Abstract
Histologic transformation (HT) of follicular lymphoma to diffuse large B-cell lymphoma (DLBCL-t) is associated with accelerated disease course and drastically worse outcome, yet the underlying mechanisms are poorly understood. We show that a network of gene transcriptional modules underlies HT. Central to the network hierarchy is a signature strikingly enriched for pluripotency-related genes. These genes are typically expressed in embryonic stem cells (ESCs), including MYC and its direct targets. This core ESC-like program was independent of proliferation/cell-cycle and overlapped but was distinct from normal B-cell transcriptional programs. Furthermore, we show that the ESC program is correlated with transcriptional programs maintaining tumor phenotype in transgenic MYC-driven mouse models of lymphoma. Although our approach was to identify HT mechanisms rather than to derive an optimal survival predictor, a model based on ESC/differentiation programs stratified patient outcomes in 2 independent patient cohorts and was predictive of propensity of follicular lymphoma tumors to transform. Transformation was associated with an expression signature combining high expression of ESC transcriptional programs with reduced expression of stromal programs. Together, these findings suggest a central role for an ESC-like signature in the mechanism of HT and provide new clues for potential therapeutic targets.
Introduction
Follicular lymphoma (FL), the most common indolent subtype of non-Hodgkin lymphoma, is clinically heterogeneous and remains largely incurable. Persons with FL frequently experience histologic transformation (HT) to a more aggressive histology, such as diffuse large B-cell lymphoma (DLBCL), usually followed by rapid disease progression, resistance to therapy, and death.1-3 Estimates of the probability of HT have varied significantly from 10% to 60%; and given biases in ascertainment, it has been difficult to discern whether potential for transformation is an inherent property of all FL tumors.4 A variety of factors have been associated with risk of HT, including histologic grade and stage of disease at diagnosis, although in most series the strongest predictor remains the Follicular Lymphoma Prognostic Index.2
A comprehensive understanding of the mechanisms underlying HT of FL is important for identifying patients at risk and guiding novel therapies. Several studies have investigated global gene expression and genetic changes associated with HT,5-9 and implicated several pathways and processes, including deregulation of MYC and its target genes,8 activation of mitogen-activated protein kinase signaling,10 activation of proliferation pathways,5 preservation of a germinal center B-cell like phenotype,5 interactions with infiltrating T cells and the FL nodal microenvironment.7,11 Given these numerous and seemingly heterogeneous findings, the most critical factors underlying HT remain obscure.
In an effort to better understand HT, we constructed a module network from gene expression profiles of patients with FL and transformed DLBCL.12 This method does not assume a uniform shift in expression patterns of genes between phenotypic classes but instead allows for more complex changes in expression that probably underlie molecular networks. Module networks have been successfully applied to dissecting regulatory networks in yeast microarray data, and in the analysis of human cancer, yielding experimentally validated models of regulatory relationships.13 Given a microarray dataset, the aim of module network construction is to deduce the most probable assignment of genes to modules (ie, groups of genes with correlated expression profiles) and regulatory programs that could have produced the observed expression data (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Defining the unit of analysis as coherent changes in collections of genes (modules) can facilitate interpretation and understanding of the biologic processes underlying expression changes.13
Here, we describe the transcriptional module hierarchy in the network underlying HT, and the relationship of constituent modules to known pathways and biologic processes (Figure 1). Strikingly, central to the hierarchy predicting HT, we found a core program highly enriched for genes typically expressed in embryonic stem cells (ESCs). This ESC-like signature overlapped with a similar expression program distinguishing the centroblast stage of normal B-cell development within germinal centers, with the exception of MYC and several of its direct targets. When assessed in separate patient cohorts and independent data, expression of this signature was associated with shorter survival of patients with FL and was predictive of their HT to DLBCL. Our results resonate with other recent studies implicating inappropriate activation of stem cell–like transcriptional programs in cancer initiation, aggressiveness, and progression.14-16
Overview of analysis. A module network was constructed from expression data on FL/DLBCL-t7 using Genomica. Modules were annotated by comparison with curated gene sets, and a core network, putatively underlying transformation, was identified that involved differentiation/stem cell–related transcriptional programs. We selected modules for incorporation into a survival model using stepwise regression. The survival model was validated in an independent set from a different group.11 Survival analysis based on core modules was performed with and without proliferation genes.
Overview of analysis. A module network was constructed from expression data on FL/DLBCL-t7 using Genomica. Modules were annotated by comparison with curated gene sets, and a core network, putatively underlying transformation, was identified that involved differentiation/stem cell–related transcriptional programs. We selected modules for incorporation into a survival model using stepwise regression. The survival model was validated in an independent set from a different group.11 Survival analysis based on core modules was performed with and without proliferation genes.
