Abstract
Carfilzomib is a proteasome inhibitor in clinical development that primarily targets the chymotrypsin-like (CT-L) subunits in both the constitutive proteasome (c20S) and the immunoproteasome (i20S). To investigate the impact of inhibiting the CT-L activity with carfilzomib, we set out to quantitate the levels of CT-L subunits β5 from the c20S and LMP7 from the i20S in normal and malignant hematopoietic cells. We found that the i20S is a major form of the proteasome expressed in cells of hematopoietic origin, including multiple myeloma (MM) CD138+ tumor cells. Although specific inhibition of either LMP7 or β5 alone was insufficient to produce an antitumor response, inhibition of all proteasome subunits was cytotoxic to both hematologic tumor cells and peripheral blood mononuclear cells. However, selective inhibition of both β5 and LMP7 was sufficient to induce an antitumor effect in MM, non-Hodgkin lymphoma, and leukemia cells while minimizing the toxicity toward nontransformed cells. In MM tumor cells, CT-L inhibition alone was sufficient to induce proapoptotic sequelae, including proteasome substrate accumulation, Noxa and caspase 3/7 induction, and phospho-eIF2α suppression. These data support a hypothesis that hematologic tumor cells are uniquely sensitive to CT-L inhibition and provide a mechanistic understanding of the clinical safety profile and antitumor activity of proteasome inhibitors.
Introduction
The proteasome is a multicatalytic protein complex that is responsible for the turnover of cytosolic and nuclear proteins.1 There are 2 forms of the proteasome: the ubiquitously expressed c20S and the i20S, predominantly found in hematopoietic cells and cells that have been exposed to inflammatory cytokines.2-4 The catalytic activities of the c20S proteasome, chymotrypsin-like (CT-L), caspase-like (C-L), and trypsin-like (T-L), are encoded by separate polypeptides, β5, β1, and β2, respectively. These subunits are replaced by LMP7, LMP2, and MECL1, respectively, in the i20S. The proteasome is responsible for the degradation of long-lived proteins that are turned over, resulting from protein “wear and tear,” such as oxidation, proteolysis, and deamination.1 The proteasome also plays a regulatory role in cell proliferation by destroying proteins that trigger endoplasmic reticulum (ER) stress (eg, misfolded and partially assembled proteins), cell-cycle progression (eg, cyclins), cell-signaling (eg, nuclear factor-κB [NF-κB]), apoptosis (eg, Noxa), and cell survival (eg, Bcl2/Bax) pathways.5-7
The success of the proteasome inhibitor (PI) bortezomib in the treatment of multiple myeloma (MM)8 and mantle cell lymphoma9 has supported the clinical development of at least 3 second-generation PIs10 : carfilzomib, NPI-0052, and CEP-18770. Given the central role of the proteasome in cellular homoeostasis, it is unclear why PI treatment results in selective tumor cell death and a therapeutic window in B-cell malignancy therapy. Specific interventions in MM and mantle cell lymphoma with PI treatment regimens probably requires accurate and specific modulation of proteasome activity to take advantage of the dependence of the tumor cell on proteasome function without causing cytotoxicity in nontransformed cells.11
Protein labeling experiments have revealed that selective inhibition of the proteasome CT-L activity leads to a minor effect on total cellular protein turnover.12 Bortezomib and the second-generation inhibitors currently undergoing clinical investigation have all shown potent inhibition of CT-L activity as a shared property. However, these PIs differ in their inhibition profile for non–CT-L subunits. Bortezomib and CEP-18770 are potent inhibitors of the C-L subunits, whereas NPI-0052 is a potent inhibitor of the T-L subunits.10 On the other hand, carfilzomib demonstrates the largest selectivity for the CT-L activity.13,14 The impact of selective inhibition of C-L and T-L activities remains to be elucidated.
