.09-4.24) was a favorable factor only in the model for all patients. In multivariable analyses, all other variables had no significant impact. However, in univariable analyses higher mutant to wild-type ITD allelic ratio and larger ITD length, both dichotomized at the median level, were associated with reduced CR rates after induction therapy for patients exhibiting a normal karyotype (P = .03 and P = .02, respectively) as well as for all patients (P = .002 and P < .001, respectively).
Survival analysis.
The median follow-up for survival was 35.4 months. Of the 237 patients, 149 (62.9%) died; the median RFS and OS were 9 months and 14 months, respectively, and the 3-year rates of RFS and OS were 27.3% (95% CI, 20.7%-34.3%) and 34.3% (95% CI, 27.6%-41.0%), respectively.
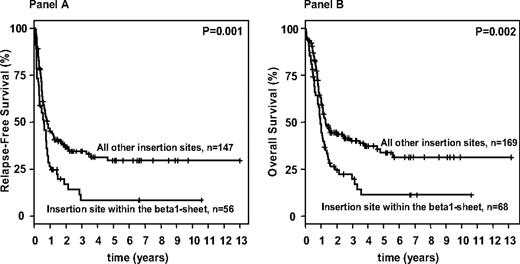
Allogeneic SCT was performed from MRD in 38 patients, from MUD in 60 patients, and from haploidentical donors in 3 patients. In addition, 34 patients received an allogeneic SC transplant after relapse (MRD, n = 7; MUD, n = 26; haploidentical donors, n = 1). To address the high proportion of patients receiving an allogeneic SC transplant, multivariable models were stratified for this variable. Survival analyses according to the 4 categorized functional FLT3-ITD groups were again performed for both groups: patients exhibiting a normal karyotype as well as for all patients. For both groups, multivariable analyses on RFS and OS revealed consistently similar results, with FLT3-ITD insertion sites in the beta1-sheet of the TKD1 being an unfavorable variable (Table 5). In OS models, additional significant variables were log (WBC), age, as well as a mutation in the CEBPA gene. Survival analyses performed according to insertion site in the beta1-sheet of the TKD1 showed that patients with FLT3-ITD insertions in the beta1-sheet had a significantly inferior RFS and OS compared with patients with “all other insertion sites.” This was true for the patients exhibiting a normal karyotype (RFS: P < .001, OS: P < .001) as well as for the whole patient population (RFS: P = .001, OS: P = .002; Figure 3). Of note, the proportions of patients receiving an allogeneic SC transplant in first CR were similar in the group of patients defined by insertion site in the beta1-sheet of the TKD1 compared with all other patients. In contrast to the end point “achievement of a CR after induction therapy,” higher mutant to wild-type ITD allelic ratio and larger ITD length, both dichotomized at the median level, did not significantly impact on RFS (P = .12 and P = .08, respectively) and OS in univariable analyses (P = .24 and P = .09, respectively). Furthermore, there was no significant difference in number of mutated ITDs for either RFS or OS for the patients exhibiting a normal karyotype, as well as for all patients or OS (P = .81 and .38; P = .39 and .84, respectively).
Survival analysis for FLT3-ITD mutated AML according to the FLT3-ITD mutation site. Kaplan-Meier curves for relapse-free (A) and overall (B) survival for FLT3-ITD–mutated AML patients according to the FLT3 insertion site.
Survival analysis for FLT3-ITD mutated AML according to the FLT3-ITD mutation site. Kaplan-Meier curves for relapse-free (A) and overall (B) survival for FLT3-ITD–mutated AML patients according to the FLT3 insertion site.
Discussion
Internal tandem duplications of the FLT3 gene in AML were reported more than 10 years ago.1 Such ITDs as well as other mutations of the gene represent attractive targets for therapy with specific FLT3 inhibitors32-34 that recently entered phase 3 clinical trials (CALGB 10603, http://clinicaltrials.gov: NCT00651261). It is generally believed that FLT3-ITDs are localized in the JMD, thereby leading to disruption of the autoinhibitory conformation of the receptor.12,35 Recently, however, Breitenbuecher et al could show that almost one-third of FLT3-ITDs have their insertion site outside the JMD, most commonly in the beta1-sheet of the TKD1.18 As integration of non-JMD ITDs may result in differential structural conformation of the TKD1, this may translate in differential FLT3 signaling and biologic behavior. This hypothesis is supported by a recent study indicating rewired signal transduction and differential responsiveness to kinase inhibitors of non–JM-ITD627E.19
In this large independent cohort of younger adult patients, we could confirm that approximately 30% of FLT3-ITDs are localized outside the JMD. A major cluster of ITD insertions was found in the beta1-sheet of the TKD1 (25.9%), and at lower frequencies in the nucleotide binding loop (2.5%), in the beta2-sheet (1.4%), and 3′ of the beta2-sheet (0.7%). In addition, in this patient cohort 34 of the 241 FLT3-ITD–positive AML cases revealed more than one ITD of different leukemic subclones. Interestingly, nearly all patients with more than one ITD had at least 1 ITD integration sited in the JMD. Of note, we found a strong association between ITD integration site and ITD length, in that the more C-terminal the location of the ITD site, the longer the size of the inserted fragment. In accordance with previous studies, we were not able to detect a significant correlation between the number of ITD-positive clones per patient and clinical outcome, either in univariable or in multivariable analyses.10,36
To date, the molecular mechanism that generates an FLT3-ITD remains unclear. Kiyoi et al hypothesized that the DNA sequence between amino acids 593 and 602 forms a palindromic intermediate that leads to a DNA replication error, thus inducing the tandem duplication.37 Other investigators showed that 95% of the patients with FLT3-ITD mutations contained at least one amino acid of the tyrosine-rich stretch Y591 to Y59930 and that the amino acid residue R595 may play an essential role in activation of STAT5.30 In addition, tyrosine to phenylalanine substitution of residues 589 and 591 abrogates STAT5 activation of FLT3-ITD, and FLT3-ITD-Y589/591F is incapable of inducing a myeloproliferative phenotype.31 Interestingly, we showed that in 96.1% of all analyzed FLT3-ITDs at least one amino acid of this motif (Y591-Y599) was affected, and this was independent of ITD length and insertion site. One may hypothesize that at least one residue of this tyrosine-rich stretch is necessary for effective transformation by ligand-independent activation of FLT3. Furthermore, the combined duplication of the tyrosine residues Y589 and Y591, which are also suggested to be required for activation of the STAT5 signaling pathways,31 were found in 42.2% of the cases.