Methods
Our overall strategy (Figure 1) was to construct a network of modules of coregulated genes, using expression data from both FL and DLBCL,7 leveraging the strongest possible signature of transformation-associated expression changes. We then applied survival analysis to the FL samples alone, to refine understanding of which combination of modules was predictive of HT. We validated the survival predictor in an independent set of FL patients, selected without bias in their propensity to undergo HT.11 Finally, we conducted additional validation of our results using a transgenic mouse model of MYC-driven lymphomas that we have described previously.17
Gene expression data used in construction of module network
We used gene expression data from 88 patient samples with unambiguous histology, generated by Glas et al on cDNA microarrays.7,18 Of these, 30 samples represented DLBCLs known to result by transformation from antecedent FL. The other 58 were classified as FL, of which 40 were known to transform to DLBCL during subsequent patient follow-up. A set of 24 samples from 12 patients represented paired pre- and post-HT tumor samples. Of the unpaired samples, 5 patients were represented by 2 FL samples each, whereas the remaining samples were from unique patients. A total of 36 diagnostic samples were obtained from patients who had not received any treatment. Detailed clinical data are given in supplemental Table 1 of Glas et al.7 We refer to FL tumors where the disease was not known to transform as FL-nt, FL that transform to DLBCL as FL-pt (“primed” to transform), and DLBCL that result by HT as DLBCL-t, to distinguish them from other types of DLBCL that arise de novo. FL-pt and FL-nt are referred to collectively as simply FL. Because transcriptional regulation can only be inferred for genes that show significant variation in their expression level, we filtered the array probes to eliminate genes with only small variance, retaining approximately 12 700 probes. Each probe was standardized across samples to have mean 0 and SD 1.
Construction and analysis of module network
We used Genomica12 to construct a module network, using a list of 2846 putative regulatory genes, including transcription factors, mRNA modification and processing, chromatin modification, canonical cellular pathways, and immune system signaling pathways. This list was compiled from the following sources. First, we included transcription factors listed in TRANSFAC, and a custom transcription factor array maintained by the University of Washington (http://hg.wustl.edu/lovett/TF_june04table.html). Entrez Gene was searched for all genes annotated as relevant to transcription (co)activation, (co)repression, (co)regulation, (co)suppression, and initiation/termination. To these we added genes involved in nonmetabolic canonical pathways, which we obtained by merging the KEGG, Biocarta, and Ingenuity Pathways Analysis (IPA; Ingenuity Systems Inc) databases. IPA was used to identify genes involved in chromatin modification, and RNA modifications and processing (splicing, decay, metabolism). Finally, we included genes related to cytokine signaling, immune system signaling, and development from IPA. The list was screened against the array annotation data, yielding 3946 probes corresponding to 2846 unique gene symbols.
Based on prior experience, module sizes of 50 to 80 genes are generally appropriate, and regulators at a tree depth of 4 levels or more are rarely statistically significant. Therefore, we limited the tree search depth to 4, with 200 modules to allow approximately 65 genes per module on average. Each inferred module was then represented across samples by the mean expression of its component genes (“metagene”). Statistically significant associations between sample types (FL-nt, FL-pt, and DLBCL-t) and modules were evaluated using prediction analysis of microarrays (PAM).19 Separate analyses were performed on FL-pt versus DLBCL-t, and FL-nt versus FL-pt. After score thresholding, 10-fold cross-validation was used to estimate misclassification errors.
For mapping module gene assignments defined in one dataset to a second expression dataset, we used official gene symbols. If multiple probes in the mapped module corresponded to the same gene, they were collapsed by averaging, before calculating a final metagene.
Biologic coherence of regulatory modules
To infer potential biologic relevance of modules, we compared the genes assigned to them with the mSigDB database of precompiled gene sets.20 Significant overlaps between modules and known gene sets were evaluated by hypergeometric test and corrected for false discovery rate (FDR; q) by permutation test. A complete list of significant (q < .05) associations is given in supplemental Tables 1 and 4. We augmented the mSigDB database of gene sets with additional ones pertaining to the immune system and stem cell regulation. Immune gene sets were obtained from the database maintained by the Staudt laboratory.21 Stem cell–related sets included expression experiments comparing stem cells with their normal counterparts, chromatin binding assays examining induced pluripotency, and cellular differentiation programs driven by specific transcription factors, such as IRF4 and PAX5 (supplemental Table 5). Mouse gene symbols were mapped to human using Homologene release 62 (http://www.ncbi.nlm.nih.gov/HomoloGene/).
Gene expression of normal germinal center B cells
Tonsils were obtained from children undergoing routine tonsillectomy with informed consent obtained in accordance with the Declaration of Helsinki and approval from the Ethics Committee of the Norwegian Radium Hospital. The tonsils were minced, and mononuclear cells were purified by standard density gradient centrifugation (Lymphoprep, 1077 mg/l; Nycomed). CD19+ B cells were purified by magnetic bead separation (Dynabeads CD19 Pan B and DETACHaBEAD CD19; Invitrogen). The CD19+ cells were stained with anti-CD38 PECy5 (Coulter Beckman), anti-IgD PE (DakoCytomation), and anti-CD77 FITC (Becton Dickinson) for 30 minutes at 4°C, washed once in RPMI with 2% fetal calf serum, followed by sorting of CD38−IgD+CD77− naive B cells, CD38+IgD−CD77− centrocytes (CCs), CD38+ IgD−CD77+ centroblasts (CBs), or CD38−IgD−CD77− memory B cells using a FACStar Plus cell sorter (BD Biosciences). The purity of CC and CB populations was reproducibly greater than 92%, and each cell population was isolated from 4 different donors. RNA was extracted using TRIzol, and RNA from the different donors was pooled together for each population. RNA amplification, labeling, and hybridization to Lymphochip cDNA microarrays were performed as described by Hystad et al.22
Survival analysis
Cox proportional hazards regression was used to build a model of survival based on metagene expression of modules in the training set of FL-nt and FL-pt samples. The model was initialized on the core ESC1 modules and stepwise AIC regression was applied to build a multivariate predictor from the 18 modules in total that were annotated as related to cellular differentiation/stemness. The resulting model is composed of a total of 5 modules. Because 2 of these modules contributed only very weakly to survival (P < .001 vs P < .001 without them), we eliminated these and retained only 3 modules. The final model (and associated linear predictor score [LPS]) was built using these 3 modules alone in the Cox regression. To obtain Kaplan-Meier curves, patients were divided into high- or low-risk groups according to whether their LPS was higher or lower than its median value. Statistical significance of survival curve separation was evaluated by log-rank test.