We set out to test the following question: Is inhibition of the CT-L activity of the proteasome sufficient to produce an antitumor response in hematologic tumor cells without causing a cytotoxic effect in nontransformed cells? We found that, in contrast to nontransformed leukocytes, hematologically derived tumor cells express both the c20S and i20S. Carfilzomib, at a dose that selectively inhibits both CT-L active sites β5 and LMP7, induced an antitumor response in MM, non-Hodgkin lymphoma (NHL), and leukemia cells with minimal cytotoxic effects in nontransformed cells. Specific chemical inhibitors of the CT-L subunits β5 or LMP7 had no antitumor effect, but combining both inhibitors produced an antitumor effect equivalent to carfilzomib. Importantly, inhibition of all proteasome subunits had a profound cytotoxic effect on nontransformed cells.
Methods
Compounds
ProCISE assay protocol
The ProCISE (proteasome constitutive/immunoproteasome subunit enzyme-linked immunosorbent) assay involves the following steps: (1) incubation of PABP with activated 20S, (2) denaturation with 6 M guanidine hydrochloride, (3) addition of streptavidin-coated beads, (4) extensive bead washing, (5) addition of proteasome subunit-specific primary antibody followed by a secondary horseradish peroxidase (HRP)–conjugated antibody, and (6) luminescence-based detection. (An extended “Methods” section can be found in the supplemental data, available on the Blood website; see the Supplemental Materials link at the top of the online article.)
Primary cells and tumor cell lines
Tumor cell lines were purchased from ATCC and were cultured in media recommended by the supplier. MM1.S cells were obtained from Steven Rosen's laboratory at Northwestern University. CD138+ cells and peripheral blood mononuclear cells (PBMCs) were purchased from Allcells. The purity of CD138+ cells was greater than 90% as determined by the supplier. HS-Sultan-GFPu cell lines were generated by transfecting the GFPu plasmid (ATCC), and cells were selected in media containing G418 (Invitrogen). Cells were grown to 70% confluence, and the green fluorescent protein (GFP) expression in the population was verified by flow cytometry after treatment with 1 μM carfilzomib.
Identification of active shRNAs against β5
Candidate shRNAs were screened for silencing activity by retroviral infection into HS-Sultan cells carrying a retroviral RNAi-reporter construct. (See the extended “Methods” section in the supplemental data.)
Cell viability, apoptosis assay, and flow cytometry
Drug treatment was performed as previously described.13 Briefly, cells were exposed to compound (see figure legends for description) for a 1-hour period followed by 3 washes in media (RPMI 1640 plus 5% fetal bovine serum) and incubated for an additional 0.5, 1, 2, 3, 4, 5, 6, 8, and 24 hours. Cell viability was assayed with Cell Titer Glo reagent at 24 hours (Promega). Apoptosis was determined by caspase 3/caspase 7 induction using caspase 3/caspase 7 Glo (Promega) at 5 or 8 hours after washout. In flow cytometry experiments to monitor GFP accumulation, cells were treated with compound and 10 000 events were collected using a Becton Dickinson flow cytometer (BD Biosciences). For each time point, a gate that captured 99% of untreated cells was set, and compound-treated cells that lay outside the gate were counted as GFP-expressing using the CellQuest software.
Western blot analysis
Cells were treated as described in “Cell viability, apoptosis assay, and flow cytometry,” and lysates were prepared as previously described.13 Clarified lysates (10 μg) were electrophoresed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with anti-ubiquitin (BIOMOL Research Laboratories), anti-NOXA (Calbiochem), anti-IκBα (Cell Signaling Technologies), anti-GADD153 (Santa Cruz Biotechnology), anti–phosphoS51-eIF2α (ABcam), anti-eIF2α (Abcam), and anti-actin (Cell Signaling Technologies). After incubation with the appropriate HRP-linked secondary antibodies (Jackson Immunoresearch Laboratories), bands were detected by Super Signal Chemiluminescent Substrate (Thermo).
Statistical analysis
For comparisons of treatment groups, 1-way and 2-way analysis of variance with Bonferroni post-hoc analysis were used. All statistical analyses were performed using GraphPad Prism Software (Version 4.01). Statistical significance was achieved when P was less than .05.
