Abstract
Natural killer (NK) cells provide innate control of infected and neoplastic cells. Multiple receptors have been implicated in natural cytotoxicity, but their individual contribution remains unclear. Here, we studied the activation of primary, resting human NK cells by Drosophila cells expressing ligands for receptors NKG2D, DNAM-1, 2B4, CD2, and LFA-1. Each receptor was capable of inducing inside-out signals for LFA-1, promoting adhesion, but none induced degranulation. Rather, release of cytolytic granules required synergistic activation through coengagement of receptors, shown here for NKG2D and 2B4. Although engagement of NKG2D and 2B4 was not sufficient for strong target cell lysis, collective engagement of LFA-1, NKG2D, and 2B4 defined a minimal requirement for natural cytotoxicity. Remarkably, inside-out signaling induced by each one of these receptors, including LFA-1, was inhibited by receptor CD94/NKG2A binding to HLA-E. Strong inside-out signals induced by the combination of NKG2D and 2B4 or by CD16 could overcome CD94/NKG2A inhibition. In contrast, degranulation induced by these receptors was still subject to inhibition by CD94/NKG2A. These results reveal multiple layers in the activation pathway for natural cytotoxicity and that steps as distinct as inside-out signaling to LFA-1 and signals for granule release are sensitive to inhibition by CD94/NKG2A.
Introduction
Natural killer (NK) cells represent a subset of cytotoxic lymphocytes that detect and kill pathogen-infected cells, tumor cells, and certain hematopoietic cells, a process known as natural cytotoxicity.1-3 Natural cytotoxicity involves a coordinated series of events encompassing contact with target cells, adhesion, synapse formation, granule polarization, and granule exocytosis.4,5 These events are induced by numerous germline-encoded receptors, which bind to ligands expressed on target cells.6,7 Recent findings have substantiated a role for NK-cell activation receptors in immunosurveillance of neoplastic cells.8,9 In addition to natural cytotoxicity, NK cells mediate antibody-dependent cellular cytotoxicity (ADCC) through Fc receptors.10 NK cells also secrete cytokines and chemokines upon receptor stimulation. These NK-cell responses are controlled by inhibitory receptors, many of which bind to MHC class I molecules on target cells.11,12
Identifying the minimal activation requirements for natural cytotoxicity is important for understanding target cell recognition by NK cells and its consequences. Previous studies demonstrated that engagement of receptors NKG2D (CD314), DNAM-1 (CD226), 2B4 (CD244), or CD2 by specific mAbs induces target cell lysis by IL-2–activated NK cells in redirected lysis assays, identifying these receptors as “activation receptors” per se.13-16 Surprisingly, in similar assays using resting NK cells, only poor lysis of target cells was observed.17 However, mAb-mediated coengagement of specific, pairwise combinations of receptors induced target cell lysis by resting NK cells.17 A limitation in these studies, however, is that mAbs do not recapitulate activation by physiologic ligands. Moreover, redirected lysis assays using FcR+ cells involve several other receptor-ligand interactions. Furthermore, because of the multiplicity of receptor-ligand interactions between NK cells and target cells, it has been difficult to assign specific functions to individual activating receptors, or to assess the outcome of simultaneous engagement of defined combinations of receptors. For example, it is not clear which receptors mediate signals for adhesion and for degranulation during natural cytotoxicity, which signaling events are targeted by inhibitory receptors, and whether sensitivity to inhibition varies among activation receptors. Moreover, the minimal receptor-ligand interactions required to elicit natural cytotoxicity are not known.
To overcome the complexity in receptor-ligand interactions and to dissect the contribution of individual receptors to natural cytotoxicity, we have developed a reconstitution system using Drosophila cells as targets.18 A notable advantage of such a system is that discrete events in the activation of primary, unmanipulated NK cells can be studied in the context of ligand-receptor interactions. Using this approach, initial studies revealed that ligands of the integrin leukocyte functional antigen 1 (LFA-1), which is composed of an αL (CD11a) and β2 (CD18) heterodimer, induce signals for adhesion and granule polarization.18,19 In addition, the combination of LFA-1 and CD16 (FcγRIIIA), the low-affinity Fc receptor expressed predominantly on CD56dim NK cells, was sufficient to trigger ADCC by resting NK cells.19
Here, using this Drosophila cell system, we have studied the minimal requirements for natural cytotoxicity, and the level(s) at which inhibitory receptor engagement intersects this process. First, we compared the potential of NKG2D, DNAM-1, 2B4, CD2, as well as LFA-1, for activation of NK cells freshly isolated from human peripheral blood, and compared results with the requirements for activation of ADCC. Then we coexpressed different combinations of ligands for activation receptors with HLA-E, a ligand for the inhibitory receptor CD94/NKG2A, to evaluate the impact of inhibition on signaling by individual activation receptors. By this approach, we have determined the requirements for inside-out signaling to LFA-1,20 for degranulation, and for natural cytotoxicity, as well as the sensitivity of these diverse signals to inhibition by CD94/NKG2A. The results provide new insights into the regulation of resting NK-cell recognition of target cells by receptors that mediate natural cytotoxicity.
Methods
Cells
Human NK cells were isolated from peripheral blood by negative selection (Miltenyi Biotec). Resting NK cells were resuspended in RPMI 1640 (Invitrogen) supplemented with 10% human serum (Valley Biomedical Inc) and used within 2 days of isolation. These cells were 95% to 99% CD3− CD56+. Where specified, biotinylated anti-NKG2C (MAB138; R&D Systems) was added to the negative selection antibodies to remove NKG2C+ NK cells. The human erythroleukemia cell line K562 (ATCC) was maintained in complete medium (RPMI 1640 medium supplemented with 2 mM l-glutamine and 10% FBS; all from Invitrogen). Transfection and maintenance of Drosophila Schneider line 2 (S2) cells was as described.18 cDNA encoding human NK-cell receptor ligands was under control of the metallothionein promoter in plasmid pRmHa3. Expression in transfected S2 cells was induced 48 hours before assays with 1 mM CuSO4 and assessed by flow cytometry. Levels of expression were comparable with that of normal human target cells. For inhibition experiments, 2 μM HLA-E–specific peptide was added 24 hours before assays. Synthetic peptides derived from signal sequences of HLA-B*27 and HLA-C*03 were provided by Dr F. Borrego (NIH, Rockville, MD).21 Approval for use of buffy coats from the blood bank for this study was acquired from the regional ethical review board in Stockholm.
S2 cell transfections
S2 cells expressing ICAM-1, CD48, and CD58, either singly or in combination, have been described.18,22 cDNA clones for CD155, ULBP1, and HLA-E were obtained by reverse-transcription–polymerase chain reaction from peripheral blood RNA. Primers were designed to incorporate restriction sites at the 5′ and the 3′ ends, respectively, for cloning into the pRmHa3 Drosophila expression vector.
Antibodies
MAbs to CD3 (UCHT1), CD56 (NCAM16.2), and CD107a (H4A3) were from Becton Dickinson, and to NKG2A (Z199), from Beckman Coulter. LFA-1 conformation-specific antibodies 327C and mAb24 were gifts from D. Staunton (ICOS Corporation, Bothell, WA) and N. Hogg (Cancer Research UK, London, United Kingdom), respectively.23,24 The LFA-1 activating antibody 240Q was from D. Staunton.24 Allophycocyanin-conjugated streptavidin (SAv)–APC was from Becton Dickinson. A rabbit serum against S2 cells was raised by immunizations with S2 cell membranes.19
Inside-out signaling assay
To evaluate LFA-1 conformation, 105 resting NK cells were washed twice in medium and added to 2 × 105 target cells in 100 μL complete medium. Cells were spun down for 1 minute at 30g and incubated for 5 minutes at 37°C in 5% CO2. Thereafter, the cells were spun down and stained with biotinylated LFA-1 conformation-specific mAbs in ice-cold PBS supplemented with 2% FBS for 45 minutes. Cells were washed and stained with fluorochrome-conjugated anti-CD56 and anti-NKG2A mAbs, in addition to fluorochrome-conjugated streptavidin (BD Bioscience) in PBS supplemented with 2% FBS for 45 minutes on ice. Finally, cells were washed, resuspended in PBS with 2% FBS, and analyzed by flow cytometry (FACSCalibur; Becton Dickinson). Data were analyzed with FlowJo software (TreeStar Inc).