Transgenic mice
The Tet system was used to generate transgenic mice that conditionally express the human MYC cDNA in T-cell lymphocytes.23 Titration of MYC levels in cultured tumor cells was performed as described previously.17 Apoptotic cells were detected by the TdT-mediated dUTP nick end labeling assay in situ death detection kit (Roche Diagnostics) and counterstained with 4,6-diamidino-2-phenylindole (Vector Laboratories). Cells were grown in their respective media requirements and then pulsed with 0.1 mM bromodeoxyuridine for 1 hour. They were then collected and fixed with 70% ethanol and stained for bromodeoxyuridine incorporation according to the manufacturer's instructions (BD Biosciences PharMingen). For gene expression profiling, 50 μg total RNA and 50 μg pooled sample reference mRNA were differentially labeled with Cy5 and Cy3, respectively, and hybridized to Stanford MEEBO oligonucleotide arrays as described.17 Data are available in the Gene Expression Omnibus (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE10200.
Results
Construction and annotation of a regulatory network of FL transformation
To identify mechanisms underlying HT, we constructed a module network24 using microarray gene expression data from patients with FL and patients with DLBCL-t that evolved from antecedent FL.7 The resulting network consists of modules of coregulated genes, with inferred regulatory relationships between them (supplemental Figure 1). To infer the biologic relevance of modules, we compared them with precompiled gene sets.20 Consistent with previous analyses of FL and DLBCL expression profiles,5,7,8,10,11 the regulatory network included coherent modules highly enriched for genes involved in cell-cycle progression, proliferation, cellular differentiation, ribosomal protein activity, T-cell activation and receptor signaling, B-cell development and differentiation, inflammatory response, proteasome components, hypoxia signaling, and mitochondrial function (supplemental Table 1).
Unexpectedly, multiple modules within the network significantly overlapped with gene sets stereotypically expressed in ESCs (supplemental Table 1). We refined associations between these modules and ESC-related transcriptional programs by comparing them with 385 additional gene sets that we curated from the literature, focusing on studies of self-renewing cell types from various developmental stages, pluripotency pathways, and genes associated with proliferation and cell cycle regulation. The most significant recurring association was between several core network modules and a set of ESC-like signatures (supplemental Table 1) that has been described as aberrantly expressed in multiple epithelial tumors, coordinately regulated by MYC, and independently associated with histologic grade, risk of metastasis, and death in patients with carcinomas of the breast and lung.16
A core network associated with HT reveals an ESC-like signature
We next identified 19 modules that were most closely associated with HT. Genes in 10 modules were induced on HT, whereas 9 modules were repressed (Figure 2A; supplemental Table 2). Strikingly, several modules induced in transformation were enriched for genes highly expressed in ESCs relative to adult stem cells and differentiated tissues (Figure 2B). Consistent with this, modules coordinately repressed on HT were enriched for genes that are repressed by the polycomb group (PcG) in ESCs relative to differentiated cells, and for genes expressed in adult tissue stem cells and differentiated cell types, including T cells (Figure 2B). This implies that DLBCL-t does not arise from an HSC progenitor, but rather from a more differentiated germinal center B cell (FL, or a progenitor of FL clones) that acquires an enhanced stem cell–like expression pattern. With the exception of one module, the 19 HT modules formed a tightly connected subnetwork within the overall module network (Figure 3A). Comparison of paired FL-pt and DLBCL-t specimens from 12 patients revealed the 19 modules to be consistently induced or repressed in HT (Figure 3B). One module (denoted as ESC1) lay at the core of the group of 19 modules (Figure 3A), being highly connected to others while itself having few upstream neighbors. ESC1 was also the single module most predictive for HT, suggesting a key role in transformation. The ESC1 module was composed of 101 genes, including MYC and RUVBL1, and highly overlapped genes previously identified as core components of ESC maintenance programs (FDR, q = 1.2 × 10−7), as well as direct transcriptional targets of MYC (q = 6 × 10−7) and IRF4 (q = 1.4 × 10−5; Figure 2B).16
Modules associated with HT are enriched for ESC-like gene expression signatures. In a sample (column), each module (row) is represented by the mean expression of its component genes (“metagene”). (A) PAM was used to compare FL-pt with DLBCL-t, and modules were ordered by significance, according to their PAM score. Also shown for comparison are the FL-nt samples. There is no coherent differential expression of these modules between transforming and nontransforming FL. (B) Selected associations between transformation-related modules learned from expression data and predefined gene sets as determined by hypergeometric test with permutation correction for multiple hypothesis testing (supplemental Tables 1,4). Except for module 3, FDR values shown here are for gene sets with cell-cycle and proliferation-related genes removed.
Modules associated with HT are enriched for ESC-like gene expression signatures. In a sample (column), each module (row) is represented by the mean expression of its component genes (“metagene”). (A) PAM was used to compare FL-pt with DLBCL-t, and modules were ordered by significance, according to their PAM score. Also shown for comparison are the FL-nt samples. There is no coherent differential expression of these modules between transforming and nontransforming FL. (B) Selected associations between transformation-related modules learned from expression data and predefined gene sets as determined by hypergeometric test with permutation correction for multiple hypothesis testing (supplemental Tables 1,4). Except for module 3, FDR values shown here are for gene sets with cell-cycle and proliferation-related genes removed.
