Abstract
Eosinophils are recruited to the lung in response to infection with pneumovirus pathogens and have been associated with both the pathophysiologic sequelae of infection and, more recently, with accelerated virus clearance. Here, we demonstrate that the pneumovirus pathogens, respiratory syncytial virus (RSV) and pneumonia virus of mice (PVM), can infect human and mouse eosinophils, respectively, and that virus infection of eosinophils elicits the release of disease-related proinflammatory mediators from eosinophils. RSV replication in human eosinophils results in the release of infectious virions and in the release of the proinflammatory mediator, interleukin-6 (IL-6). PVM replication in cultured bone marrow eosinophils (bmEos) likewise results in release of infectious virions and the proinflammatory mediators IL-6, IP-10, CCL2, and CCL3. In contrast to the findings reported in lung tissue of RSV-challenged mice, PVM replication is accelerated in MyD88 gene-deleted bmEos, whereas release of cytokines is diminished. Interestingly, exogenous IL-6 suppresses virus replication in MyD88 gene-deleted bmEos, suggesting a role for a MyD88-dependent cytokine-mediated feedback circuit in modulating this response. Taken together, our findings suggest that eosinophils are targets of virus infection and may have varied and complex contributions to the pathogenesis and resolution of pneumovirus disease.
Introduction
Eosinophils are unique proinflammatory leukocytes that are easily recognized but poorly understood. Although eosinophil expansion in the bone marrow and recruitment from the bloodstream to peripheral tissues are prominent findings that are clearly associated with asthma, allergic diseases, and infection with helminthes, there is no full consensus about the physiologic or pathophysiologic roles played by eosinophils in any of these disease states.1-3 Interestingly, among the many points that have emerged from the clinical studies featuring therapeutic administration of anti–interleukin 5 (IL-5) monoclonal antibody (mepolizumab),4-7 it has become evident that we have a very limited understanding of the subtleties of eosinophil recruitment and function; this limited understanding may relate to a long-standing focus on numbers of eosinophils recruited rather than on the nature, quality, and extent of eosinophil activation.
With this in mind, we have focused our studies on the role of eosinophils and of eosinophil activation in acute respiratory virus infections,8 specifically, infections with respiratory syncytial virus (RSV) and pneumonia virus of mice (PVM), a mouse pneumovirus pathogen that replicates the sequelae of the more severe forms of RSV infection in inbred strains of mice.9,10 Several converging lines of evidence suggest that eosinophils recruited to the lungs in response to pneumovirus infection may have a direct effect on the outcome of disease. Among these findings, several groups have detected immunoreactive eosinophil degranulation products in bronchial washings of patients with the most severe forms of RSV infection,11,12 and we and others have reported direct recruitment of eosinophils to the lungs of PVM-infected mice.13-15 RSV and PVM infections of their respective target cells result in the production of numerous proinflammatory cytokines, including the eosinophil chemoattractants RANTES and macrophage inflammatory protein-1α/CCL3,16-19 and RSV-infected target cells can activate eosinophils directly by virus-induced expression of ICAM-1.20 Most intriguing, children who have recovered from severe RSV infection frequently go on to develop prolonged postinfectious wheezing21 ; likewise, mice that have recovered from acute PVM infection experience persistent respiratory dysfunction with features similar to those described in mouse allergic asthma models.22
Interestingly, in many of the earliest studies, eosinophils were perceived as contributing only to the inflammatory pathophysiology and related negative sequelae typical of RSV disease. Our group was among the first to consider the possibility that eosinophils might be capable of promoting antiviral host defense, and thereby function in a positive as well as a negative sense (ie, a double-edged sword) in the setting of respiratory virus infection.23,24 Toward this end, we demonstrated that eosinophils and eosinophil granule proteins reduced the infectivity of RSV for their target epithelial cells in culture.25 Likewise, Adamko et al26 demonstrated eosinophil-mediated reduction in lung titers of Sendai virus (parainfluenza virus 1) in guinea pig infection and more recently, Phipps et al27 demonstrated toll-like receptor–dependent eosinophil-mediated clearance of RSV virions in a mouse challenge model.
Here, we explore another role for eosinophils and their interactions with respiratory virus pathogens, as we present evidence showing that both mouse and human eosinophils can be infected by pneumoviruses, serving as target cells supporting pneumovirus replication. We explore replication of RSV and PVM in human and in wild-type and MyD88 gene-deleted (MyD88−/−) mouse eosinophils, respectively, and consider the effect of cytokine release from eosinophils on the overall pathogenesis of pneumovirus disease.
Methods
Cell culture
BALB/c and C57BL/6 wild-type mice were purchased from Taconic Laboratories and MyD88−/− mice were a kind gift of Dr Shizuo Akira (Osaka University, Japan).28 All mice were handled according to the approved National Institute of Allergy and Infectious Diseases (NIAID) Animal Care and Use Committee protocol LAD-8E. Total bone marrow cells were collected from the femurs and tibiae of mice and cultured in 100 ng/mL stem cell factor and 100 ng/mL FLT-3L (PeproTech) for 3 days followed by 10 ng/mL IL-5 (R&D Systems Inc) for 7 to 10 days, a protocol that generates phenotypically mature eosinophils at high purity (∼ 99%) as previously described.29 RAW 264.7 (ATCC TIB-71; ATCC) cells were maintained in IMDM containing 10% FBS, 2 mM glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin. HEp-2 (ATCC CCL23; ATCC) cells were maintained in DMEM containing 10% FBS, 2 mM glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin.