Degranulation assays
Resting NK cells (105) were washed twice in PBS and added to 2 × 105 target cells in 200 μL complete medium. Cells were mixed by gentle pipetting, spun down for 3 minutes at 30g, and incubated for 2 hours at 37°C in 5% CO2. Thereafter, the cells were spun down and stained with fluorochrome-conjugated mAbs to CD56, CD107, and NKG2A in PBS supplemented with 2% FBS and 2 mM EDTA for 45 minutes on ice. The cells were washed, resuspended in PBS supplemented with 2% FBS and 2 mM EDTA, and analyzed by flow cytometry.
Cytotoxicity assays
NK-cell cytotoxicity toward S2 cells was determined as described,22 with the following modification: S2 cells were labeled with 4 μg/mL Cell Tracker Green (Molecular Probes) for 30 minutes at room temperature. To induce ADCC, S2 cells were preincubated with rabbit anti-S2 serum diluted 1:10000.
Statistics
Statistical analysis was performed using paired Student t test.
Results
Multiple NK-cell activating receptors induce inside-out signals for LFA-1
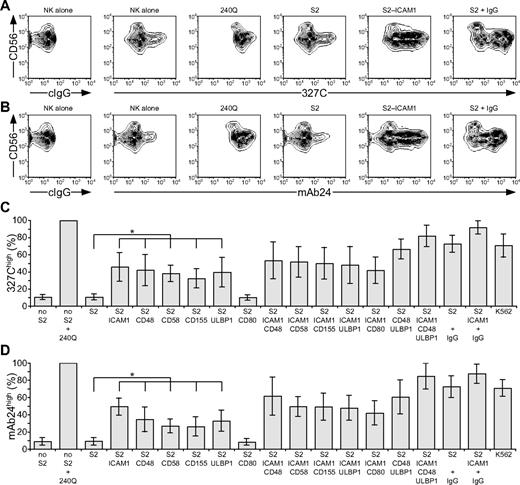
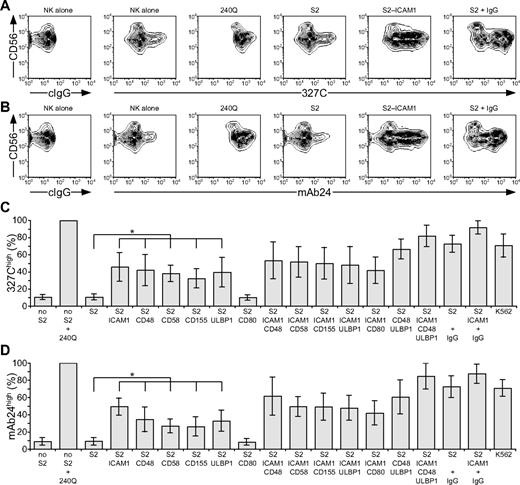
LFA-1 is a central mediator of NK-cell adhesion.25,26 Apart from LFA-1,18 receptors that provide initial signals promoting NK cell–target cell interactions are poorly defined.5 LFA-1 binding to ligands is dynamically regulated by changes in conformation and spatial distribution, which modify receptor affinity and avidity.27 Signals that augment affinity by inducing an open conformation of LFA-1 are termed “inside-out” signals. Freshly isolated, resting NK cells were stained with mAbs 327C and mAb24, which bind selectively to open conformations of LFA-1.23,24 Whereas LFA-1 is expressed at uniformly high levels on resting NK cells, as shown with conformation-independent mAbs, staining with 2 conformation-dependent mAbs was heterogeneous (Figure 1A-B). Analysis was performed on CD56dim NK cells, as CD56bright NK cells represent a discrete NK-cell subset with distinct receptor expression.28 Approximately 10% of CD56dim NK cells displayed an open conformation of LFA-1 (Figure 1C-D). After addition of 240Q, a mAb that stabilizes LFA-1 in the open conformation,24 more than 99% of NK cells displayed an open conformation of LFA-1 (Figure 1).
Engagement of individual activation receptors on resting NK cells induces inside-out signals. Resting NK cells were mixed with target cells as indicated. Where indicated, S2 cells were preincubated with a rabbit anti-S2 serum (+ IgG). Cells were incubated for 5 minutes at 37°C, stained with conformation-specific, biotinylated anti–LFA-1 mAbs, washed, and stained with fluorochrome-conjugated anti-CD56 and streptavidin. (A-B) NK cells were gated on forward scatter/side scatter plots, and the profiles show CD56 versus 327C or mAb24 mAb staining, as indicated. Gates indicate the percentage of 327Chigh or mAb24high NK cells. The profiles are representative of 3 or more independent experiments. (C-D) The percentage of 327Chigh or mAb24high CD56dim NK cells is presented as the mean of 6 donors. Bars indicate SD. *P < .01.
Engagement of individual activation receptors on resting NK cells induces inside-out signals. Resting NK cells were mixed with target cells as indicated. Where indicated, S2 cells were preincubated with a rabbit anti-S2 serum (+ IgG). Cells were incubated for 5 minutes at 37°C, stained with conformation-specific, biotinylated anti–LFA-1 mAbs, washed, and stained with fluorochrome-conjugated anti-CD56 and streptavidin. (A-B) NK cells were gated on forward scatter/side scatter plots, and the profiles show CD56 versus 327C or mAb24 mAb staining, as indicated. Gates indicate the percentage of 327Chigh or mAb24high NK cells. The profiles are representative of 3 or more independent experiments. (C-D) The percentage of 327Chigh or mAb24high CD56dim NK cells is presented as the mean of 6 donors. Bars indicate SD. *P < .01.
To probe which NK-cell activation receptors can provide inside-out signals for LFA-1, Drosophila S2 cells expressing CD48, a ligand for 2B4; CD58, a ligand for CD2; CD155, a ligand for DNAM-1; and ULBP1, a ligand for NKG2D, either alone or in combination with intercellular adhesion molecule 1 (ICAM-1; CD54), a ligand for LFA-1, were generated (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Great care was taken to select S2 cells with a matched expression level of any given ligand among different transfectants. S2 cells expressing CD80 (B7.1) were also generated, as it is not clear whether human NK cells express a receptor for CD80 (B7.1).29,30 Resting NK cells were mixed with S2 cells expressing individual ligands for 5 minutes at 37°C, followed by 327C and mAb24 mAb staining. Incubation of NK cells with untransfected S2 cells did not alter the frequency of NK cells displaying an open conformation of LFA-1. However, expression of CD48, CD58, CD155, or ULBP1 on S2 cells significantly increased the frequency of NK cells with open LFA-1, even though these S2 cell transfectants did not express ICAM-1 (Figure 1C-D). Expression of ICAM-1 on S2 cells increased the frequency of NK cells displaying open LFA-1 (Figure 1), whereas expression of CD80 on S2 cells did not.
Next, the impact of activation receptor coengagement on inside-out signals for LFA-1 was assessed. Incubation of NK cells with S2 cells coexpressing ICAM-1 with CD48, CD58, CD155, or ULBP1 did not significantly alter the frequency of NK cells with open LFA-1, relative to incubation with S2 cells expressing individual ligands (Figure 1C-D). However, a modest increase in the frequency of NK cells displaying open LFA-1 was seen after incubation with S2 cells coexpressing CD48 and ULBP1 relative to cells expressing either ligand alone (Figure 1C-D). The highest frequency of NK cells displaying open conformations of LFA-1 was observed after incubation with S2 cells coexpressing ICAM-1, CD48, and ULBP1, similar to the frequency seen after incubating NK cells with the highly NK-sensitive cell line K562 (Figure 1C-D). For comparison, incubation of NK cells with IgG-coated S2 cells also induced a high frequency of NK cells with open LFA-1 (Figure 1C-D), providing evidence for inside-out signals by CD16 engagement on resting NK cells. Together, the results demonstrate that engagement of single NK-cell activation receptors (2B4, CD2, DNAM-1, or NKG2D) by their ligand is sufficient to induce inside-out signals for LFA-1 in resting NK cells. Because inside-out signaling occurred in the absence of ligands for LFA-1 on the target cells, it is clear that these 4 NK-cell receptors can signal upstream of LFA-1.