A core network of ESC-related modules defines expression changes in HT. (A) A core network representing HT was constructed by extracting 19 modules that distinguished DLBCL-t from FL-pt from the full network of 200 modules. Nodes in red indicate modules that are typically up-regulated in DLBCL-T, whereas blue nodes are modules more highly expressed in FL-pt. Stronger coloring and larger size of modules represent the mean fold change in expression between FL-pt and DLBCL-T. White nodes are modules that were not part of the 19-module classifier but are connected to them in the network. Unnumbered modules did not have a significant gene set annotation. Module 9 distinguishes FL-pt from DLBCL-T but is not directly connected to the other core modules in the network. (B) Fold changes for expression of each of the 19 core HT modules were obtained by comparing 12 paired (before and after HT) patient samples in the Glas et al dataset.7 ESC indicates embryonic stem cell signature; HSC, hematopoietic stem cell; and SSS, stromal stem cell.
A core network of ESC-related modules defines expression changes in HT. (A) A core network representing HT was constructed by extracting 19 modules that distinguished DLBCL-t from FL-pt from the full network of 200 modules. Nodes in red indicate modules that are typically up-regulated in DLBCL-T, whereas blue nodes are modules more highly expressed in FL-pt. Stronger coloring and larger size of modules represent the mean fold change in expression between FL-pt and DLBCL-T. White nodes are modules that were not part of the 19-module classifier but are connected to them in the network. Unnumbered modules did not have a significant gene set annotation. Module 9 distinguishes FL-pt from DLBCL-T but is not directly connected to the other core modules in the network. (B) Fold changes for expression of each of the 19 core HT modules were obtained by comparing 12 paired (before and after HT) patient samples in the Glas et al dataset.7 ESC indicates embryonic stem cell signature; HSC, hematopoietic stem cell; and SSS, stromal stem cell.
Given the unexpected enrichment of HT modules for genes distinguishing self-renewing stem cells of embryonic derivation, we considered the behavior of transcriptional regulators of B-cell development,25 focusing on paired samples from patients whose disease transformed (supplemental Figure 2). Genes frequently induced on HT included MYC, SOX2, and STAT4. MYC and SOX2 are 2 of 4 master regulatory transcription factors with central rolesin self-renewal, capable of pluripotency induction in terminally differentiated tissues.26 Oct4 (POU5F1) and NANOG, the other 2 reprogramming factors, were not represented in the microarray data. Genes frequently repressed on HT included PAX5, CEBPA, and STAT3. Notably, repression of PAX5 is known to be central to reprogramming of mature B lymphocytes to pluripotency,27 and inactivating somatic mutations within PAX5 are recurrent events in precursor B-lymphoblastic leukemias.28 Another module consistently down-regulated on HT (Module 13 in Figure 2) was enriched for targets of the PcG members SUZ12 (q = 3 × 10−4), EED (q = 8 × 10−4), and RNF2 (q = 8 × 10−3). These genes are repressed by PcG activity in ES cells.29-32 Aberrant methylation of PcG targets has been implicated recently as a factor in ESC-like expression signatures in other aggressive B-cell lymphomas.15
ESC-like signature underlying HT is independent of proliferation
Because programs of self-renewal and differentiation are intimately connected with proliferation and the cell cycle, we verified that core modules predictive of HT do not just reflect proliferative differences between FL and DLBCL-t tumors. We compiled a list of 844 genes (supplemental Table 3) related to proliferation and cell cycle.33,34 Even after removing these genes from our analysis, HT modules were still significantly enriched for genes expressed in ESCs (supplemental Table 4). Given the known correlation between histologic grade and proliferative indices in FL,35 we also examined the relationship between histologic grade and expression of the ESC1 module. Grades 1/2 FL tumors showed similar expression of this module (even though they differ in their proportion of rapidly proliferating centrocytes), with both being lower than FL3b/DLBCL-t samples (P < .001, t test; supplemental Figure 3). In addition, expression of the ESC1 module correlated only weakly with Ki-67 proliferation index in an independent set of 175 DLBCL cases (Pearson correlation ρ = 0.34, supplemental Figure 4).36 We conclude that core signatures underlying transformation are enriched for genes expressed in ESCs and that their induction on HT is not a proxy for proliferative differences between FL and DLBCL-t.
ESC-like signature underlying HT is distinct from normal B-cell transcriptional programs
The modules distinguishing FL from DLBCL-t could represent gene expression differences between histologies, rather than the underlying HT mechanisms. They might also reflect distinctions between NHL variants beyond FL and DLBCL-t. We therefore examined the expression of the HT modules across normal B-cell developmental stages and across diverse NHL histologies,22,37,38 with specific emphasis on the core ESC1 module. Given its role in ESC-related cancer signatures,16 we separately evaluated MYC, a component gene of the ESC1 module, in the same datasets.