Transmission electron microscopy
Cell pellets of bone marrow–derived eosinophils (bmEos) cultured as described from bone marrow cells from BALB/c mice were fixed overnight at 4°C with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.2. Samples were postfixed for 30 minutes with 0.5% osmium tetroxide/0.8% potassium ferricyanide, 1 hour with 1% tannic acid and overnight with 1% uranyl acetate at 4°C. Samples were dehydrated with a graded ethanol series and embedded in Spurr resin. Thin sections were cut with an RMC MT-7000 ultramicrotome (Ventana) and stained with 1% uranyl acetate and Reynold lead citrate before viewing at 80 kV on a Hitachi H-7500 transmission electron microscope (Hitachi). Digital images were acquired with a Hammamatsu XR-100 bottom mount CCD system (Advanced Microscopy Techniques; AMT) and processed with the use of Adobe Photoshop Version 7 (Adobe Systems Inc).
Virus stocks
PVM strain 15 was obtained from ATCC and was amplified in the mouse monocyte/macrophage RAW 264.7 cell line. Briefly, cells at 50% confluence were inoculated with PVM; 6 days after inoculation, cells were scraped into medium and sonicated to release cell-bound virions, and particulates were removed by centrifugation. Aliquots of clarified supernatant were flash frozen in dry ice–methanol bath and stored in liquid nitrogen until use. Sucrose gradient–purified RSV-A was purchased from Advanced Biotechnologies.
Challenge of bmEos with PVM
Before bmEos challenge, PVM stocks were thawed and dialyzed against 1000 volumes of phosphate buffered saline, pH 7.5, at 4°C to eliminate contaminating cytokines from RAW cell culture. BmEos at day 10 of culture were diluted to 106/mL in media as described29 that includes 10 ng/mL recombinant mouse (rm)IL-5 and challenged with PVM or heat-inactivated PVM (hiPVM; PVM virions heated to 95°C and then flash frozen in a dry ice–methanol bath 3 times) at a multiplicity of infection (MOI) of 1. The cells and virus were incubated together for 2 hours at 37°C in a humidified 5% CO2 incubator, after which the cells were washed twice and placed in fresh media containing 10 ng/mL rmIL-5. At the time points indicated, aliquots of culture media were collected for analysis of cytokine release, and aliquots of cells were collected for determination of virus titer. The volume of removed media was replaced at each harvest with media containing rmIL-5.
Challenge of human peripheral eosinophils with RSV-A
Human eosinophils were isolated from 40- to 60-mL samples of heparinized whole blood with a human eosinophil isolation kit (Miltenyi Biotec Inc). Purity was assessed by visual inspection of modified Giemsa-stained cytospin preparations. One million eosinophils maintained in 25 ng/mL recombinant human IL-5, used to maintain viability in long-term eosinophil cultures, were left unchallenged (control) or challenged with either RSV-A or equivalent quantities of heat-inactivated RSV-A (hiRSV-A; heat-inactivated as described for PVM) for 24 hours at 37°C, and supernatants were collected for detection of cytokine release. Virus titer was evaluated by qualitative reverse transcription polymerase chain reaction (Q-RT-PCR) on total RNA from eosinophils challenged with virus (MOI = 1) for 2 hours, washed, resuspended in complete medium with 25 ng/mL IL-5 (day 0); additional aliquots of cells remained in culture for 4 days, after which virus titer was determined. Recombinant green RSV (rgRSV,30 passage 4) was graciously provided by Drs Mark Peeples (Ohio State University) and Peter Collins (National Institutes of Health). Stocks provided were amplified in HEp-2 cells and used to infect isolated human eosinophils at an MOI less than 2 pfu/cell. Fluorescent cells were detected after 4 days and photographed at 7 days.
Determination of virus titer by Q-RT-PCR
RNA samples were isolated from bmEos challenged with PVM and from human eosinophils challenged with RSV with the use of the SuperArray RT2 qPCR-Grade RNA isolation kit as per the manufacturer's directions; RNA samples were treated with DNase I as part of the isolation procedure. Titers of PVM virus stocks were determined by isolation of RNA directly from virions in suspension with the RNazol reagent (RNazol B). Isolated RNA was reverse transcribed with a First-Strand cDNA Synthesis Kit for RT-PCR (AMV; Roche Diagnostics). Primers, probes, and standard curves for PVM small hydrophobic (SH) and mouse GAPDH genes were reported in previous publications.31,32 Reactions were performed in an ABI 7500 Sequence detector, cycling parameters including 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. PVM titer is reported as copies PVM genome per copy mouse GAPDH, based on quantitative detection of the virus SH gene (SHPVM/GAPDH). RSV titer is reported as copies RSV genome per copy human GAPDH, based on quantitative detection of the virus nucleoprotein (N) gene (NRSV/GAPDH). Primers used to amplify RSV N gene includes the following: forward primer, 5′-TGA TAC ACT CAA CAA AGA TCA ACT TCT G-3′, and reverse primer, 5′-TGA TAC ACT CAA CAA AGA TCA ACT TCT G-3′; the probe used to detect the RSV N gene is 5′-6FAM CAT CCA GCA AAT ACA C MGBNFQ-3′. The primers/probe set used to detect the human GAPDH gene was purchased from ABI (4326317E). Copy numbers are determined with respect to standard curves determined from serial 10-fold dilutions of plasmids containing the RSV N gene (GenBank accession no. DQ780565, nucleotides 1-1176) and human GAPDH gene (GenBank accession no. NM002046, nucleotides 41-1246), each cloned into the PCR 2.1 backbone, respectively. Cycling parameters are as described for the PVM Q-RT-PCR assay.