Individual activation receptors for natural cytotoxicity do not induce degranulation
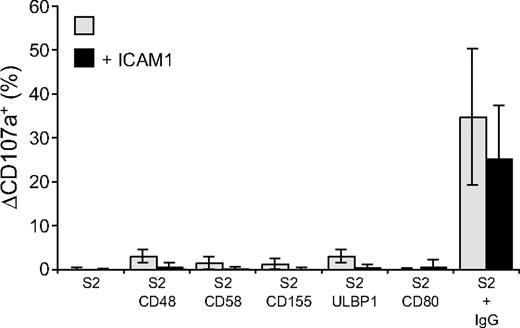
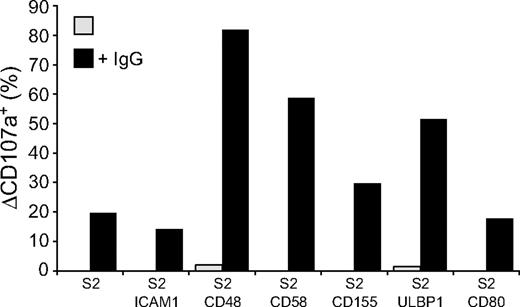
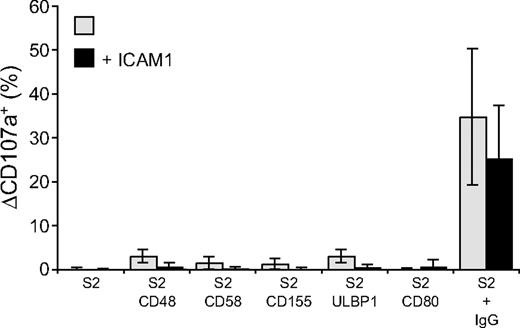
Perforin-dependent NK-cell cytotoxicity occurs through exocytosis of secretory lysosomes. The ability of NK-cell activation receptors to induce degranulation by resting NK cells was examined. Degranulation was assessed by the appearance of CD107a, also known as lysosome-associated membrane protein 1 (LAMP-1), at the cell surface.31 NK cells were incubated with S2 cells for 2 hours at 37°C and stained with fluorochrome-conjugated anti-CD107a mAbs (Figure 2). CD48, CD58, CD155, ULBP1, or CD80 on S2 cells induced very little or no degranulation (Figure 2). The lack of activation by single receptors on NK cells could result from poor adhesion of NK cells to target cells, as individual ligands for receptors 2B4, CD2, DNAM-1, and NKG2D did not induce stable NK-S2 cell conjugates (data not shown). Whereas NK cells did not form stable conjugates with S2 cells expressing CD48, CD58, CD155, or ULBP1, coexpression of these ligands with ICAM-1 markedly augmented LFA-1–dependent conjugate formation (Barber and Long18 and data not shown). However, the frequency of degranulating NK cells was, if anything, even lower when S2 cells coexpressed ICAM-1 with the ligands of activating receptors (Figure 2). Likewise, the number of degranulating NK cells after mixing with IgG-coated S2 cells was reduced in the presence of ICAM-1 (Figure 2).
Engagement of individual activation receptors on resting NK cells does not induce degranulation. Resting NK cells were mixed with target cells as indicated. Where indicated, S2 cells were preincubated with a rabbit anti-S2 serum (+ IgG). Cells were incubated for 2 hours at 37°C, then stained with fluorochrome-conjugated anti-CD56 and anti-CD107a mAbs. The percentage increase of CD107a+ NK cells after incubation with target cells relative to CD107a+ NK cells upon incubation of NK cells alone (ΔCD107a+) is presented as the mean of 8 donors.  represents target cells as indicated, whereas ■ represents target cells coexpressing ICAM-1 in addition to the other ligands, as indicated. Bars denote SD.
represents target cells as indicated, whereas ■ represents target cells coexpressing ICAM-1 in addition to the other ligands, as indicated. Bars denote SD.
Engagement of individual activation receptors on resting NK cells does not induce degranulation. Resting NK cells were mixed with target cells as indicated. Where indicated, S2 cells were preincubated with a rabbit anti-S2 serum (+ IgG). Cells were incubated for 2 hours at 37°C, then stained with fluorochrome-conjugated anti-CD56 and anti-CD107a mAbs. The percentage increase of CD107a+ NK cells after incubation with target cells relative to CD107a+ NK cells upon incubation of NK cells alone (ΔCD107a+) is presented as the mean of 8 donors.  represents target cells as indicated, whereas ■ represents target cells coexpressing ICAM-1 in addition to the other ligands, as indicated. Bars denote SD.
represents target cells as indicated, whereas ■ represents target cells coexpressing ICAM-1 in addition to the other ligands, as indicated. Bars denote SD.
Whereas engagement of NK cell activation receptors for natural cytotoxicity did not induce degranulation, it might still contribute to signals for degranulation from other receptors, such as CD16. Therefore, the influence of receptors for natural cytotoxicity on CD16-mediated degranulation was evaluated. Expression of CD48, CD58, CD155, and ULBP1, but not ICAM-1 or CD80, on IgG-coated S2 cells enhanced NK cell degranulation (Figure 3). Thus, although engagement of the individual activating receptors 2B4, CD2, DNAM-1, and NKG2D is sufficient for inside-out signaling in resting NK cells, engagement of these receptors alone or in combination with LFA-1 is not sufficient to induce strong degranulation. Yet, 2B4, CD2, DNAM-1, or NKG2D engagement can augment CD16-mediated signals for degranulation.
Activation receptor coengagement on resting NK cells enhances CD16-induced degranulation. Resting NK cells were mixed with target cells as indicated. Where indicated, S2 cells were preincubated with a rabbit anti-S2 serum (+ IgG). Cells were incubated for 2 hours at 37°C, thereafter stained with fluorochrome-conjugated anti-CD56 and anti-CD107a mAbs. (A) The percentage of ΔCD107a+ CD56dim NK cells is presented and values are representative of more than 3 independent experiments.
Activation receptor coengagement on resting NK cells enhances CD16-induced degranulation. Resting NK cells were mixed with target cells as indicated. Where indicated, S2 cells were preincubated with a rabbit anti-S2 serum (+ IgG). Cells were incubated for 2 hours at 37°C, thereafter stained with fluorochrome-conjugated anti-CD56 and anti-CD107a mAbs. (A) The percentage of ΔCD107a+ CD56dim NK cells is presented and values are representative of more than 3 independent experiments.