HT modules exhibited heterogeneous expression across diverse normal/malignant histologies (supplemental Figure 5). By measuring gene expression on Lymphochip cDNA microarrays, we determined that the ESC1 module was variably expressed across normal B-cell development, but particularly highly expressed in CBs and CCs of germinal centers (Figure 4A). MYC was highly expressed in early B-cell stages but was down-regulated in CBs/CCs, in direct contrast to the ESC1 module (Figure 4A-B). We observed an identical pattern of ESC1 module/MYC expression in an independent dataset (Figure 4C-D). MYC expression generally mirrored ESC1 module expression, but with overlap in the expression ranges across histologies and between normal/malignant B cells (Figure 4C-D). ESC1 module expression was lower in FL than in de novo DLBCL and normal CBs/CCs. In contrast, MYC was expressed at similar levels in FL to CBs/CCs, but at a higher level in de novo DLBCL (Figure 4C-D). In an independent dataset, expression of the ESC1 module and MYC was similar in transformed and de novo DLBCLs (supplemental Figure 6), although both showed a tendency to higher expression in ABC subtype de novo DLBCL. Moreover, expression of ESC1 module/MYC was independent of the presence or absence of BCL2 translocations (present in > 90% of FL cases) in germinal center B–like de novo DLBCL cases.
ESC1 module and MYC show similar expression variation, except in normal centroblasts/centrocytes. Expression of the core ESC1 module and MYC (a component of ESC1) was mapped to independent datasets via gene symbols and compared across normal and malignant B cells. Each row represents a separate expression dataset, and the 2 columns show mean expression of ESC1 genes (left, white) and MYC (right, gray). (A-B) Variation across stages of normal B-cell development from bone marrow and tonsil samples,22 showing high expression of ESC1 module but low expression of MYC in CBs/CCs relative to other normal B cells. (C-D) Normal (right of dotted line) and malignant (left) B-cell expression.37 ESC1 module is less highly expressed in FL than in de novo DLBCL (dDLBCL) and normal CBs/CCs, whereas MYC is higher in dDLBCL than in FL or normal CBs/CCs. HSC indicates hematopoietic stem cell; EB, early B cell; ProB, Pro-B cell; PreB, Pre-B cell; ImB, immature B cell; NB, naive B cell; MB, memory B cell; CLL, chronic lymphocytic leukemia; HCL, hairy cell leukemia; MCL, mantle cell lymphoma; FL, follicular lymphoma; dDL, de novo DLBCL; PEL, primary effusion lymphoma; and BL, Burkitt lymphoma.
ESC1 module and MYC show similar expression variation, except in normal centroblasts/centrocytes. Expression of the core ESC1 module and MYC (a component of ESC1) was mapped to independent datasets via gene symbols and compared across normal and malignant B cells. Each row represents a separate expression dataset, and the 2 columns show mean expression of ESC1 genes (left, white) and MYC (right, gray). (A-B) Variation across stages of normal B-cell development from bone marrow and tonsil samples,22 showing high expression of ESC1 module but low expression of MYC in CBs/CCs relative to other normal B cells. (C-D) Normal (right of dotted line) and malignant (left) B-cell expression.37 ESC1 module is less highly expressed in FL than in de novo DLBCL (dDLBCL) and normal CBs/CCs, whereas MYC is higher in dDLBCL than in FL or normal CBs/CCs. HSC indicates hematopoietic stem cell; EB, early B cell; ProB, Pro-B cell; PreB, Pre-B cell; ImB, immature B cell; NB, naive B cell; MB, memory B cell; CLL, chronic lymphocytic leukemia; HCL, hairy cell leukemia; MCL, mantle cell lymphoma; FL, follicular lymphoma; dDL, de novo DLBCL; PEL, primary effusion lymphoma; and BL, Burkitt lymphoma.
Thus, the ability of modules to distinguish FL from DLBCL-t appears to reflect HT mechanisms, not histology. The core transcriptional program in HT (ESC1 module) and in normal CBs/CCs cells overlaps (supplemental Figure 7), but the MYC-related component is aberrantly expressed in HT.
Correlation between the ESC program (implicated in HT) and MYC-driven tumors in transgenic mouse models
To confirm whether the core ESC-related program implicated in HT is directly associated with tumor maintenance and aggressiveness, we examined its behavior in transgenic mouse models of MYC-driven lymphomas. We used this approach because FL tumors do not grow in culture and FL cell lines representative of the indolent stages of the disease are not available.
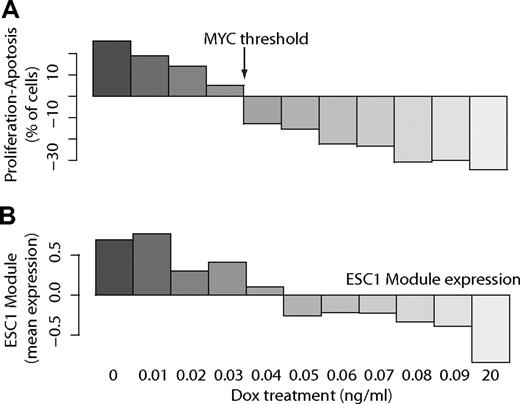
In a transgenic mouse model, conditional MYC overexpression using the Tet-system results in development of aggressive T-cell lymphomas.23 The levels of MYC in the tumors are comparable with human Burkitt lymphomas. When mice with tumor burden are treated with doxycycline, transgenic MYC expression is inhibited, and tumors regress. By titrating the level of MYC expression by various levels of doxycycline, we have shown previously that there is a specific threshold level of MYC required to maintain a tumor phenotype.17 As MYC decreases below the threshold, the balance between growth and apoptosis of tumor cells switches, so that tumors begin to regress and die (Figure 5A).
