To the editor:
Despite major improvements of treatments, chronic lymphocytic leukemia (CLL) remains an incurable disease. One of the reasons for the inability to completely eliminate the leukemic clone is survival signals from the microenvironment. Coculture models demonstrated that various stromal cells can support CLL cells by induction of survival proteins, such as Mcl-1.1 Therefore, stromal cell cocultures are used to model the in vivo microenvironment and to develop strategies to overcome stroma-derived drug resistance.
Vogler and colleagues2 examined the impact of microenvironment-mediated drug resistance on the BCL-2 antagonist ABT-737 in CLL. They designed a coculture model, using fibroblasts transfected with CD154 (CD40 ligand). Their key finding is that CLL cells cocultured with CD154-expressing fibroblasts (and interleukin-4 [IL-4]) become resistant to ABT-737, which was accompanied by the induction of antiapoptotic proteins (BCL-xL, BCL2A1). The authors postulate that treatment of CLL patients with ABT-737 is likely to result in residual disease within lymph nodes, where CLL cells can receive drug resistance signals via CD154 and IL-4.
Vogler's paper is in line with previous publications that demonstrate protection of CLL cells by nurselike cells (NLCs),3 marrow stromal cells (MSCs),3,4 and follicular dendritic cells.5 Based on the in vitro observation that CD154 induces prosurvival and drug resistance pathways, and the notion that T cells in the CLL microenvironment can express CD154,6 they propose that their model mimics the lymph node microenvironment. However, this CD154/IL-4 model is simplifying the complex cellular and molecular interactions that CLL cells encounter in vivo (Figure 1). In vitro stimulation assumes that CD154 functions as a survival and drug resistance factor in vivo, which remains controversial. Regulation of pro- and antiapoptotic molecules after CD154 stimulation is a dynamic process in which ratios of the different molecules, rather than their absolute levels, determine the fate of the cells.7 CD154 stimulation also can increase the immunogenicity of CLL cells, turning nonimmunogenic CLL cells into effective T-cell stimulators,8 which can overcome immune tolerance in vivo.9 Herishanu et al compared gene expression profiles of CLL cells obtained from lymph nodes versus peripheral blood, demonstrating up-regulation of cell-cycle, proliferation-related, and nuclear factor–κB target genes in lymphatic tissues.10 These up-regulated genes are strikingly similar to the genes induced in CLL cells by NLCs,11 suggesting that NLC-CLL cocultures are a solid in vitro model of the lymph node microenvironment.
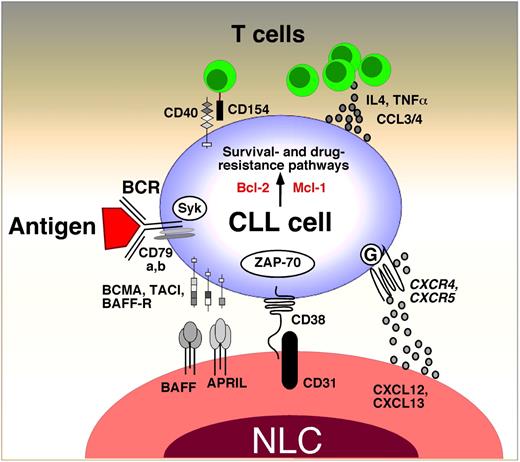
Cross-talk between CLL cells and the lymph node microenvironment. This figure displays the molecules involved in cross-talk between CLL cells and accessory cells in the lymphoid tissue microenvironments. Contact between CLL cells and nurselike cells (NLCs) is established and maintained by chemokine receptors and adhesion molecules. NLCs express the chemokines CXCL12 and CXCL13. NLCs attract CLL cells via the G protein–coupled chemokine receptors CXCR4 and CXCR5, which are expressed at high levels on CLL cells. NLCs also express the tumor necrosis factor family members BAFF and APRIL, providing survival signals to CLL cells via corresponding receptors (BCMA, TACI, BAFF-R). CD38 expression allows CLL cells to interact with CD31, the ligand for CD38, expressed by stromal and nurselike cells. Ligation of CD38 activates ZAP-70 and downstream survival pathways. Self and/or environmental antigens (Ags) are considered a key factor in stimulation and expansion of the CLL clone. Stimulation of the B-cell antigen receptor (BCR) complex (BCR and CD79a,b) induces downstream signaling by recruitment and activation of Syk and ZAP-70. BCR stimulation and coculture with NLC also induces CLL cells to secrete high levels of the chemokines CCL3 and CCL4, which are potent T cell–attracting chemokines. Through this mechanism, CLL cells can actively recruit T cells for cognate T-cell interactions with CLL cells. CD154+ T cells are preferentially found in CLL pseudofollicles and can interact with CLL cells via CD40. Cytokine secreted by T cells or CLL cells, such as IL-4 or tumor necrosis factor α, are considered important regulators of CLL cell survival. Collectively, this cross-talk between CLL cells and accessory cells results in activation of survival and drug resistance pathways, such as those provided by Bcl-2 and Mcl-1.