Results
ProCISE
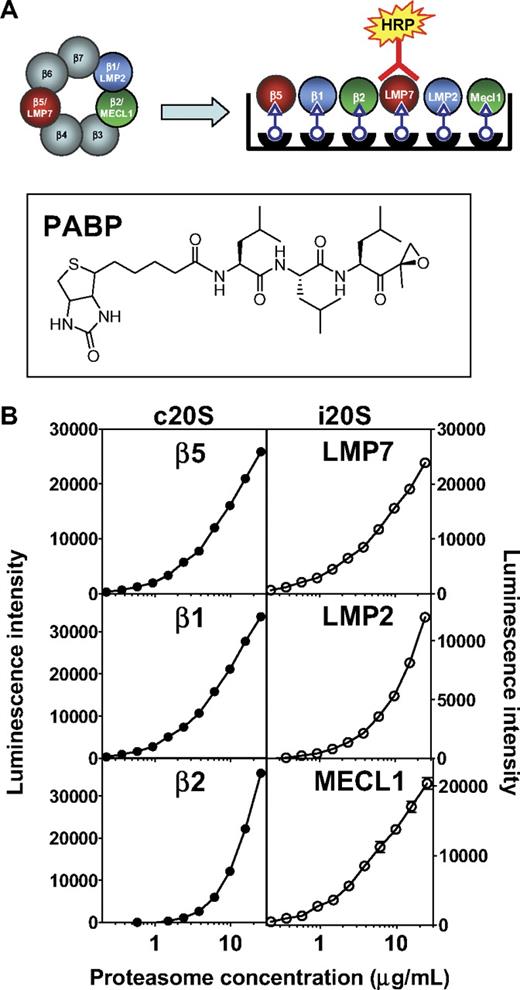
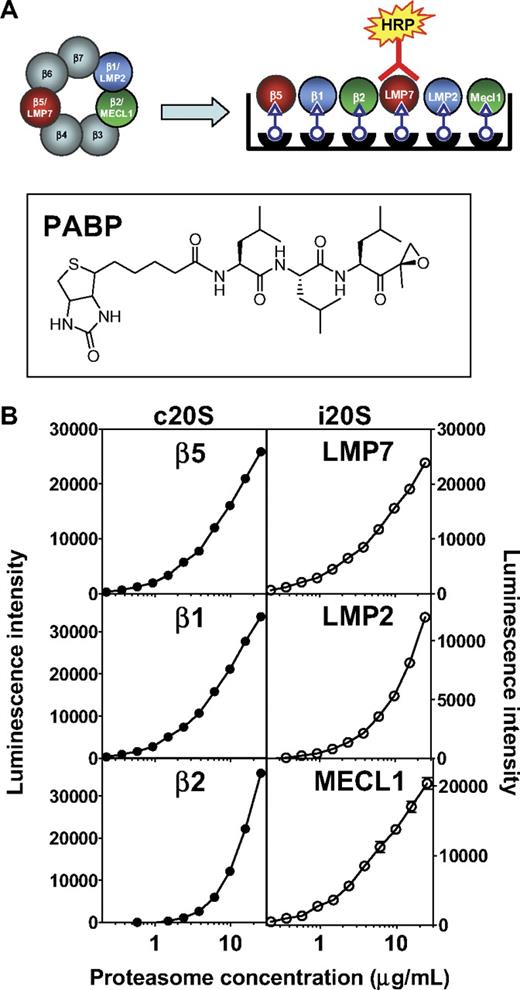
To uncover the role of specific proteasome subunits on antitumor activity, we first determined the abundance of both forms of CT-L subunits, β5 and LMP7, as well as C-L and T-L subunits in relevant cell types. This was accomplished using the ProCISE assay (Figure 1A), an enzyme-linked immunosorbent assay-based method that allows for the accurate quantitation of each proteasome active-site subunit in a cell homogenate. The precision of the ProCISE assay was measured using standard curves of the c20S or i20S (Figure 1B). A variable slope 4 parameter model predicted the relationship between luminescence signal and proteasome concentrations for each active site subunit. The relative error of each point on the c20S and i20S standard curve was calculated (data not shown) and used to calculate the upper and lower limits of quantitation, ULOQ and LLOQ, respectively (Table 1). Next, we empirically determined the optimal concentration of MM1.S and HS-Sultan lysate for use in the ProCISE assay by quantifying c20S and i20S subunits using the standard curves described in Figure 1. When cell lysates were assayed at 2 mg/mL, the level of proteasome active site subunits could be accurately quantitated between 100% and 6.5% activity (for β2, 100%-25% activity is quantifiable; Table 1). For each subunit, the dynamic window and interday coefficient of variation (Table 1) are within acceptable parameters.18
ProCISE assay schematic and 20S standard curves. (A) Schematic representation of an artificial hybrid constitutive proteasome and immunoproteasome followed by chemiluminescence detection of LMP7 by the ProCISE. The chemical structure of the proteasome active site probe, PABP, is shown. (B) c20S (●) and i20S (○) standard curves (0.23-25 μg/mL) were prepared by probing with anti-20S subunit primary antibodies followed by HRP-coupled secondary antibodies and by chemiluminescence detection. The c20S standard curve is used to quantitate the β5, β1, and β2 subunits, whereas the i20S standard curve is used to quantitate the LMP7, LMP2, and MECL1 subunits. Each panel shows the relationship between luminescence intensity and c20S or i20S concentration for each individual 20S subunit.