Core HT module correlates with MYC-driven tumor growth in transgenic FVB/N mouse. Gene expression analysis of MYC and expression of ESC1 module in tumor cells from FVB/N mice with conditional overexpression of human MYC that were treated with different doses of doxycycline to down-regulate MYC. At 0.04 to 0.05 ng/mL doxycycline, the level of MYC has dropped below the threshold required for tumor maintenance. (A) The difference between the proportion of tumor cells undergoing growth versus apoptosis shifts in favor of the latter at a specific MYC level (threshold) required to maintain a tumor phenotype. (B) Mean expression of genes in the core FL transformation module, as determined from microarray analysis of mouse tumors at each level of doxycycline.
Core HT module correlates with MYC-driven tumor growth in transgenic FVB/N mouse. Gene expression analysis of MYC and expression of ESC1 module in tumor cells from FVB/N mice with conditional overexpression of human MYC that were treated with different doses of doxycycline to down-regulate MYC. At 0.04 to 0.05 ng/mL doxycycline, the level of MYC has dropped below the threshold required for tumor maintenance. (A) The difference between the proportion of tumor cells undergoing growth versus apoptosis shifts in favor of the latter at a specific MYC level (threshold) required to maintain a tumor phenotype. (B) Mean expression of genes in the core FL transformation module, as determined from microarray analysis of mouse tumors at each level of doxycycline.
We hypothesized that genes in the core ESC module implicated in FL transformation should show significant expression changes at the threshold level of MYC in the mouse MYC-dependent tumors. By measuring gene expression using microarrays, we found 1904 genes that are statistically significantly (FDR = 5%) down-regulated as MYC expression drops below the threshold required for tumor maintenance.17 Fifty-two of the genes from the core HT ESC1 module mapped to orthologous loci on the mouse arrays, of which 36 were among those down-regulated at the MYC threshold (P < .001, hypergeometric test). Changes in expression of the ESC1 module (mapped to homologous mouse genes) as MYC levels were progressively reduced are shown in Figure 5B. Hence, the ESC1 module was highly enriched for genes whose expression changes decisively when MYC levels are reduced below those required to maintain a tumor phenotype in our mouse model system. Separately, we verified that ESC1 module genes were rapidly down-regulated on MYC inactivation in the mouse model in a time-dependent fashion (supplemental Figure 8). Finally, we used the Nextbio search engine (Nextbio Inc), to identify experiments in the NCBI Gene Expression Omnibus where significant changes in gene expression between experimental conditions were correlated with the up-regulation of ESC1 module genes in HT. The most significant association (P < .001) was between ESC1 module genes and genes that are up-regulated in B-cell lymphomas generated by MYC overexpression in the EμMYC transgenic mouse model, relative to normal tissue.39
Thus, the core ESC-like module induced in FL transformation was strongly enriched for genes that are induced specifically by MYC overexpression in transgenic mouse lymphoma models. Furthermore, many of these genes are associated with MYC-dependent tumor maintenance, as we have previously established.17
A 3-module ESC model stratifies survival in FL and predicts transformation
Having identified a module enriched for ESC-associated genes at the core of HT, we hypothesized that, if ESC-like processes drive HT, rather than being consequences of it, there might be an ESC-like signature present in pretransformation FL-pt. Given that HT has a major influence on patient outcomes, we reasoned that analysis of survival data would provide additional insight.
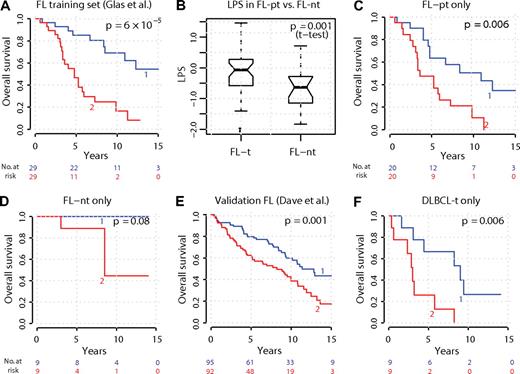
The ESC1 module on its own was only weakly predictive of survival (P = .02, log-rank test) in the FL patients of Glas et al.7 We therefore constructed a multivariate predictor from all modules annotated by gene sets associated with cellular differentiation processes. The resulting model defined a score based on 3 modules: LPS = 1.143 × (ESC1) − 1.354 × (stromal module) + 0.722 × (ESC2 module) (supplemental Table 1), which was predictive of FL survival (P < .001, log-rank test; hazard ratio = 2.72, 95% confidence interval = 1.71-4.32). Stratification of the training set into high- or low-risk, based on the median of the LPS, robustly separated survival curves (Figure 6A). High expression of modules ESC1 and ESC2 (annotated for genes sharing a similar pattern of up-regulation in ESCs and induced pluripotent stem [iPS] cells; q = 2 × 10−4, supplemental Table 4) was associated with worse outcome. Conversely, high expression of a third module (stromal, enriched for targets of transforming growth factor-β [TGF-β] signaling and stromal genes) was associated with improved prognosis. Strikingly, LPS was higher in FL that were known to transform (FL-pt) than FL that did not transform (FL-nt; Figure 6B), indicating that the combined expression signature of these 3 modules was associated with propensity to transform (P < .001, t test), even though none of them individually distinguished FL-pt from FL-nt. Separation of survival curves based on LPS was also significant within FL-pt only (P = .006, Figure 6C), but not within FL-nt (P = .08, Figure 6D).