Purification of anti-NPVM antibody and Western analysis
Protein from bmEos was collected by lysing 106 cells in 5 μL triple detergent lysis buffer (50 mM Tris, pH 8.0, 1 mM EDTA, 100 mM NaCl, 1% NP-40, 0.5% Triton-100, 0.05% SDS). We previously detected PVM replication by Western analysis with rabbit antisera specific for a linear epitope of the N protein (aa 379-393; GenBank no. AAS8736017 ). The antibody fraction of this antisera was isolated and concentrated on a HiTrap Protein A column (Sigma-Aldrich) according to the manufacturer's instructions, and the protein concentration was adjusted to 1 mg/mL. The purified antibody fraction was used to detect PVM in bmEos extracts by Western blotting at a 1:100 dilution followed by a goat anti–rabbit IgG-AP secondary (Bio-Rad). The membranes were developed with BCIP and NBT in alkaline development buffer (Bio-Rad). Chicken anti-GAPDH (US Biological) was used in conjunction with anti–IgY-AP secondary antibody (US Biological) to confirm loading.
Cytokine measurements
Cytokine concentrations (with the exception of IFNα and IFNβ) were determined with 16-plex and 22-plex bead assays (Linco Research). The assays were read on a Bioplex (Bio-Rad) plate reader, and the values of the experimental data points were extrapolated from a standard curve generated by the instrument software with a 5-parameter logistic method. Concentrations of IFNα and IFNβ were determined by enzyme-linked immunoabsorbent assay (PBL Biomedical Laboratories).
Detection of infectious virions released by cultured eosinophils
Supernatants from infected eosinophil cultures (mouse bmEos infected with PVM or human eosinophils infected with RSV) were collected at the time points indicated and stored at −80°C until use. Aliquots of these culture supernatants were used to inoculate either RAW264.7 cells (for assessing the presence of infectious PVM virions) or HEp-2 cells (for assessing the presence of infectious RSV virions). Supernatants from eosinophil culture (200 μL bmEos culture supernatant or 100 μL human eosinophil culture supernatant) were added to RAW264.7 or HEp-2 cell cultures, respectively, in 1 mL media. After 5 days (RAW 264.7 cells challenged with PVM-infected bmEos supernatants) or 7 days (HEp-2 cells challenged with RSV-infected human eosinophils supernatants), total cellular RNA was isolated, and the absolute copy number of PVM (SHPVM/GAPDH) or RSV (NRSV/GAPDH), respectively, was determined by Q-RT-PCR.
Statistical analysis
Datasets were analyzed with statistics package provided in GraphPad Prism 5 (GraphPad Software Inc). All data are reported as the mean plus or minus SEM. Statistical outliers were detected with the use of the Grubbs test (extreme studentized deviate) and excluded from further analysis.
Results
Electron microscopic characterization of bmEos
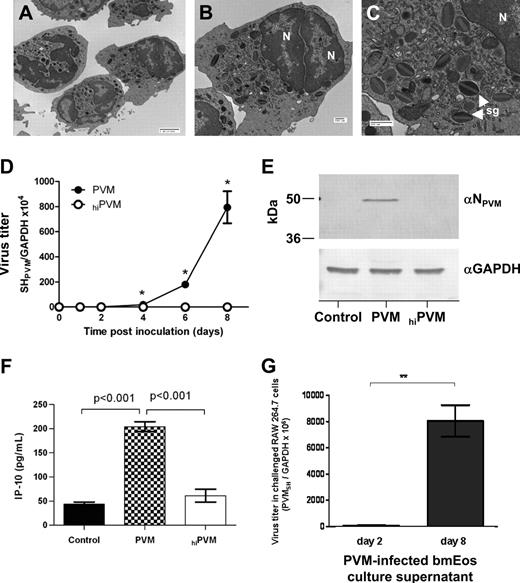
In a previous study, we described an original ex vivo culture method for generating phenotypically mature, functional eosinophils at high purity from normal BALB/c mouse bone marrow progenitors (bmEos)29 ) Mouse bmEos express immunoreactive major basic protein, IL-5 receptor α chain, and Siglec F; produce characteristic cytokines; and undergo chemotaxis in response to the eosinophil-specific ligand, eotaxin-1. Here, we document the subcellular morphology of mouse bmEos and identify features typical of mature eosinophils, including multilobed eccentric nuclei (Figure 1A-B) and distinct lucent-to-dense layered specific granules found throughout the cytoplasm (Figure 1C).