ProCISE assay schematic and 20S standard curves. (A) Schematic representation of an artificial hybrid constitutive proteasome and immunoproteasome followed by chemiluminescence detection of LMP7 by the ProCISE. The chemical structure of the proteasome active site probe, PABP, is shown. (B) c20S (●) and i20S (○) standard curves (0.23-25 μg/mL) were prepared by probing with anti-20S subunit primary antibodies followed by HRP-coupled secondary antibodies and by chemiluminescence detection. The c20S standard curve is used to quantitate the β5, β1, and β2 subunits, whereas the i20S standard curve is used to quantitate the LMP7, LMP2, and MECL1 subunits. Each panel shows the relationship between luminescence intensity and c20S or i20S concentration for each individual 20S subunit.
Quantitation of the CT-L subunits, β5 and LMP7, in hematopoietic cells
The levels of β5 and LMP7 in primary CD138+ bone marrow cells from 3 MM patients and 2 normal volunteers as well as PBMCs from healthy volunteers were quantitated (Table 2). In CD138+ cells, the total amount of proteasome per total protein varied (0.2-2.4 ng/μg total protein) and was consistently lower compared with proteasome levels in PBMCs (7.6 ng/μg protein). LMP7 was the predominant form of the CT-L-like subunit expressed in both normal (85%-89%) and malignant (60%-80%) CD138+ cells as well as PBMCs from healthy volunteers (97%). Next, we extended our studies to include cell lines derived from hematopoietic and solid tumors. In hematopoietically derived cell lines, the i20S represents a major form of the proteasome, constituting 37% to 69% of the total proteasome pool in the cell lines tested (Table 3). Interestingly, solid tumor HT-29 cells (colon adenocarcinoma) and A549 cells (lung carcinoma) express relatively low amounts of i20S (18% and 6%, respectively). This analysis confirms that the i20S is a major constituent of the total proteasome pool in cells of hematopoietic origin, including MM (primary CD138+, MM1.S, and RPMI-8226), plasmocytoid lymphoma-type (Arh77), T-cell lymphoma (Molt-4), and NHL (HS-Sultan) cells. We also profiled cells for levels of the C-L subunits β1 and LMP2 and the T-L subunits β2 and MECL1. We consistently observed that hematopoietically derived cells express lower levels of MECL1 (15%-76%) and LMP2 (22%-84%) relative to LMP7 (37%-94%; Table 3, supplemental Table 1). These data are consistent with previous reports that describe mixed proteasome populations in cells (ie, LMP7-LMP2-β2 or LMP7-β1-MECL1 or LMP7-β1-β2).19,20 To our knowledge, this is the first report of a quantitative measure of all 6 proteasome catalytic subunits in tumor cells. In sum, hematologically derived tumor cells express significant amounts of both the β5 and LMP7 subunits, and the activity of both subunits must be monitored to ensure complete CT-L subunit inhibition.