Three-module stemness model stratifies survival in FL and DLBCL-t and predicts transformation. A linear predictor score (LPS) based on 3 stemness modules was defined that predicted survival across FL patients in the training set.7 (A) Training set FL patients were split into high-risk/low-risk groups according to whether their LPS was more or less than its median value. (B) Patients with high LPS were more likely to have subsequent transformation of their FL to DLBCL-t, compared with patients with low LPS. (C-D) LPS stratified survival within FL-pt, but not within FL-nt. (E) LPS performance in validation set of 187 FL patients.11 (F) Surprisingly, the LPS defined in FL also stratified patient survival when applied to DLBCL-t gene expression, despite survival times being from initial diagnosis of FL, not time of DLBCL-t biopsy.
Three-module stemness model stratifies survival in FL and DLBCL-t and predicts transformation. A linear predictor score (LPS) based on 3 stemness modules was defined that predicted survival across FL patients in the training set.7 (A) Training set FL patients were split into high-risk/low-risk groups according to whether their LPS was more or less than its median value. (B) Patients with high LPS were more likely to have subsequent transformation of their FL to DLBCL-t, compared with patients with low LPS. (C-D) LPS stratified survival within FL-pt, but not within FL-nt. (E) LPS performance in validation set of 187 FL patients.11 (F) Surprisingly, the LPS defined in FL also stratified patient survival when applied to DLBCL-t gene expression, despite survival times being from initial diagnosis of FL, not time of DLBCL-t biopsy.
We validated the survival predictor in an independent dataset of 187 FL patient gene expression samples,11 where it robustly stratified survival (P = .001, Figure 6E). Surprisingly, this predictor also stratified patient survival when evaluated within DLBCL-t samples (P = .006, Figure 6F), an unexpected finding because survival times were from time of initial diagnosis of FL, not from histologic transformation. This suggested that the relative strength of the 3-module “HT signature” might be preserved between patients before and after HT. Indeed, we found that 10 of 12 patients with paired pre- and post-HT samples were similarly grouped (low- vs high-risk) based on the LPS evaluated in their FL-pt or DLBCL-t sample (P = .03, hypergeometric test).
Although we previously demonstrated that individual modules associated with stem cell programs were not simply proxies for proliferation, we tested the possibility that the multivariate predictor of survival might still correlate to proliferative differences. We retained only genes associated with self-renewal, stem cells, and pluripotency, and eliminated genes known to be closely tied to cell cycle and proliferation. We then rederived the survival predictor and found that it remained predictive of overall survival (P < .001, hazard ratio = 2.72, 95% confidence interval = 1.6-4.5), with little difference in the model parameters (supplemental Table 7). Moreover, proliferation genes alone were not able to stratify FL survival (supplemental Figure 9).
Hence, a 3-module model stratified survival in FL, with 2 ESC-related modules contributing to poor prognosis and 1 stromal signature related module contributing to good prognosis. The LPS power to discriminate between FL-nt and FL-pt supports that an ESC-related mechanism underlies HT, is present before HT, and persists after HT.
Discussion
The mechanisms driving lymphoma transformation are undoubtedly complex and heterogeneous.40 Previous gene expression studies have revealed underlying changes related to proliferation, deregulation of signaling pathways and oncogenes, such as Myc.5-9 The combinatorial nature of transcriptional regulation poses a challenge to interpreting such data. We performed a network-based analysis that captures the interconnected nature of regulation. The module network successfully recapitulated known aspects of the transformation process, such as shifts in gene expression resulting from loss of infiltrating T cells in most DLBCL-t relative to FL,8,18 increased proliferative drive, and deregulation of the cell cycle.5-7 The hierarchy suggested by the module network implies that a group of core ESC-like transcriptional programs, previously associated with MYC deregulation in epithelial cancers,16 is central to HT (Figure 3). Notably, the core network module (ESC1, Figure 3) showed distinct down-regulation at a threshold level of MYC required for tumor maintenance, in a transgenic mouse lymphoma model that we have described previously.17 In addition, the same module of genes was coordinately up-regulated in a different mouse model of MYC-driven B-cell lymphoma.39
A survival model built around ESC1 stratifies FL survival and is validated in independent data. Furthermore, this survival predictor is associated with propensity of FL to transform, supporting that expression of these modules underlies HT, and is not merely a consequence of it (Figure 5). Given the centrality of ESC1 in the network, a tempting interpretation is that it provides the “engine” for transformation, with probability of HT being increased by high expression of a second ESC-like module (ESC2) and reduced by a third (stromal), which is enriched for stromal genes and TGF-β signaling. TGF-β signaling has been shown previously to repress MYC activity through the SMAD pathway.41 Importantly, our use of survival analysis was intended specifically to elucidate possible mechanisms of HT. Other approaches may be better suited to deriving optimal predictors of FL survival.11,42 However, it is important that our model validates in an unbiased FL patient population consisting entirely of diagnostic samples,11 to alleviate the possibility that our analysis is affected by selection bias in the training data,7 which contain a high proportion of FL-pt.