Synergy between activation receptors for degranulation
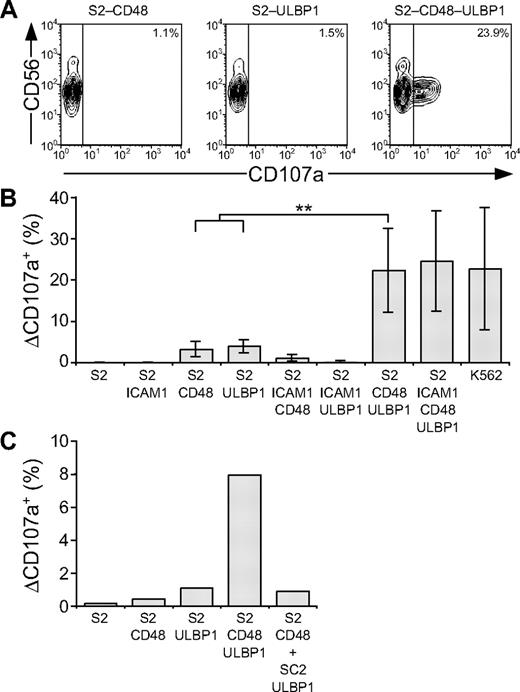
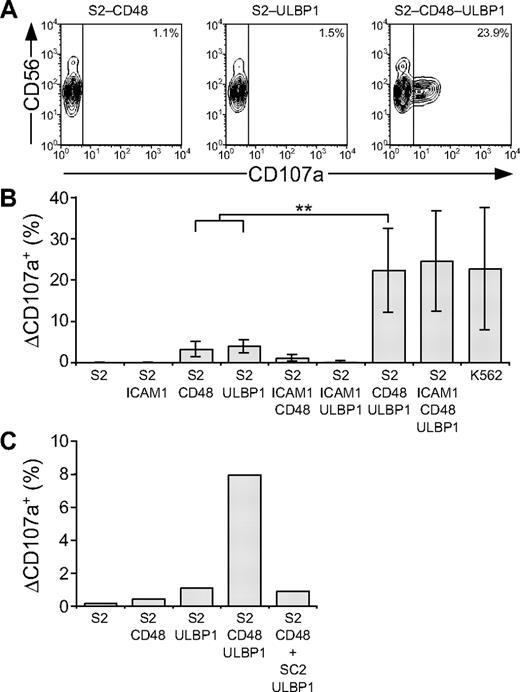
Whereas engagement of individual NK-cell activation receptors was insufficient, coengagement of combinations of such receptors might cooperate to induce degranulation. We thus assessed whether activation of NK cells by ligands for NKG2D and 2B4 could cooperate for induction of resting NK-cell degranulation. To this end, S2 cells expressing both ligands (S2-CD48-ULBP1) were generated. Incubation of resting NK cells with S2-CD48-ULBP1 cells resulted in degranulation, whereas incubation with either S2-CD48 or S2-ULBP1 cells alone did not (Figure 4A-B). Degranulation induced by 2B4 and NKG2D synergy was not enhanced by coexpression of ICAM-1 (Figure 4B). By comparison, K562 cells induced a similar frequency of degranulating NK cells as did S2-CD48-ULBP1 and S2–ICAM-1–CD48–ULBP1 cells (Figure 4B). To assess whether degranulation induced by NKG2D and 2B4 synergy required coexpression of ligands for 2B4 and NKG2D on the same cell or whether expression of ligands for NKG2D and 2B4 on adjacent target cells could suffice, resting NK cells were incubated with either S2-CD48-ULBP1 or a mixture of S2-CD48 and S2-ULBP1 cells (Figure 4C). Incubation of NK cells with a mixture of S2-CD48 and S2-ULBP1 cells did not increase the low degranulation obtained with either S2 cell separately (Figure 4C). Therefore, coexpression of ligands for NKG2D and 2B4 on the same target cell is required to induce synergistic degranulation signals.
Resting NK-cell degranulation is facilitated by synergy among activation receptors. Resting NK cells were mixed with target cells as indicated. Cells were incubated for 2 hours at 37°C, then stained with fluorochrome-conjugated anti-CD56 and anti-CD107a mAbs. (A) NK cells were gated on forward scatter/side scatter plots, and the profiles show CD56 versus CD107a mAb staining. Gates indicate the percentage of ΔCD107a+ NK cells. (B) The percentage of ΔCD107a+ NK cells is presented as the mean of 7 donors. Bars indicate SD. **P < .005. (C) The percentage of ΔCD107a+ NK cells from 1 representative experiment is shown. Experiments are representative of at least 3 independent experiments.
Resting NK-cell degranulation is facilitated by synergy among activation receptors. Resting NK cells were mixed with target cells as indicated. Cells were incubated for 2 hours at 37°C, then stained with fluorochrome-conjugated anti-CD56 and anti-CD107a mAbs. (A) NK cells were gated on forward scatter/side scatter plots, and the profiles show CD56 versus CD107a mAb staining. Gates indicate the percentage of ΔCD107a+ NK cells. (B) The percentage of ΔCD107a+ NK cells is presented as the mean of 7 donors. Bars indicate SD. **P < .005. (C) The percentage of ΔCD107a+ NK cells from 1 representative experiment is shown. Experiments are representative of at least 3 independent experiments.
Coengagement of LFA-1, 2B4, and NKG2D defines a minimal requirement for NK-cell natural cytotoxicity
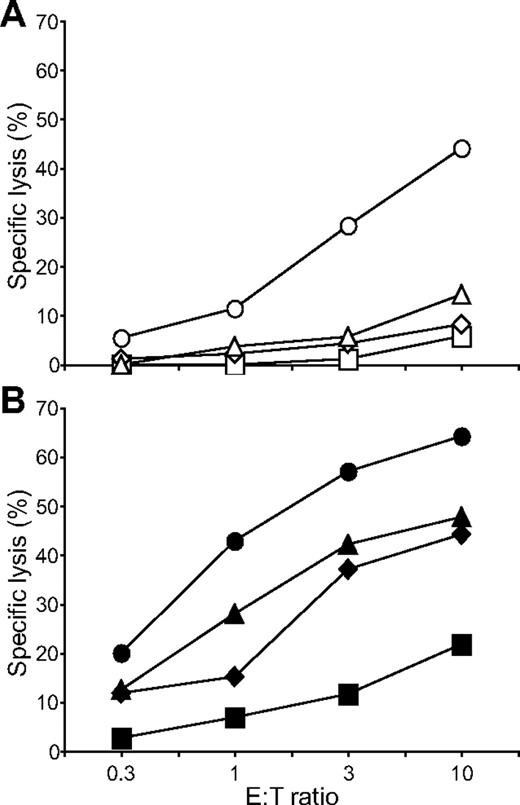
Having identified a combination of activation receptors that synergized to induce degranulation, we examined the minimal requirements for natural cytotoxicity. ICAM-1 on S2 cells is sufficient to induce polarization of cytolytic granules.19 NK cells were incubated with S2, S2–ICAM-1, S2-CD48-ULBP1, and S2–ICAM-1–CD48–ULBP1 cells and lysis was determined by the uptake of propidium iodide in S2 cells (Figure 5). Modest S2 cell lysis was observed with S2-CD48-ULBP1 cells, barely over the lysis obtained with untransfected S2 cells and S2–ICAM-1 cells (Figure 5A). In contrast, strong lysis of S2–ICAM-1–CD48–ULBP1 cells was observed (Figure 5A). By comparison, IgG-coated S2–ICAM-1 cells were lysed to a similar extent as S2–ICAM-1–CD48–ULBP1 cells (Figure 5B). S2–ICAM-1–CD48 and S2–ICAM-1–ULBP1 cells were not lysed (Bryceson et al19 and data not shown). Therefore, the combined engagement of 2B4, NKG2D, and LFA-1 defines a minimal requirement for natural cytotoxicity by resting NK cells.
Coengagement of NKG2D, 2B4, and LFA-1 induces natural cytotoxicity by resting NK cells. Resting NK cells were mixed with (A) S2 (□), S2–ICAM-1 ( ), S2-CD48-ULBP1 (△), or S2–ICAM-1–CD48–ULBP1 (○) cells, or (B) S2 cells as in panel A preincubated with a rabbit serum raised against S2 cells (■, ♦, ▲, ●). Cells were incubated for 3 hours at 37°C. Specific lysis of S2 cells was calculated from the percentage of propidium iodide–positive S2 cells in duplicate samples, as determined by flow cytometry. The experiment is representative of 3 or more independent experiments.
), S2-CD48-ULBP1 (△), or S2–ICAM-1–CD48–ULBP1 (○) cells, or (B) S2 cells as in panel A preincubated with a rabbit serum raised against S2 cells (■, ♦, ▲, ●). Cells were incubated for 3 hours at 37°C. Specific lysis of S2 cells was calculated from the percentage of propidium iodide–positive S2 cells in duplicate samples, as determined by flow cytometry. The experiment is representative of 3 or more independent experiments.