Selective inhibition of CT-L activity
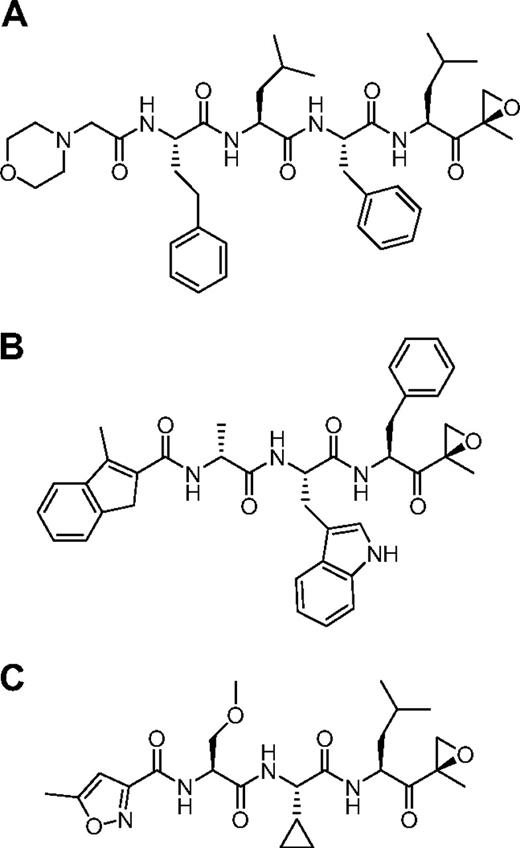
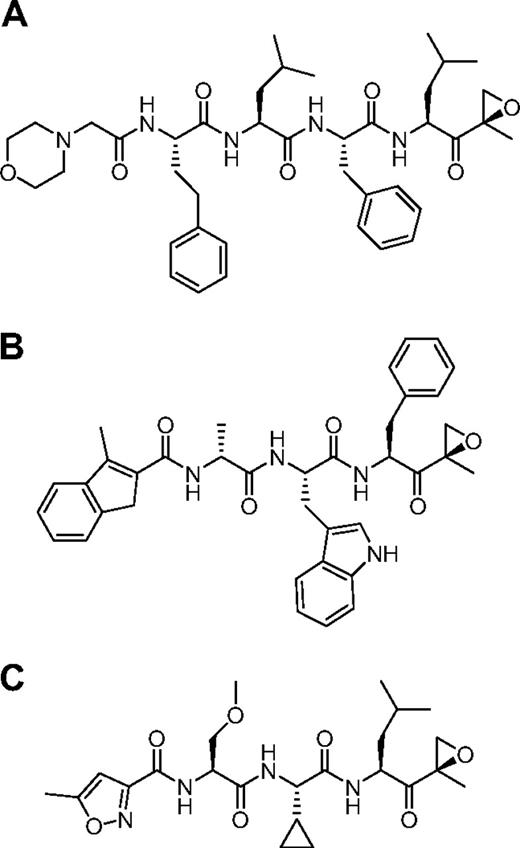
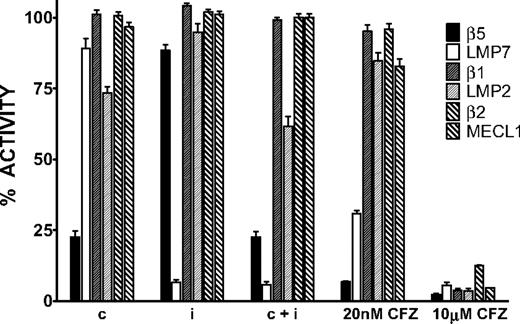
We used the ProCISE assay to titrate the amount of carfilzomib (Figure 2A) required to selectively inhibit the CT-L subunits and not the C-L nor T-L subunits (Figure 3). A 20 nM dose of carfilzomib was found to inhibit β5 and LMP7 by 93% and 70%, respectively, with minimal effects on the other subunits (Figure 3). Because carfilzomib strongly inhibits both β5 and LMP7, we generated subunit selective compounds to determine the effect of individual CT-L subunit inhibition in hematopoietic tumor cells. Through a medicinal chemistry effort to discover carfilzomib analogs that preferentially target β5 or LMP7, we discovered an IPSI (Figure 2B) with 130-fold selectively for LMP7 compared with β5. IPSI shows weak activity toward LMP2 and no detectable activity toward β1, β2, or MECL1 (Table 4). A CPSI (Figure 2C) was discovered in a similar effort to identify orally bioavailable analogs of carfilzomib16 and demonstrated a 21-fold and 13-fold selectivity for β5 over LMP7 and LMP2 subunits, respectively (Table 4). CPSI shows weak inhibition of the β1, β2, and MECL1 subunits (Table 4). Doses of CPSI and IPSI were identified that primarily inhibit β5 and LMP7, respectively (Figure 3).