PVM replicates in mouse bone marrow–derived eosinophils. Electron micrographs of mouse bone marrow–derived eosinophils (bmEos) documenting morphologic features typical of eosinophils, including the bilobed nucleus (N) and specific granules in the cytoplasm (sg). Magnification: (A) ×2500, bar = 2 μm; (B-C) ×5000 and ×12000, respectively; bar = 500 nm. (D) Q-RT-PCR detection of virus replication in bmEos. bmEos were inoculated with PVM (●) or heat-inactivated PVM (hiPVM; ○) at a multiplicity of infection (MOI) of 1; *P < .05. Data shown are representative of 2 experiments performed in triplicate. (E) PVM N protein is detected in infected bmEos at day 8 after inoculation. Total protein extracts from control, PVM-infected, or hiPVM-challenged bmEos were probed with anti-PVM N peptide antibody (αNPVM). Anti–human GAPDH (αGAPDH) antibody was used as a control for protein loading. (F) Interferon-γ–induced protein (IP-10/CXCL-10) released from bmEos on day 4 after inoculation in response to PVM infection, hiPVM-challenge, or control challenge. Results are representative of 2 experiments performed in triplicate, P values as indicated. (G) Infectious virions are released from PVM-infected bmEos. Supernatants from bmEos cultures at 2 or 8 days after inoculation were used to challenge cells of the RAW 264.7 cell line, which is highly permissive for PVM replication.14 Total RNA from the RAW 264.7 cells was harvested at 5 days after inoculation, and Q-RT-PCR was performed to determine virus copy number. Results are data combined from 3 experiments, each performed in triplicate, **P < .01. Data represent mean ± SEM.
PVM replicates in mouse bone marrow–derived eosinophils. Electron micrographs of mouse bone marrow–derived eosinophils (bmEos) documenting morphologic features typical of eosinophils, including the bilobed nucleus (N) and specific granules in the cytoplasm (sg). Magnification: (A) ×2500, bar = 2 μm; (B-C) ×5000 and ×12000, respectively; bar = 500 nm. (D) Q-RT-PCR detection of virus replication in bmEos. bmEos were inoculated with PVM (●) or heat-inactivated PVM (hiPVM; ○) at a multiplicity of infection (MOI) of 1; *P < .05. Data shown are representative of 2 experiments performed in triplicate. (E) PVM N protein is detected in infected bmEos at day 8 after inoculation. Total protein extracts from control, PVM-infected, or hiPVM-challenged bmEos were probed with anti-PVM N peptide antibody (αNPVM). Anti–human GAPDH (αGAPDH) antibody was used as a control for protein loading. (F) Interferon-γ–induced protein (IP-10/CXCL-10) released from bmEos on day 4 after inoculation in response to PVM infection, hiPVM-challenge, or control challenge. Results are representative of 2 experiments performed in triplicate, P values as indicated. (G) Infectious virions are released from PVM-infected bmEos. Supernatants from bmEos cultures at 2 or 8 days after inoculation were used to challenge cells of the RAW 264.7 cell line, which is highly permissive for PVM replication.14 Total RNA from the RAW 264.7 cells was harvested at 5 days after inoculation, and Q-RT-PCR was performed to determine virus copy number. Results are data combined from 3 experiments, each performed in triplicate, **P < .01. Data represent mean ± SEM.
PVM replicates in mouse bmEos
Mouse bmEos were challenged with PVM (MOI = 1) or an equivalent number of virions of hiPVM. After 2 hours, virus was removed, the cells were provided fresh medium, and virus replication was assessed at subsequent time points by Q-RT-PCR. Elevated virus titers were detected in PVM-infected mouse bmEos cultures beginning on day 4 and continuing through day 8 (Figure 1D), with replication occurring at a constant rate. No replication was detected in mouse bmEos inoculated with hiPVM. Of note, virus infection has no effect on the total cell number nor did it alter cell viability in culture (supplemental Figure 1A-B, available on the Blood website; see the Supplemental Materials link at the top of the online article). Furthermore, by day 4 of virus infection, 100% of the cells in culture were phenotypically mature eosinophils (supplemental Figure 1C). Thus, although we cannot rule out the possibility that other cells become infected when PVM virions are first introduced to the culture on day 0, by day 4, we are observing accelerated replication within a homogenous culture consisting of 100% eosinophils. Consistent with the Q-RT-PCR results, immunoreactive PVM N protein was detected in Western blot of total protein extracts from bmEos at day 8 after inoculation with PVM, but not in extracts from cells that were inoculated with hiPVM or from control cells (Figure 1E).
PVM infection elicits release of IP-10 from mouse bmEos
In previous work, we demonstrated replication-dependent release of IL-6 from bmEos in response to PVM infection.29 Here, we find the same to be true for the chemoattractant cytokine, IP-10/CXCL-10 (Figure 1F). IP-10 release over background levels was not observed in response to challenge with hiPVM, suggesting that intracellular replication or challenge or both in response to an increasing number of released extracellular virions is crucial to elicit IP-10 release.
PVM infection of bmEos results in production and release of infectious virions
BmEos cultures were inoculated with PVM by standard method (virus challenge for 2 hours, virus removed, and fresh medium added). Culture supernatants were sampled at 2 days and 8 days after initial challenge and used to infect cells of the highly susceptible mouse monocyte/macrophage RAW 264.7 cell line17 to detect release of infectious PVM virions from bmEos. As shown in Figure 1G, there are 130-fold more infectious virions in the supernatants of day 8 bmEos cultures than can be detected in the day 2 cultures. These data indicate that cultured mouse eosinophils support productive virus replication ex vivo.