Coengagement of NKG2D, 2B4, and LFA-1 induces natural cytotoxicity by resting NK cells. Resting NK cells were mixed with (A) S2 (□), S2–ICAM-1 ( ), S2-CD48-ULBP1 (△), or S2–ICAM-1–CD48–ULBP1 (○) cells, or (B) S2 cells as in panel A preincubated with a rabbit serum raised against S2 cells (■, ♦, ▲, ●). Cells were incubated for 3 hours at 37°C. Specific lysis of S2 cells was calculated from the percentage of propidium iodide–positive S2 cells in duplicate samples, as determined by flow cytometry. The experiment is representative of 3 or more independent experiments.
), S2-CD48-ULBP1 (△), or S2–ICAM-1–CD48–ULBP1 (○) cells, or (B) S2 cells as in panel A preincubated with a rabbit serum raised against S2 cells (■, ♦, ▲, ●). Cells were incubated for 3 hours at 37°C. Specific lysis of S2 cells was calculated from the percentage of propidium iodide–positive S2 cells in duplicate samples, as determined by flow cytometry. The experiment is representative of 3 or more independent experiments.
CD94/NKG2A engagement inhibits inside-out signals for LFA-1
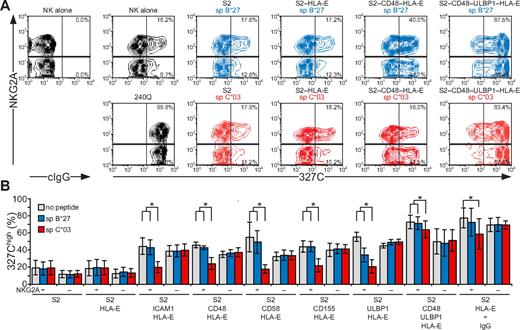
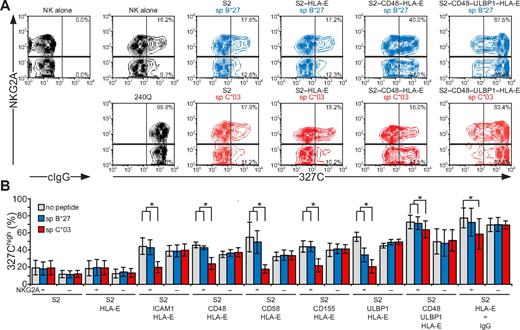
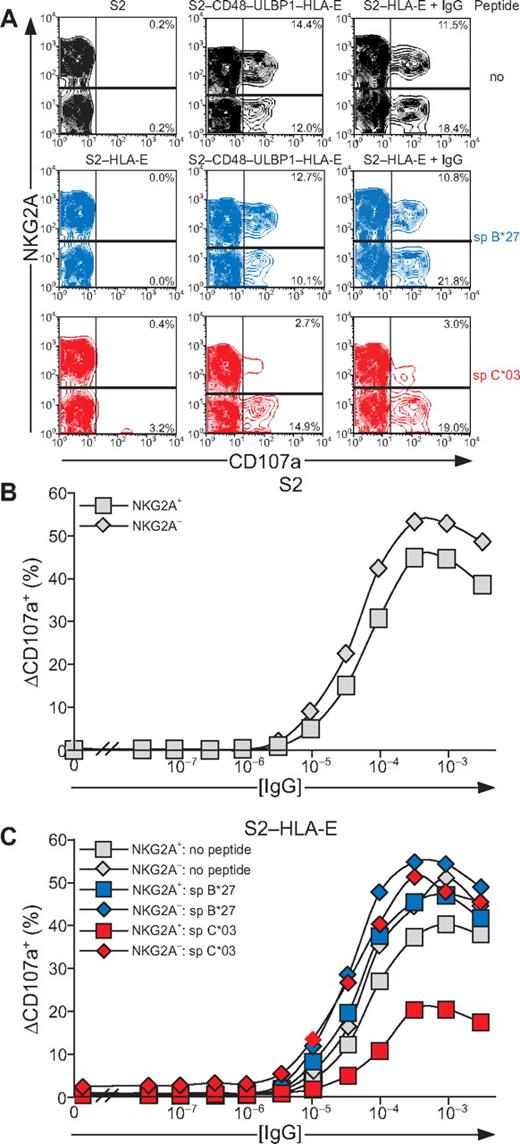
NK-cell responses are tightly controlled by inhibitory receptors. As shown in the first and second sections in “Results,” activation receptors 2B4, CD2, DNAM-1, and NKG2D are each sufficient to induce inside-out signals for LFA-1, but not degranulation. Whether inhibitory receptors can block NK cell activation at a step as proximal as inside-out signaling and whether signaling by various activating receptors is equally sensitive to inhibition are not known. Engagement of CD94/NKG2A by HLA-E21,32 was thus used to evaluate the impact of inhibitory receptors on initial signals for NK-cell activation. HLA-E binds a restricted peptide repertoire derived mainly from leader peptides of other MHC class I molecules.33 As Drosophila cells cannot load peptides onto MHC class I molecules,34 exogenous peptide had to be provided for HLA-E folding. S2 cells expressing HLA-E and β2-microglobulin (S2–HLA-E) together with ligands for NK-cell activation receptors were generated (supplemental Figure 2). Care was taken to isolate S2 cells expressing matched levels of any given ligand among the different S2 transfectants. For inhibition, HLA-E was loaded with a peptide corresponding to the signal peptide of HLA-C*03 (spC*03). As a control, a peptide corresponding to the signal peptide of HLA-B*27 (spB*27), which binds to HLA-E but is not recognized by CD94/NKG2A,21,32 was used. Among resting NK cells, a higher frequency of NKG2A+ cells (18.2%) displayed LFA-1 in an open conformation, compared with NKG2A− cells (9.7%; Figure 6). Addition of mAb 240Q stabilized the open LFA-1 conformation in all of the NKG2A+ and NKG2A− NK cells (Figure 6A). Incubation of NK cells with S2 cells expressing spC*03-loaded HLA-E did not influence the low frequency of NKG2A+ or NKG2A− NK cells displaying an open conformation of LFA-1 (Figure 6B). In contrast, S2 cells coexpressing spC*03-loaded HLA-E with ICAM-1, CD48, CD58, CD155, or ULBP1 significantly reduced the frequency of NKG2A+ NK cells with an open LFA-1 conformation, compared with the same cells either without peptide or loaded with spB*27. The frequency of NKG2A− NK cells displaying an open LFA-1 conformation was not altered by the presence of spC*03 (Figure 6B). S2–CD48–ULBP1–HLA-E cells or IgG-coated S2–HLA-E cells provided strong inside-out signals to LFA-1. Loading of S2–CD48–ULBP1–HLA-E or IgG-coated S2–HLA-E cells with spC*03 partially reduced the frequency of NKG2A+ NK cells with an open LFA-1 conformation, relative to cells either without peptide or loaded with spB*27 (Figure 6B). Remarkably, these results demonstrate that inside-out signals for LFA-1 from diverse activating receptors were all inhibited by engagement of CD94/NKG2A. Notably, whereas CD94/NKG2A coengagement inhibited NKG2D, DNAM-1, 2B4, CD2, or LFA-1 inside-out signals by at least 80%, the signals delivered by the combination of NKG2D and 2B4 or by CD16 were inhibited by only 16% or 32%, respectively. Thus, strong inside-out signals may overcome inhibition by CD94/NKG2A (Figure 6B).