The fact that MYC and SOX2, 2 key drivers of iPS, were generally more highly expressed in DLBCL-t relative to FL whereas PAX5 was generally repressed, is provocative. Recent investigations have demonstrated the possibility of conversion of mature cells into pluripotent cells.43,44 In particular, in B cells, a key requirement for iPS is abrogation of PAX5 activity.45 PAX5 is a master regulator of B-cell development whose continued expression is required for maintenance of B-cell identity.44 However, overexpression of PAX5 can also induce lymphomagenesis via activation of B-cell receptor signaling.46 Its involvement in HT deserves further investigation. Furthermore, our observation that a significant number of PcG targets are down-regulated in HT is consistent with other recent findings suggesting that aberrant methylation of PcG targets might represent an initiating event of lymphomagenesis in de novo mature aggressive DLBCLs.15
Wong et al have demonstrated that MYC is capable by itself of inducing expression of ESC-like transcriptional programs.16 In this context, it is notable that the core module of genes we identified in HT strongly overlaps with genes that maintain a tumor phenotype in MYC-driven transgenic mouse models of lymphoma.17 How the ability of MYC to drive cellular transformation relates to PcG activity is a topic of considerable current interest, with efforts to understand their relationship in ESC processes still at an early stage.47 Moreover, the fact that the HT program overlaps with normal CC/CB expression patterns resonates with findings in mouse models of myeloid leukemias, which suggest that self-renewal capability in leukemic stem cells is acquired by deregulation of normal midmyeloid cell transcriptional processes.48
Recently, analyses of somatic hypermutation region sequence evolution and mutations of the switch μ region introduced by activation-induced cytidine deaminase have been reported.49,50 By constructing genealogic trees of clones from paired pretransformation and posttransformation samples, both groups found 2 possible routes through which HT could occur. In the first (direct route), HT occurred directly by transformation of an FL clone to DLBCL-t. In the second (indirect), an ancestral/progenitor cell with a less-differentiated phenotype apparently gave rise to FL and DLBCL-t as separate events in the same patient. Both are consistent with the results we present here regarding a role for ESC-like signatures.
In one possible scenario, FL-nt could arise from a less-differentiated self-renewing ancestral cell by asymmetric division, resulting in clones that do not express an ESC-like transcriptional program, and lack the ability to self-renew and seed subsequent DLBCL-t. In contrast, some FL clones may retain at least a partially active ESC-like expression program. Their transformation to DLBCL-t would then proceed by acquisition of further mutational events and reexpression of a full self-renewal program. In this model, we reason that the barrier to HT is lower in FL-pt than in FL-nt because they already exhibit an ESC-like expression signature. In the indirect HT route, the ancestral progenitor producing an initial FL tumor has full self-renewal capability and could continue to acquire mutations, eventually leading to enhanced proliferative ability and generation of DLBCL-t. Because various cell populations can coexist in tumors, it is not possible to distinguish these scenarios by gene expression data from bulk tumor samples. Future studies will be required using purified subsets of tumor cells and single-cell methods of analysis. It would also be of interest to determine whether the strength of the HT signature in FL correlates with time to transformation. Unfortunately, this information was not available for the datasets studied.
Further validation of our findings will require independent data where the occurrence or absence of HT is known subsequent to FL biopsy. Remaining questions include whether the HT signature is present in the majority of cells in FL-pt tumors that undergo HT or only a small subpopulation of clones. Additional study of MYC's role in HT is also warranted. Previous studies found variable induction/repression of MYC expression in HT,8 although up-regulation is most frequent.7,11 A key question, therefore, is how changes in MYC at the protein level relate to changes at the mRNA level. In addition, MYC activity is probably modulated through changes in activity of its cofactors (eg, MAX) or antagonists (eg, MAD), leading to changes in transcriptional programs that are not directly correlated with the expression of MYC itself. Finally, our approach may offer clues for therapeutic targets. Using the Connectivity Map, we identified drugs, including trichostatin-A and sirolimus (rapamycin), that induce expression changes in cells in the opposite direction to the core modules we implicated in HT (supplemental Table 8). Hence, they potentially obstruct changes in expression that may drive transformation. Interestingly, trichostatin-A is a histone deacetylase inhibitor that has been shown to induce cell differentiation in some contexts.51 Investigations of these aspects represent several potential directions for future research.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Daphne de Jong (Netherlands Cancer Institute), Lou Staudt (Center for Cancer Research, National Institutes of Health), Andrew Davies (Cancer Research UK), Kojo Elenitoba-Johnson (University of Michigan), and Andreas Rosenwald (University of Wuerzburg) for making data and clinical information available for this study as well as Denis Bronnikov (Nextbio Inc) for technical assistance with the Nextbio search engine.
This work was supported by National Institutes of Health grants U56CA112973 (S.K.P.) and P01-CA34233 (R.L.), a T32 Hematology training grant (A.A.A.), and a Leukemia & Lymphoma Society SCORE grant (R.L.). R.L. is an American Cancer Society Clinical Research Professor.
National Institutes of Health
Authorship
Contribution: A.J.G., A.A.A., S.-I.L., C.M.S., B.S., S.K.P., and D.K. designed and/or performed computational analyses; J.H.M. performed experiments on normal tonsil B-cell subsets; and A.J.G., A.A.A., C.M.S., R.L., and S.K.P. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew J. Gentles, Department of Radiology, Stanford University School of Medicine, Lucas Center for MR Spectroscopy and Imaging, Mailcode 5488, Stanford, CA 94305; e-mail: andrewg@stanford.edu; or Sylvia K. Plevritis, Department of Radiology, Stanford University School of Medicine, Lucas Center for MR Spectroscopy and Imaging, Mailcode 5488, Stanford, CA 94305; e-mail: sylvia.plevritis@stanford.edu.
References
Author notes
A.J.G. and A.A.A. contributed equally to this study.
Supplemental data
Two class SAM analysis was performed in TIGR MeV http://www.tm4.org/mev.html.