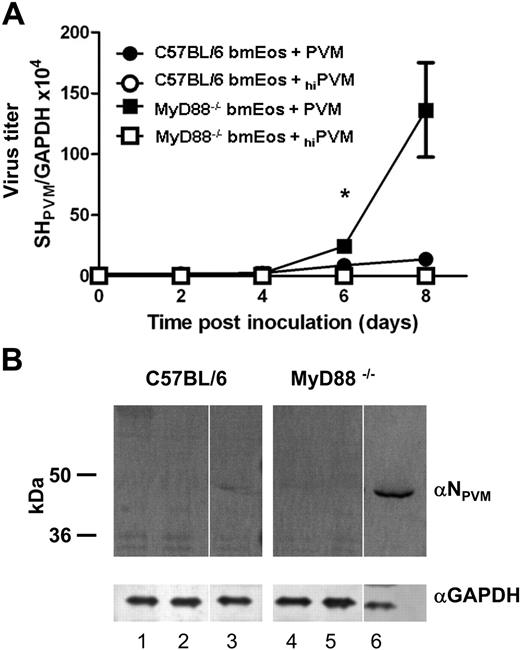
PVM replication is accelerated in the absence of MyD88
Bone marrow progenitors from wild-type C57BL/6 and MyD88−/− mice develop into high-purity cultures of phenotypically mature eosinophils in a manner indistinguishable from those from BALB/c (K.D.D., unpublished data, July 2009). Both wild-type and MyD88−/− mouse bmEos were inoculated with PVM (MOI = 1) as described in “Challenge of bmEos with PVM.” Compared with bmEos from the C57BL/6 parent strain, accelerated rates of virus replication were detected in the MyD88−/− mouse bmEos cultures; specifically, 10- to 20-fold higher virus titers were detected by Q-RT-PCR in the MyD88−/− bmEos on day 8 after inoculation (Figure 2A). Differential detection of immunoreactive PVM N protein in total protein extracts from mouse bmEos was consistent with the Q-RT-PCR findings (Figure 2B).
Accelerated replication of PVM in mouse bmEos from MyD88−/− mice. (A) Q-RT-PCR detection of virus titer in bmEos. Virus titer was determined in PVM-infected (filled symbols) and hiPVM-challenged (open symbols) wild-type (C57BL/6; circles) and MyD88−/− mouse bmEos (squares). Values shown are representative of 2 experiments performed in triplicate (data shown represent mean ± SEM); *P < .05. (B) Immunodetection of PVM in extracts of bmEos. PVM was detected at 9 days after inoculation of bmEos cultures on a Western blot probed with rabbit polyclonal anti-PVM N peptide antibody (αNPVM). Lanes 1 to 3 are extracts from bmEos from wild-type C57BL/6 mice; lanes 4 to 6, extracts from bmEos from MyD88−/− mice; lanes 1 and 4, from unchallenged bmEos (controls); lanes 2 and 5, from hiPVM-challenged bmEos; lanes 3 and 6, from PVM-infected bmEos. After probing with anti-PVM N antibody the blot was stripped and probed with anti–mouse GAPDH (αGAPDH) as a control for protein loading. Blot shown is single membrane, divided to omit intervening protein marker lanes.
Accelerated replication of PVM in mouse bmEos from MyD88−/− mice. (A) Q-RT-PCR detection of virus titer in bmEos. Virus titer was determined in PVM-infected (filled symbols) and hiPVM-challenged (open symbols) wild-type (C57BL/6; circles) and MyD88−/− mouse bmEos (squares). Values shown are representative of 2 experiments performed in triplicate (data shown represent mean ± SEM); *P < .05. (B) Immunodetection of PVM in extracts of bmEos. PVM was detected at 9 days after inoculation of bmEos cultures on a Western blot probed with rabbit polyclonal anti-PVM N peptide antibody (αNPVM). Lanes 1 to 3 are extracts from bmEos from wild-type C57BL/6 mice; lanes 4 to 6, extracts from bmEos from MyD88−/− mice; lanes 1 and 4, from unchallenged bmEos (controls); lanes 2 and 5, from hiPVM-challenged bmEos; lanes 3 and 6, from PVM-infected bmEos. After probing with anti-PVM N antibody the blot was stripped and probed with anti–mouse GAPDH (αGAPDH) as a control for protein loading. Blot shown is single membrane, divided to omit intervening protein marker lanes.
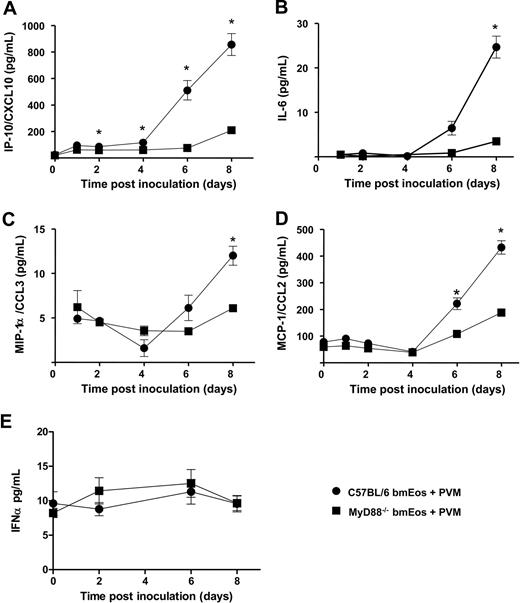
PVM infection elicits MyD88-dependent cytokine release
Supernatants from PVM-infected mouse wild-type and MyD88−/− bmEos were evaluated for cytokine release at days 0 to 8 after inoculation. Diminished release of IP-10/CXCL10 from MyD88−/− bmEos was a most prominent finding, evident as early as day 2 after virus inoculation, and remaining significant through day 8 (Figure 3A). Diminished release of IL-6 (Figure 3B) and CC chemokines MIP-1α/CCL3 (Figure 3C) and MCP-1/CCL2 (Figure 3D) was also evident. Because virus replication does not induce transcription of these mediators (supplemental Figure 2), we conclude that they are most likely released from preformed stores in both wild-type and MyD88−/− bmEos. Interestingly, PVM infection did not induce differential release or transcription of IFNα (Figure 3E; supplemental Figure 2); no IFNβ or IFNγ was detected.