Inside-out signals on resting NK cells are inhibited by CD94/NKG2A engagement. Resting, NKG2C-depleted NK cells were mixed with target cells as indicated. Where indicated, S2 cells were preincubated with a rabbit anti-S2 serum (+ IgG). Cells were incubated for 5 minutes at 37°C, stained with conformation-specific, biotinylated anti–LFA-1 mAb 327C, washed, and then stained with fluorochrome-conjugated anti-CD56, anti-NKG2A, and streptavidin. (A) NK cells were gated on forward scatter/side scatter plots and CD56dim expression. Where indicated, S2 cells were preincubated with peptides sp B*27 (blue) or sp C*03 (red). The profiles show NKG2A versus 327C mAb staining on CD56dim NK cells. Numbers indicate the percentage of 327Chigh CD56dim NK cells within the respective gates. (B) The percentage of 327Chigh CD56dim NK cells is presented as the mean of 4 donors. Bars indicate SD. *P < .05. Experiments are representative of at least 3 independent experiments.
Inside-out signals on resting NK cells are inhibited by CD94/NKG2A engagement. Resting, NKG2C-depleted NK cells were mixed with target cells as indicated. Where indicated, S2 cells were preincubated with a rabbit anti-S2 serum (+ IgG). Cells were incubated for 5 minutes at 37°C, stained with conformation-specific, biotinylated anti–LFA-1 mAb 327C, washed, and then stained with fluorochrome-conjugated anti-CD56, anti-NKG2A, and streptavidin. (A) NK cells were gated on forward scatter/side scatter plots and CD56dim expression. Where indicated, S2 cells were preincubated with peptides sp B*27 (blue) or sp C*03 (red). The profiles show NKG2A versus 327C mAb staining on CD56dim NK cells. Numbers indicate the percentage of 327Chigh CD56dim NK cells within the respective gates. (B) The percentage of 327Chigh CD56dim NK cells is presented as the mean of 4 donors. Bars indicate SD. *P < .05. Experiments are representative of at least 3 independent experiments.
Inhibition of degranulation by CD94/NKG2A
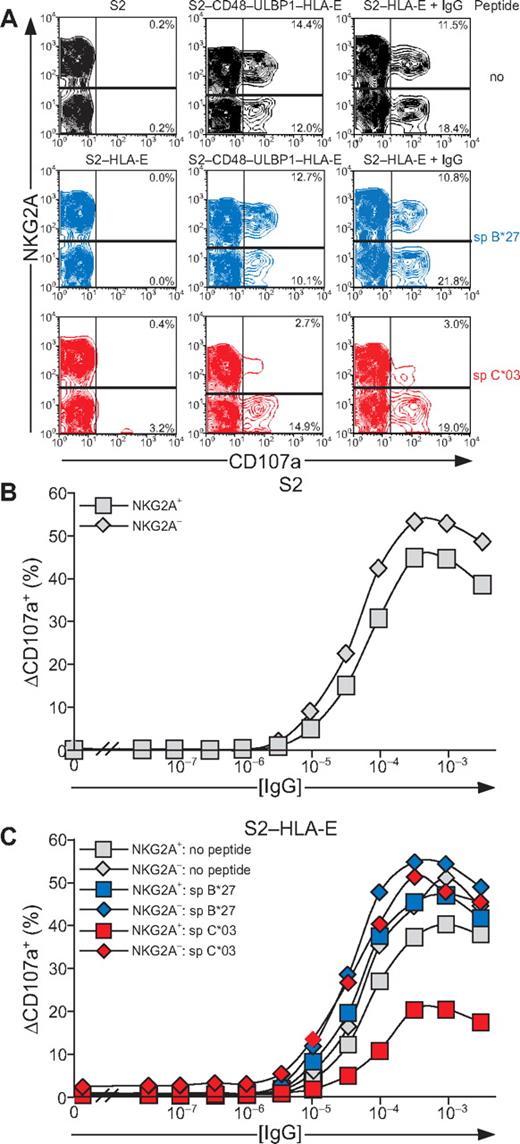
As inside-out signals provided by synergy of NKG2D and 2B4 or by CD16 showed resistance to inhibition by CD94/NKG2A, it was of interest to determine whether degranulation induced by these receptors was sensitive to inhibition. Inhibition of signals for degranulation was assessed with S2 cells expressing HLA-E, either alone or in combination with CD48 and ULBP1. Loading of peptide spC*03 onto S2–CD48–ULBP1–HLA-E cells selectively reduced degranulation by NKG2A+ NK cells (Figure 7A). Moreover, CD16-dependent degranulation by NKG2A+ NK cells was also reduced upon incubation with IgG-coated, spC*03-loaded S2–HLA-E cells (Figure 7A). Loading of HLA-E with spB*27 did not alter the frequency of degranulating NK cells relative to cells that had not been loaded with peptide (Figure 7A). We noted that incubation of NK cells with spC*03-loaded S2–HLA-E cells induced degranulation in a few NKG2A− NK cells (3.2% in Figure 7A) from several donors, whereas S2–HLA-E cells without peptide or loaded with spB*27 did not (Figure 7A and data not shown). Further examination revealed that degranulation was confined to an NKG2A− subset of NK cells that expressed the activating HLA-E–specific receptor CD94/NKG2C. Depletion of NKG2C+ NK cells abolished the degranulation observed with S2–HLA-E cells loaded with sp*03 (data not shown). Thus, in resting NK cells, engagement of CD94/NKG2A inhibited the degranulation induced by NKG2D and 2B4 synergy or by CD16, even though inside-out signals from these receptors were resistant to inhibition in similar settings. Of note, NKG2A+ NK cells degranulated more than NKG2A− NK cells in response to NKG2D and 2B4 synergy, whereas the opposite was the case for CD16-mediated stimulation.
Resting NK-cell degranulation induced by 2B4 and NKG2D synergy or CD16 is inhibited by CD94/NKG2A engagement. Resting NK cells were mixed with S2, S2–HLA-E, or S2–CD48–ULBP1–HLA-E that had been preincubated with peptides as specified. For Fc-receptor stimulation, S2 cells were also preincubated with a rabbit anti-S2 serum (+ IgG). Cells were incubated for 2 hours at 37°C, then stained with fluorochrome-conjugated anti-CD56, anti-CD107a, and anti-NKG2A mAbs. NK cells were gated on forward scatter/side scatter plots and CD56dim expression. (A) The profiles show NKG2A versus CD107a mAb staining. (B-C) S2 cells were preincubated with serial dilutions of a rabbit anti-S2 serum. The percentage of ΔCD107a+ NK cells from 1 representative experiment is shown. Experiments are representative of 3 independent experiments.
Resting NK-cell degranulation induced by 2B4 and NKG2D synergy or CD16 is inhibited by CD94/NKG2A engagement. Resting NK cells were mixed with S2, S2–HLA-E, or S2–CD48–ULBP1–HLA-E that had been preincubated with peptides as specified. For Fc-receptor stimulation, S2 cells were also preincubated with a rabbit anti-S2 serum (+ IgG). Cells were incubated for 2 hours at 37°C, then stained with fluorochrome-conjugated anti-CD56, anti-CD107a, and anti-NKG2A mAbs. NK cells were gated on forward scatter/side scatter plots and CD56dim expression. (A) The profiles show NKG2A versus CD107a mAb staining. (B-C) S2 cells were preincubated with serial dilutions of a rabbit anti-S2 serum. The percentage of ΔCD107a+ NK cells from 1 representative experiment is shown. Experiments are representative of 3 independent experiments.
The potency of the inhibition of degranulation was assessed by a titration of IgG on S2 cells. At maximal stimulation through CD16, slightly fewer NKG2A+ NK cells than NKG2A− NK cells degranulated (Figure 7B). A similar pattern was observed in a titration of IgG on S2–HLA-E cells (Figure 7C). Inhibition of degranulation by spC*03-loaded S2–HLA-E was observed over a wide range of IgG concentrations. Remarkably, even at IgG concentrations that induced maximal degranulation, a marked reduction of degranulation by NKG2A+ NK cells was observed (Figure 7C and data not shown). Thus, engagement of NKG2A on resting NK cells by HLA-E on target cells inhibited degranulation induced either by coactivation receptors for natural cytotoxicity or by CD16. Moreover, substantial inhibition of NK-cell degranulation persisted even at a saturating dose of IgG on target cells.