Regarding the ITD size, the published data are fairly contradictory, with reports of a worse overall prognosis in cases with both longer10,15 and shorter16 FLT3-ITDs, but also of no prognostic significance at all.17 However, the populations analyzed in these trials are very heterogeneous with respect to age, variation of cutoff values from 33 to 70 base pairs, relatively small number of FLT3-ITD–mutated cases, and inclusion of acute promyelocytic leukemia patients in one study. In our analysis, the ITD size varied from 15 to 180 nucleotides, with a median of 48 nucleotides. Importantly, we were able to show that insertion site within the beta1-sheet of the TKD1 was an significant prognostic marker within the FLT3-ITD AML population for all clinical end points analyzed, such as achievement of CR, RFS, and OS. And this was true for the subset of patients exhibiting a normal karyotype as well as for the whole study population. Of note, in our population neither length of ITD nor mutant to wild-type allelic ratio showed this consistent impact on all clinical end points. However, in univariable analysis, longer ITD lengths as well as higher mutant to wild-type allelic ratios were significantly associated with lower CR rates. In contrast, for neither RFS nor OS such a prognostic impact was detected in univariable analyses for these 2 variables, which is in discordance with previous work.8,9,15 In multivariable models for patients exhibiting a normal karyotype as well as the whole patient population, insertion site within the beta1-sheet of the TKD1 was a significant unfavorable parameter for all 3 analyzed clinical end points, underlining its independent importance. The same results were obtained when the Cox regression models on RFS and OS were stratified for treatment with allogeneic SCT in first CR, indicating the independence of this significant parameter from treatment strategy.
In summary, the comprehensive sequence analysis of FLT3-ITD adds to previous knowledge in that insertion site within the beta1-sheet of the TKD1 appears to be an important prognostic parameter in patients with FLT3-ITD–positive AML, even outweighing the impact of FLT3 mutant to wild-type allelic ratio. Because disruption of the beta1-sheet of the TKD1 might result in a conformation change of the protein structure, one might speculate that in these cases response to distinct chemotherapeutic agents but also tyrosine kinase inhibitors might be impaired. Therefore, we propose to prospectively analyze not only the FLT3-ITD mutation status but also the ITD integration site in future clinical trials, in particular in the context of treatment with FLT3 tyrosine kinase inhibitors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to all members of the German-Austrian AML Study Group (AMLSG) for their participation in this study and providing patient samples.
This work was supported in part by grants 01GI9981 (Competence Net for Acute and Chronic Leukemias) and 01KG0605 (IPD-Meta-Analysis: A model-based hierarchical prognostic system for adult patients with acute myeloid leukemia [AML]) from the Bundesministerium für Bildung und Forschung (BMBF), Germany, the Deutsche José Carreras Leukämie-Stiftung (DJCLS R 08/23v).
Authorship
Contribution: S.K. designed research, analyzed data, and wrote the paper; R.F.S. analyzed data and wrote the paper; M.C.L. performed research and contributed intellectually; F.B. contributed intellectually and critically reviewed the paper; K.W., J.D., S.G., and D.S. performed research; J.K. and A.G. critically reviewed the paper; and H.D., T.F., and K.D. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
For a complete list of the German-Austrian AML Study Group (AMLSG) participants, see the supplemental Appendix, available on the Blood website (see the Supplemental Materials link at the top of the online article).
Correspondence: Konstanze Döhner, Department of Internal Medicine III, University Hospital of Ulm, Albert-Einstein-Allee 23, 89081 Ulm, Germany; e-mail: konstanze.doehner@uniklinik-ulm.de.