Cytokine release is MyD88-dependent. Differential release of (A) IP-10/CXCL10, (B) IL-6, (C) MIP-1α/CCL3, (D) MCP-1/CCL2, and (E) IFNα from PVM-infected wild-type (●) and MyD88−/− (■) mouse bmEos cultures. (A-D) Data were determined by multiplex bead assay, n = 3; *P < .05. (E) Data were results from enzyme-linked immunoabsorbent assay, n = 4. Data shown represent mean ± SEM.
Cytokine release is MyD88-dependent. Differential release of (A) IP-10/CXCL10, (B) IL-6, (C) MIP-1α/CCL3, (D) MCP-1/CCL2, and (E) IFNα from PVM-infected wild-type (●) and MyD88−/− (■) mouse bmEos cultures. (A-D) Data were determined by multiplex bead assay, n = 3; *P < .05. (E) Data were results from enzyme-linked immunoabsorbent assay, n = 4. Data shown represent mean ± SEM.
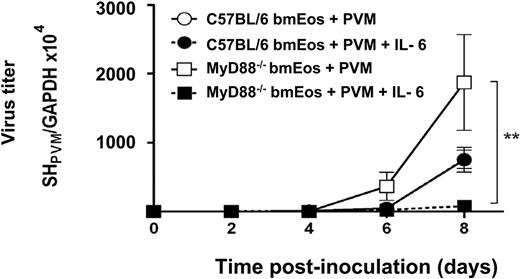
IL-6 suppresses virus replication in MyD88−/− bmEos
In an attempt to relate the augmented virus replication (Figure 2A) and diminished cytokine release (Figure 3) observed in bmEos in the absence of MyD88, the effect of exogenous IL-6 on virus replication on both wild-type and MyD88−/− bmEos was examined (Figure 4).
Recombinant IL-6 suppresses virus replication MyD88−/− bmEos. Cells were infected at MOI of 1 and resuspended in media with or without IL-6 (20 ng/mL). At time points indicated, RNA was isolated from the cells, and RT-PCR was performed to determine absolute virus copy number. Experimental conditions are as indicated, points for the wild-type C57BL/6 with and without IL-6 are superimposed over one another; n = 3 mice per experimental condition, *P < .01. Data shown represent mean ± SEM.
Recombinant IL-6 suppresses virus replication MyD88−/− bmEos. Cells were infected at MOI of 1 and resuspended in media with or without IL-6 (20 ng/mL). At time points indicated, RNA was isolated from the cells, and RT-PCR was performed to determine absolute virus copy number. Experimental conditions are as indicated, points for the wild-type C57BL/6 with and without IL-6 are superimposed over one another; n = 3 mice per experimental condition, *P < .01. Data shown represent mean ± SEM.
IL-6 (20 ng/mL) had no effect on virus replication in infected wild-type bmEos, but it diminished the extent of replication observed in MyD88−/− bmEos by nearly 25-fold; indeed, the addition of exogenous IL-6 reduced virus titers in MyD88−/− mice to less then that of wild-type bmEos. Note the differences in virus copy number comparing the results from the wild-type C57BL/6 bmEos alone here with those observed in the set of experiments reported in Figure 2A. We are not certain what factors contributed to the differences in copy number observed between experiments.
RSV replicates in human eosinophils
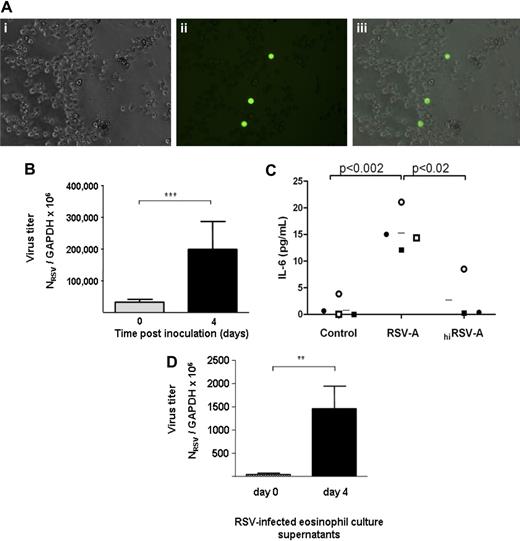
For the following experiments, human eosinophils were isolated from peripheral blood by negative selection (see “Challenge of human peripheral eosinophils with RSV-A”). Eosinophil viability ex vivo is sustained with the addition of recombinant IL-5 (reviewed in Hogan et al1 ). Without cytokine support, isolated eosinophil cultures sustain 30% to 40% loss of viability within 24 hours and are 80% nonviable within 5 days (data not shown). Isolated human eosinophils were challenged with recombinant GFP-tagged RSV at an MOI of approximately 2, and green fluorescent protein was readily detected within these cells at 4 to 7 days in culture (Figure 5A). Virus replication within human eosinophils was measured over time (Figure 5B). At day 0, eosinophils were challenged with RSV (MOI = 1) for 2 hours, washed, resuspended, and evaluated immediately (0 days after infection) or maintained in culture for 4 days, after which the increase in virus titer over that observed at day 0 was 7-fold. Analogous to cultured mouse eosinophils,29 virus replication, RSV infection, but not challenge with hiRSV elicits release of IL-6 from human eosinophils (Figure 5C). Other cytokines released in response to both RSV and hiRSV include IL-1α, IL-13, IL-15, G-CSF, and GM-CSF; CXCL-10/IP-10, CCL3/MIP-1α, CCL2/MCP-1, and IFN-γ were not detected. Finally, analogous to cultured mouse eosinophils, human eosinophils produce and release infectious virions (Figure 5D). Culture supernatants collected on day 0 and day 4 were used to challenge the HEp-2 human epithelial cell line; 6-fold more infectious virus copies were detected in the eosinophil culture supernatants at day 4 than at day 0, indicative of a productive infection.