Cross-talk between CLL cells and the lymph node microenvironment. This figure displays the molecules involved in cross-talk between CLL cells and accessory cells in the lymphoid tissue microenvironments. Contact between CLL cells and nurselike cells (NLCs) is established and maintained by chemokine receptors and adhesion molecules. NLCs express the chemokines CXCL12 and CXCL13. NLCs attract CLL cells via the G protein–coupled chemokine receptors CXCR4 and CXCR5, which are expressed at high levels on CLL cells. NLCs also express the tumor necrosis factor family members BAFF and APRIL, providing survival signals to CLL cells via corresponding receptors (BCMA, TACI, BAFF-R). CD38 expression allows CLL cells to interact with CD31, the ligand for CD38, expressed by stromal and nurselike cells. Ligation of CD38 activates ZAP-70 and downstream survival pathways. Self and/or environmental antigens (Ags) are considered a key factor in stimulation and expansion of the CLL clone. Stimulation of the B-cell antigen receptor (BCR) complex (BCR and CD79a,b) induces downstream signaling by recruitment and activation of Syk and ZAP-70. BCR stimulation and coculture with NLC also induces CLL cells to secrete high levels of the chemokines CCL3 and CCL4, which are potent T cell–attracting chemokines. Through this mechanism, CLL cells can actively recruit T cells for cognate T-cell interactions with CLL cells. CD154+ T cells are preferentially found in CLL pseudofollicles and can interact with CLL cells via CD40. Cytokine secreted by T cells or CLL cells, such as IL-4 or tumor necrosis factor α, are considered important regulators of CLL cell survival. Collectively, this cross-talk between CLL cells and accessory cells results in activation of survival and drug resistance pathways, such as those provided by Bcl-2 and Mcl-1.
Vogler's study, using CD154 and IL-4 stimulation, focuses on the effects of these molecules that may represent important signals from T cells in the lymphatic tissues, rather than providing a more complex mode of stimulation present in NLC cocultures. The overarching question is which model approximates the in vivo microenvironment. The current consensus is that multiple cells and signaling pathways provide drug resistance in cross-talk between the CLL cells and their milieu, not only in lymph nodes, but also in other tissue sites, such as the marrow. ABT-737 (as ABT-263) is under clinical investigation,12 and its cytoreductive effects in the marrow versus lymph nodes will be of major interest.
This study also emphasizes the challenge to identify, target, and validate the key pathways of microenvironment-CLL interactions. While ABT-737 targets Bcl-2 and Bcl-xL with high potency, a panantagonist of Bcl-2 family antiapoptotic proteins is a desirable molecule. Gossypol analogs act as pan–Bcl-2 antagonists, but they have not reached the desired potency. Alternatively, CXCR4 antagonists13 and spleen tyrosine kinase (Syk) inhibitors14 therapeutically target and disrupt the leukemia microenvironment cross-talk in CLL and now are entering the clinical stage. These targets were developed and tested in the above-mentioned models,13,14 emphasizing the importance and validity of in vitro models for dissecting the tumor microenvironment in leukemia and beyond.
Authorship
Acknowledgment: This work was supported by CLL Global Research Foundation grants to J.A.B. and V.G.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan A. Burger, MD, PhD, Department of Leukemia, Unit 428, The University of Texas M. D. Anderson Cancer Center, PO Box 301402, Houston, TX 77230-1402; e-mail: jaburger@mdanderson.org.