Proteasome active-site subunit inhibition profiles of CPSI, IPSI, and carfilzomib in MM1.S cells. Cells were treated with a 1-hour pulse of 0.18 μM CPSI (c), 0.27 μM IPSI (i), 0.18 μM CPSI plus 0.27 μM IPSI (c + i), 20 nM carfilzomib (20 nM CFZ), or 10 μM carfilzomib (10 μM CFZ), and the ProCISE assay was performed to determine the level of available (uninhibited) subunits. For each compound, the level of residual activity compared with DMSO-treated cells is plotted for each proteasome subunit. Mean ± SD are shown (n = 24).
Proteasome active-site subunit inhibition profiles of CPSI, IPSI, and carfilzomib in MM1.S cells. Cells were treated with a 1-hour pulse of 0.18 μM CPSI (c), 0.27 μM IPSI (i), 0.18 μM CPSI plus 0.27 μM IPSI (c + i), 20 nM carfilzomib (20 nM CFZ), or 10 μM carfilzomib (10 μM CFZ), and the ProCISE assay was performed to determine the level of available (uninhibited) subunits. For each compound, the level of residual activity compared with DMSO-treated cells is plotted for each proteasome subunit. Mean ± SD are shown (n = 24).
Inhibition of the CT-L subunits leads to an antitumor effect
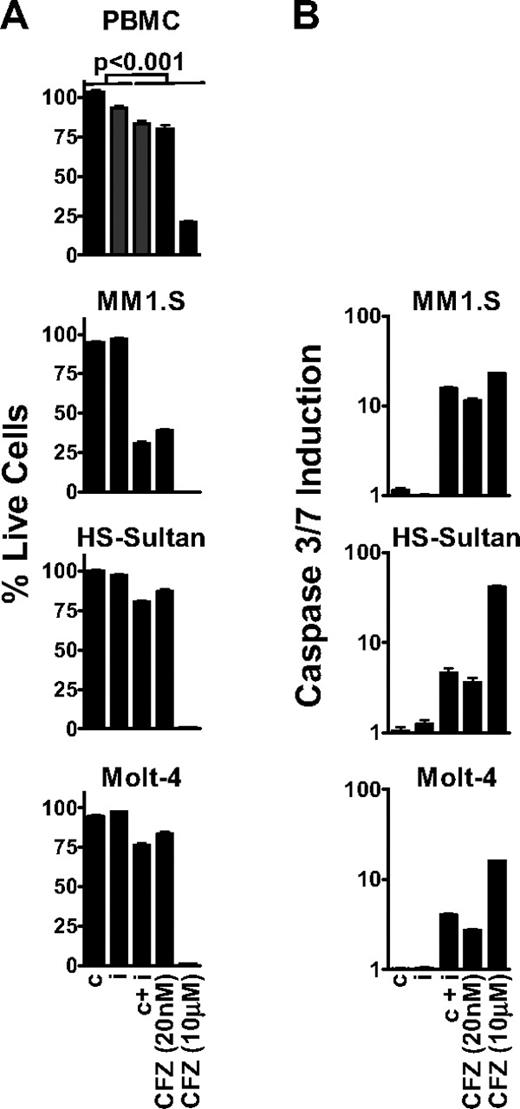
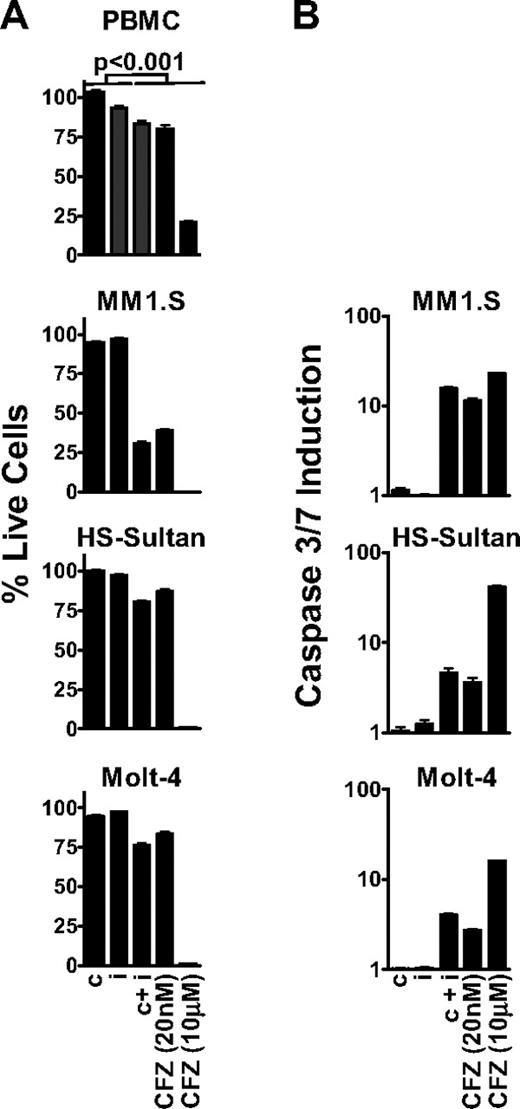
Because ketoepoxide-based proteasome inhibitors exhibit time-dependent inhibition,13 we performed all of our biologic assays using a 1-hour pulsatile exposure of carfilzomib, IPSI, and CPSI. After treatment, metabolic activity (ie, adenosine triphosphate concentration) was assayed at 24 hours as a surrogate marker for cell viability.21 We observed that inhibition of all 6 proteasome active-site subunits with 10 μM carfilzomib (termed pan-proteasome inhibition; Figure 3) had a profound antitumor effect in MM1.S, HS-Sultan, and Molt-4 cells (Figure 4A). However, pan-proteasome inhibition displayed an equally pronounced cytotoxic effect on nontransformed cells (80% cell death), suggesting that cell death was not tumor selective. On the other hand, inhibition of either the β5 or LMP7 subunit by CPSI or IPSI treatment did not affect viability of either tumor cells or PBMCs.