Respiratory syncytial virus replicates in human eosinophils. (A) Green fluorescent protein detected in eosinophils after challenge with rgRSV. Shown are (i) phase contrast, (ii) fluorescent, and (iii) composite of panels i and ii, showing expression of green protein in eosinophils. Photographed with 32× objective in 6-well culture dish in media. (B) Replication of RSV in human eosinophils. Cells were challenged with respiratory syncytial virus (RSV; MOI = 1) for 2 hours, washed, and resuspended, and total RNA was isolated immediately (day 0) or 4 days later for determination of virus copy number. Results are pooled data from 3 separate experiments, ***P < .001. (C) IL-6 is released by RSV-infected eosinophils. Detection of IL-6 by multiplex bead assay in culture supernatants from unchallenged (control), RSV-infected, and hiRSV-challenged eosinophils. Each symbol represents an independent eosinophil donor; n = 3 to 4; *P < .02 and **P < .002. (D) Infectious virions are released from RSV-infected human eosinophils. Supernatants from human eosinophil cultures at 0 or 4 days after inoculation were used to challenge cells of the HEp-2 line, which is highly permissive for RSV replication. Total RNA from the HEp-2 cells was harvested at 7 days after inoculation, and Q-RT-PCR was performed to determine virus copy number. Results shown are a single experiment performed in triplicate, *P < .01. Data shown represent mean ± SEM.
Respiratory syncytial virus replicates in human eosinophils. (A) Green fluorescent protein detected in eosinophils after challenge with rgRSV. Shown are (i) phase contrast, (ii) fluorescent, and (iii) composite of panels i and ii, showing expression of green protein in eosinophils. Photographed with 32× objective in 6-well culture dish in media. (B) Replication of RSV in human eosinophils. Cells were challenged with respiratory syncytial virus (RSV; MOI = 1) for 2 hours, washed, and resuspended, and total RNA was isolated immediately (day 0) or 4 days later for determination of virus copy number. Results are pooled data from 3 separate experiments, ***P < .001. (C) IL-6 is released by RSV-infected eosinophils. Detection of IL-6 by multiplex bead assay in culture supernatants from unchallenged (control), RSV-infected, and hiRSV-challenged eosinophils. Each symbol represents an independent eosinophil donor; n = 3 to 4; *P < .02 and **P < .002. (D) Infectious virions are released from RSV-infected human eosinophils. Supernatants from human eosinophil cultures at 0 or 4 days after inoculation were used to challenge cells of the HEp-2 line, which is highly permissive for RSV replication. Total RNA from the HEp-2 cells was harvested at 7 days after inoculation, and Q-RT-PCR was performed to determine virus copy number. Results shown are a single experiment performed in triplicate, *P < .01. Data shown represent mean ± SEM.
Discussion
In this work, we have demonstrated that the pneumoviruses RSV and PVM are capable of replicating and producing infective virus in human and mouse eosinophils, respectively. To the best of our knowledge, this is the first demonstration of pneumovirus replication in eosinophils, cells with limited biosynthetic capacity (reviewed in Hogan et al1 ). Interestingly, Kimpen et al33 have previously demonstrated RSV uptake by human eosinophils and localized virions in intracellular vesicles as early as 2 hours after challenge, although no active replication or infection was shown. More recently, Kolokoltsov et al34 have demonstrated pH-independent uptake and endosome formation as a crucial first step in RSV infection of human epithelial cells. Intracellular replication leading to the production of infectious virions within eosinophils has not been tracked morphologically. Similarly, the endosomal compartment has not been characterized extensively in eosinophils, although Lei and Martinez-Moczygemba35 have explored IL-5 receptor internalization by clathrin-coated endocytic vesicles, thus documenting the existence and functionality of this mechanism in this lineage. As various TLRs have likewise been localized in this compartment,36 one or more that are expressed on eosinophils and that transduce signals from pneumoviruses, such as TLR7,27 may be activated there as well as at the cell surface.