Discussion
NK-cell recognition of susceptible target cells comprises several activation events. Whether they occur in a sequential fashion, with different receptors contributing to distinct steps, or whether signals from multiple receptors have to be integrated to result in an activation program was still not well defined.5,7 Using Drosophila cells that express ligands of NK-cell activation receptors, in the absence of antireceptor antibodies and exogenous cytokines, we have defined coengagement of receptors NKG2D, 2B4, and LFA-1 as a minimal requirement for natural cytotoxicity mediated by normal, freshly isolated, resting NK cells. Furthermore, by demonstrating that individual activation receptors contribute to the initiation of interaction with target cells, as well as degranulation, our study offers further insight into NK-cell activation by defined ligands on target cells.
Here, we have assessed the ability of individual receptors to induce inside-out signals for LFA-1 by use of a sensitive and rapid flow cytometric assay, which quantified the frequency of NK cells with LFA-1 in an open, ligand-binding conformation. Drosophila cells alone did not induce inside-out signals for NK-cell activation. Remarkably, Drosophila cells expressing individual ligands for NKG2D, DNAM-1, 2B4, or CD2, one at a time, all induced inside-out signals for LFA-1, as did the LFA-1 ligand ICAM-1 itself. It is likely that other receptors, such as members of the selectin family,5 can also enhance NK-cell adhesion. Animal models have provided evidence for NK-cell receptors, such as NKG2D and DNAM-1, in mediating immunosurveillance of spontaneously developing tumors.8,9,35 Our results suggest that NK-cell activating receptors NKG2D, DNAM-1, 2B4, and CD2 provide early signals for interaction with target cells, thereby contributing to immunosurveillance. The inside-out signal delivered by NKG2D and DNAM-1 binding to their respective ligand resulted in enhanced adhesion of NK cells to insect cells expressing ICAM-1, as shown previously with 2B4 and CD2.18 Expanding the realm of receptors capable of inducing inside-out signals for LFA-1, these receptors might provide important initial cues for NK-cell immunosurveillance of infected or neoplastic cells.
IgG-mediated engagement of CD16 induced strong inside-out signals for LFA-1. HLA-E–mediated engagement of CD94/NKG2C, a receptor associated with the immunoreceptor tyrosine-based activation motif (ITAM)–containing adaptor DAP12,36 also induced strong inside-out signals (data not shown). In mouse NK cells, engagement of activating Ly49 receptors, which are also associated with DAP12, conveys inside-out signals for LFA-1.37 Thus, a wide range of NK-cell receptors induce inside-out signals, promoting LFA-1–dependent adhesion, despite their different cytoplasmic domains and signaling properties. In T cells and B cells, inside-out signals for LFA-1 are triggered by the antigen-specific T- and B-cell receptors, respectively, and by chemokine receptors. Inside-out signaling involves Src-family kinases, phosphatidyl inositol-3-kinase, and phospholipase C-γ which in turn induce downstream RAPL and talin recruitment to the cytoplasmic tail of LFA-1.38-41 In future studies, it will be interesting to explore how signaling from such disparate receptors in NK cells converges into an inside-out signal.
Unexpectedly, LFA-1 conformation-specific mAbs displayed a heterogeneous staining pattern on resting NK cells, indicating that a subset of resting NK cells exhibits LFA-1 in an open conformation. Moreover, treatment of resting NK cells with pharmacologic inhibitors of Src-family kinases, phosphatidyl inositol-3-kinase, and phospholipase C-γ diminished the basal level of “open” LFA-1 (Y.T.B., unpublished data, February 2005). These results suggest that tonic signals maintain an active LFA-1 conformation in a subset of NK cells, consistent with the signaling-dependent ability of resting NK cells to bind ICAM-1.18 The presence of open LFA-1 on resting NK cells, a feature not shared by resting T cells (data not shown), may be part of the process by which NK cells acquire responsiveness to target cells and become primed to perform immunosurveillance.42,43
NK-cell cytotoxicity requires exocytosis of secretory lysosomes, as illustrated in certain immunodeficient patients with defective degranulation.44,45 Our results revealed that receptors NKG2D, DNAM-1, 2B4, and CD2 do not individually induce degranulation by resting NK cells. In an earlier study, redirected lysis assays with the Fc receptor–positive mouse cell line P815 and resting NK cells were used to show that mAbs for different NK-cell activating receptors could not induce degranulation, target cell lysis, or secretion of TNF-α and IFN-γ on their own.17 Instead, specific synergistic, pairwise combinations of mAbs induced effector functions in resting NK cells.17 However, mAbs do not recapitulate activation processes induced by physiologic ligands. Furthermore, P815 cells express molecules that are recognized by human NK-cell receptors, such as mouse ICAM-1,46 which is required for redirected lysis of P815 cells.47 To circumvent these limitations, natural ligands of the NK-cell receptors NKG2D and 2B4 were used here to stimulate degranulation. Coexpression of ULBP1 and CD48 on Drosophila S2 cells was sufficient to induce degranulation by resting NK cells. Notably, this synergistic induction of degranulation required expression of ligands on the same target cell, because mixing of NK cells with separate S2 cells expressing either ULBP1 or CD48 did not induce NK-cell degranulation. These results suggest that synergy involves integration of proximal signals delivered by NKG2D and 2B4 at the NK-cell immune synapse.
NK cells express several different inhibitory receptors that control their function. The point at which inhibitory signals intersect activating receptor signaling is not clear. Here, CD94/NKG2A was selected as a prototypical inhibitory receptor for MHC class I because its interaction with the conserved class I molecule HLA-E eliminated the need for donor genotyping. Upon engagement by their ligands, inhibitory receptors recruit the tyrosine phosphatases SHP-1 and SHP-2 to immunoreceptor tyrosine-based inhibition motifs (ITIMs) in their cytoplasmic tail.48,49 In turn, SHP-1 dephosphorylates the guanine nucleotide exchange factor Vav1.50,51 In our experiments, coengagement of CD94/NKG2A by HLA-E on Drosophila S2 cells inhibited early inside-out signals for LFA-1 induced by NKG2D, DNAM-1, 2B4, and CD2, and by LFA-1 itself. Phosphorylation of Vav1 can be induced by cross-linking 2B4, NKG2D, LFA-1, or CD16 on NK cells,52-56 and a role of Vav1 in inside-out signaling to LFA-1 by the T-cell receptor has been reported.57 Therefore, phosphorylation of Vav1 could represent a point of convergence for different activating signals leading to actin reorganization and adhesion. Intersection by inhibitory receptors of proximal signals for actin reorganization and NK-cell adhesion to target cells would thus represent an efficient and versatile mechanism for terminating NK-cell activation at an early step.