Inhibition of the CT-L subunits is sufficient to cause an antitumor effect in MM, NHL, and leukemia tumor cells. (A) PBMC, MM1.S, HS-Sultan, and Molt-4 cells were pulse-treated with 0.18 μM CPSI (c), 0.27 μM IPSI (i), 0.18 μM CPSI plus 0.27 μM IPSI (c + i), 20 nM carfilzomib (CFZ 20 nM), or 10 μM carfilzomib (CFZ 10 μM) for 1 hour, and cell viability was measured at 24 hours. For all datasets, significant differences between the first 2 columns (c or i) and last 3 columns (c + i, CFZ 20 nM, or CFZ 10 μM) were observed (P < .001). (B) MM1.S, HS-Sultan, and Molt-4 cells were 1-hour pulse-treated with compounds as in panel A, and caspase3/caspase 7 activation was monitored at 6 hours for HS-Sultan cells or 9 hours for Molt-4 and MM1.S cells. Representative data from 1 of 4 independent experiments are shown.
Inhibition of the CT-L subunits is sufficient to cause an antitumor effect in MM, NHL, and leukemia tumor cells. (A) PBMC, MM1.S, HS-Sultan, and Molt-4 cells were pulse-treated with 0.18 μM CPSI (c), 0.27 μM IPSI (i), 0.18 μM CPSI plus 0.27 μM IPSI (c + i), 20 nM carfilzomib (CFZ 20 nM), or 10 μM carfilzomib (CFZ 10 μM) for 1 hour, and cell viability was measured at 24 hours. For all datasets, significant differences between the first 2 columns (c or i) and last 3 columns (c + i, CFZ 20 nM, or CFZ 10 μM) were observed (P < .001). (B) MM1.S, HS-Sultan, and Molt-4 cells were 1-hour pulse-treated with compounds as in panel A, and caspase3/caspase 7 activation was monitored at 6 hours for HS-Sultan cells or 9 hours for Molt-4 and MM1.S cells. Representative data from 1 of 4 independent experiments are shown.