Although the intracellular cytokines stored in and released from human eosinophils have been characterized extensively,37 the pool of preformed cytokines stored in mouse eosinophils have been subjected to only limited analysis. Although morphologically similar to one another, human and mouse eosinophils have some profound differences, such as distinct responses to chemotactic cytokines, differential expression of IgE receptors and sialic acid binding lectins, and highly divergent specific granule proteins (reviewed in Rosenberg et al38 ). As such, there may be substantial differences in intracellular cytokine content. That said, one of the most prominent responses observed in mouse eosinophils is the replication-dependent release of the chemotactic cytokine, IP-10 (CXCL10). IP-10 is a chemoattractant for monocytes, T cells, and natural killer cells and has been detected in activated neutrophils39 ; this is the first report of IP-10 release from eosinophils. Likewise, no published reports have documented IP-10 in clinical samples from patients infected with RSV, but IP-10 has been detected in RSV-infected epithelial cell cultures40,41 and in response to RSV challenge in mice.42,43 Transcript-encoding IP-10 has also been detected in response to PVM infection; IP-10 is among the cytokines differentially expressed in PVM-infected wild-type versus IFNαβR gene–deleted mice.32
The finding that PVM replication is accelerated in eosinophils derived from MyD88−/− mice is particularly intriguing in light of the fact that Rudd et al44 as well as Bhoj et al45 have found no significant differences in virus titer in lung tissue of RSV-challenged wild-type versus MyD88−/− mice. There are several explanations for these seemingly discordant findings. It is certainly possible that eosinophils represent a unique niche for pneumovirus replication; likewise, it is possible that PVM has interactions with TLR signaling pathways that are distinct from those of RSV. However, the most important distinction between these observations is that in this work we are exploring responses to PVM in a mouse model that supports substantial virus replication. In contrast, RSV inoculation in mice is a challenge-clearance model27,46,47 (reviewed in Rosenberg and Domachowske10 ). As such, our findings suggest that MyD88 has more of a profound effect on regulating the rate of intracellular virus replication than has previous been recognized.
In a series of studies that address this issue directly, Xiong et al48 and Lin et al49 have determined that overexpression of MyD88 inhibited replication of hepatitis B virus. In this work, we found that PVM infection elicited the release of IL-6 from bmEos (Figure 3B). In the absence of the TLR signaling adapter, MyD88, release of IL-6 was diminished substantially, and virus replication proceeded at an accelerated rate. Interestingly, supplementation with IL-6 resulted in the suppression of virus replication in MyD88−/− bmEos, suggesting the existence of a MyD88-dependent IL-6–mediated feedback mechanism. The details of this mechanism, the specificity for eosinophils, and/or its existence as a more general feature of pneumovirus infection in other target cells remain to be determined. We also show that RSV infection of human eosinophils results in the replication-dependent release of IL-6, a cytokine localized to specific granules50 and released in response to mast cell chymase, IL-23, and various TLR ligands.51-53 Although the precise role of IL-6 in the overall clinical picture of RSV disease has not been discerned, Bennett et al54 and Oh et al55 independently evaluated intranasal samples from patients with RSV and other virus infections and found elevated levels of proinflammatory cytokines, including IL-6.
Among the questions resulting from this study: because eosinophils are targets of virus infection, can support genome replication, and can release infectious virions, yet at the same time, more or less in association with these activities, eosinophils release mediators (such as IL-6) that suppress these responses in themselves and probably in other infected cells, what is the ultimate outcome of these events vis-à-vis health and disease? Are eosinophils contributing to the disease process or are they promoting host defense? This question has no simple answer. Eosinophils themselves, depending on factors that we do not yet understand, have antiviral properties in vitro23-25 and can clear virus from infected lung tissue in vivo,26,27 although not in all circumstances.56 At the same time, respiratory virus infection is not only a disease of virus replication, because we and others have shown that inflammatory mediators produced locally have a profound effect on the clinical course.22,57,58 Thus, although eosinophils as targets of virus infection may be releasing a limited number of infectious virions, this might be outweighed by the release of important immunomodulatory cytokines. The interplay of these factors is difficult to predict but potentially intriguing.
In summary, we document for the first time the observation that human and mouse eosinophil leukocytes might serve as targets for pneumovirus replication and infection. We find that infection of eosinophils with the human pneumovirus, RSV, is associated with replication-dependent release of the pleiotropic cytokine IL-6, which has been detected in clinical RSV samples and in numerous experimental settings. The natural rodent pneumovirus pathogen, PVM, replicates in mouse eosinophils, and infection of MyD88−/− mouse eosinophils results in accelerated rates of virus replication accompanied by diminished release of cytokines, including IL-6, IP-10, CCL3, and CCL2. Furthermore, we document the MyD88-dependent role of IL-6 in suppressing virus replication. Taken together, our findings suggest that eosinophils, which are recruited to the lungs in response to virus infection in vivo, may have varied and mechanistically complex contributions to the pathogenesis of severe pneumovirus disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Mark Peeples and Dr Peter Collins for sharing recombinant rgRSV, Dr Shizuo Akira for his generous gift of MyD88−/− mice, and Dr Brian Kelsall (NIAID/National Institutes of Health) for facilitating their transfer to us. We also thank all of the members of the Eosinophil Biology Section, Laboratory of Allergic Diseases (LAD), NIAID for their thoughtful comments and suggestions.
This work is supported by Division Intramural Research (DIR) funding from the NIAID (H.F.R.).
National Institutes of Health
Authorship
Contribution: K.D.D. designed the primary experiments, executed most of the technical work, designed the figures, and wrote the manuscript; C.M.P. contributed major sections of the RSV infection of human eosinophils experiment; E.R.F. executed the ultrastructural analysis of the mouse bone marrow–derived eosinophils; S.J.G. executed technical work that contributed toward the revised version of the manuscript; and H.F.R. provided direction for the study and wrote and edited the manuscript for submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kimberly Dyer, 10 Center Dr, Bldg 10 Rm 11C216, Bethesda, MD 20892-1883; e-mail: kdyer@niaid.nih.gov.