Another mechanism by which inhibitory receptors may interfere with inside-out signaling is through inactivation of the Crk-C3G–mediated activation of Rap1, a small GTPase known to promote inside-out signals to LFA-1 via RAPL.51,58 Engagement of ITIM-containing inhibitory receptors on NK cells by MHC class I on target cells induces phosphorylation of the small adapter Crk and its dissociation from the scaffold proteins c-Cbl and p130CAS and from C3G.51 “Outside-in” LFA-1 signals are also sensitive to inhibition by ITIM-containing receptors. Furthermore, LFA-1–dependent lysis of insect cells expressing ICAM-1 and HLA-C was blocked by engagement of HLA-C–specific KIR on NK clones, as shown by direct loading of empty HLA-C with peptide.22
Although inhibition of inside-out signals for LFA-1 could be overcome by NKG2D and 2B4 synergy, the synergistic signals for degranulation remained sensitive to inhibition by CD94/NKG2A. It is noteworthy that the synergistic signals of NKG2D and 2B4 for degranulation are more sensitive to inhibition by CD94/NKG2A than those for inside-out signaling. A simple reason could be that stronger signals are required for degranulation. Alternatively, the point at which inhibitory signals intervene may not be at the level of receptor phosphorylation, but at a step further downstream, beyond a bifurcation of signals for degranulation and for inside-out signaling. In fact, the direct substrate of the tyrosine phosphatase SHP-1 recruited by phosphorylated ITIMs in inhibitory receptors is the guanine exchange factor Vav1.50,51 Thus, inside-out signaling could be less sensitive to inhibition if it is not as dependent on Vav1 activity as degranulation is. Underscoring the potency of CD94/NKG2A inhibitory receptor signaling, inhibition of degranulation persisted even at IgG concentrations that induced maximal CD16-mediated degranulation. These results are in line with experiments in which engagement of CD94/NKG2A by HLA-E potently inhibited natural cytotoxicity and ADCC by NKG2A+ NK-cell clones.21,32,59
Coengagement of receptors NKG2D, 2B4, and LFA-1 by ligands on Drosophila cells defined a minimal requirement for natural cytotoxicity by resting NK cells. Of note, robust degranulation in response to HLA-E was also observed on a subset of NKG2A−NKG2C+ NK cells. This suggests that, although NKG2C signaling is subject to inhibition in NKG2A+ NK cells, the small subset of NKG2A−NKG2C+ NK cells likely mediates NKG2C-dependent natural cytotoxicity. Thus, ITAM-associated NK receptors, such as CD16 and NKG2C, might be less dependent on coactivation than ITAM-independent NK cell receptors. Although coengagement of NKG2D and 2B4 was sufficient for NK cell degranulation, Drosophila cells expressing ligands for these 2 receptors were poorly lysed. Efficient killing occurred only when granule polarization was also induced, through coengagement of LFA-1. Patients with leukocyte adhesion disorder (LAD), an immunodeficiency caused by mutations affecting β2-integrin function, suffer severe recurrent bacterial infections and impaired immunity.60 Interestingly, a recent study reported that IL-2–activated NK cells from 2 patients diagnosed with leukocyte adhesion disorder (LAD) type 1, caused by mutations in the β2-integrin (CD18) gene, displayed only marginally impaired lysis of tumor cells.61 However, studies of LFA-1–deficient mouse NK cells have shown that LFA-1 is necessary for efficient cytotoxicity by IL-2 or IL-12–activated NK cells.62,63 Moreover, antibody-blocking of LFA-1–ICAM interaction impairs ADCC and natural cytotoxicity by human NK cells.64,65 The assessment of resting NK-cell cytotoxicity in LAD patients could provide more insight into the role of LFA-1 in natural cytotoxicity.
The assays described here for sensitive and rapid quantification of parameters of lymphocyte activation may be useful for diagnosis of human immunodeficiencies. Assessment of inside-out signals for LFA-1 may be useful for diagnosis of patients with clinical symptoms resembling LAD, but where LFA-1 expression is not perturbed, as observed in LAD type 3 patients.66 Furthermore, this assay could be useful for identifying defects in specific NK-cell receptors and their associated signaling pathways.
In conclusion, the data presented here show that individual NK- cell activating receptors may contribute to different activation events, but not necessarily in a given order. For example, coactivation receptors NKG2D, DNAM-1, 2B4, and CD2 facilitate contact and signals for LFA-1–dependent adhesion. At the same time, these receptors may cooperate with each other and with CD16 to induce degranulation. Moreover, inhibitory receptors block early signals for adhesion, as well as signals for degranulation. Therefore, our results revealed that target cell recognition by NK cells is a highly dynamic process controlled by the integration of signals from multiple receptors, many of which promote adhesion, some of which synergize to induce degranulation, and all of which are sensitive to inhibitory signaling by receptor CD94/NKG2A.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank D. Barber, M. Peterson, and S. Rajagopalan for the S2-CD80, S2–ICAM-1–CD80, S2-ULBP1, and S2–ICAM-1–ULBP1 cells, D. Staunton for the gift of 327C and 240Q mAbs, N. Hogg for the gift of mAb24 mAb, F. Borrego for peptides, and S. Wood for critical reading of the paper.
Y.T.B. was supported by the NIH–Karolinska Institutet Graduate Partnership Program, the Mary Beve Foundation, and the David and Astrid Hagelen Foundation. This research was supported by the Intramural Research Program of the NIH, NIAID (E.O.L.) and the Swedish Foundation for Strategic Research, Research Council and Cancer Society (H.-G.L.).
National Institutes of Health
Authorship
Contribution: Y.T.B. designed and performed the experimental work, analyzed and interpreted data, and drafted the paper; and H.-G.L. and E.O.L. designed research, analyzed and interpreted data, and drafted the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yenan T. Bryceson, Center for Infectious Medicine, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Huddinge, S-14186 Stockholm, Sweden; e-mail: yenan.bryceson@ki.se; or Eric O. Long, Laboratory of Immunogenetics, NIAID, NIH, 12441 Parklawn Dr, Rockville, MD 20852; e-mail: elong@nih.gov.


 represents target cells as indicated, whereas ■ represents target cells coexpressing ICAM-1 in addition to the other ligands, as indicated. Bars denote SD.
represents target cells as indicated, whereas ■ represents target cells coexpressing ICAM-1 in addition to the other ligands, as indicated. Bars denote SD.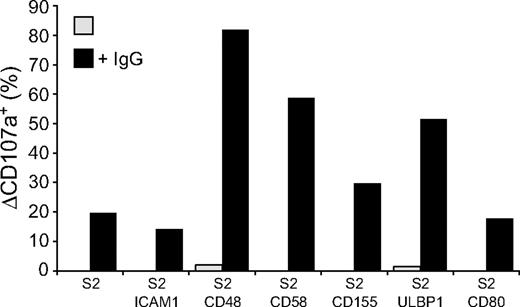
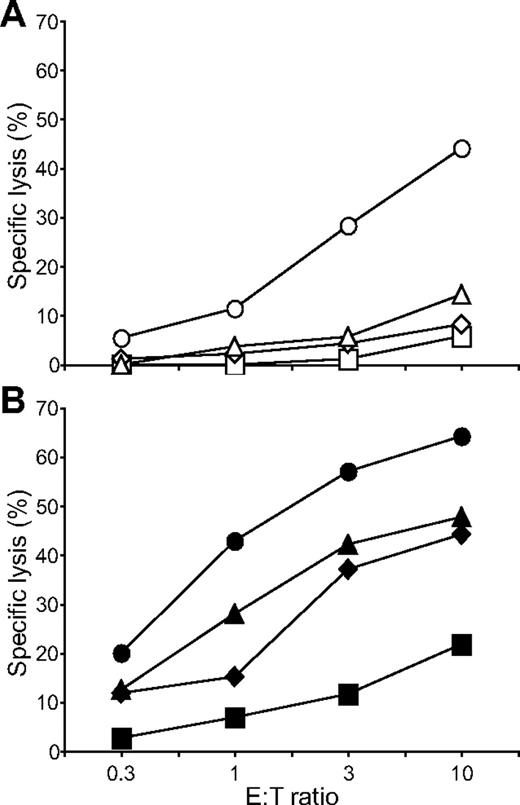
 ), S2-CD48-ULBP1 (△), or S2–ICAM-1–CD48–ULBP1 (○) cells, or (B) S2 cells as in panel A preincubated with a rabbit serum raised against S2 cells (■, ♦, ▲, ●). Cells were incubated for 3 hours at 37°C. Specific lysis of S2 cells was calculated from the percentage of propidium iodide–positive S2 cells in duplicate samples, as determined by flow cytometry. The experiment is representative of 3 or more independent experiments.
), S2-CD48-ULBP1 (△), or S2–ICAM-1–CD48–ULBP1 (○) cells, or (B) S2 cells as in panel A preincubated with a rabbit serum raised against S2 cells (■, ♦, ▲, ●). Cells were incubated for 3 hours at 37°C. Specific lysis of S2 cells was calculated from the percentage of propidium iodide–positive S2 cells in duplicate samples, as determined by flow cytometry. The experiment is representative of 3 or more independent experiments.