Subsequently, we examined the antitumor activity of selective inhibition of both CT-L subunits β5 and LMP7. This was accomplished with either a dose of 20 nM carfilzomib or the combination of CPSI and ISPI. Dual inhibition of β5 and LMP7 led to amoderate, but statistically meaningful, antitumor effect (∼ 20% cell death) in HS-Sultan and Molt-4 cells. However, CT-L inhibition in MM1.S cells resulted in a profound antitumor response (∼ 70% cell death). Carfilzomib (20 nM) and combined treatment with CPSI and IPSI had minimal effects on PBMCs. Inhibition of both β5 and LMP7 resulted in activation of caspase 3 and caspase 7 in tumor cells, whereas inhibition of either β5 or LMP7 alone produced no effect (Figure 4B). Consistent with the cell viability findings, caspase 3/caspase 7 activation after CT-L inhibition was greater in MM1.S cells (17-fold) than in Molt-4 or HS-Sultan cells (4.0- and 4.6-fold, respectively). These data suggest that in hematologic tumor cells, inhibition of both CT-L subunits is required to induce cell death.
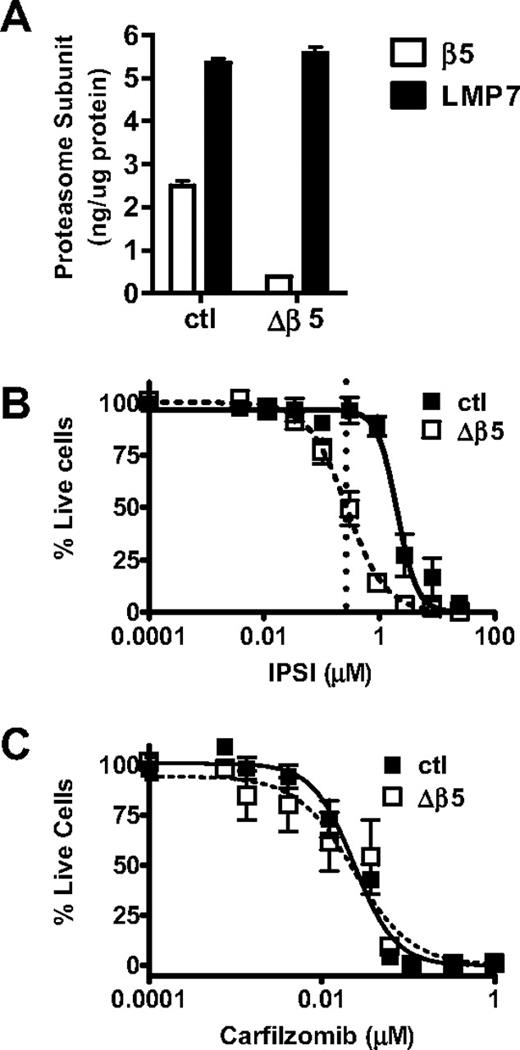
To confirm these results, we generated shRNA-expressing retroviral vectors to knockdown proteasome subunit expression in tumor cells. A β5-targeting shRNA vector was transduced into HS-Sultan cells generating the Δβ5 HS-Sultan cell line. In the Δβ5 HS-Sultan cells, β5 represents 6.5% of total CT-L-like activity compared with 31% in the parental line (Table 3; Figure 5A). We observed a pronounced antitumor effect (Figure 5B, ∼ 50% cell death) with IPSI treatment of the Δβ5 HS-Sultan cells. To verify that the Δβ5 HS-Sultan cells are not more sensitive to proteasome inhibition than the parental cell line, we exposed both cell lines to increasing doses of carfilzomib and observed that both parental and Δβ5 HS-Sultan cells were similarly sensitive (Figure 5C). We were unable to generate vectors capable of mediating significant reduction of LMP7 expression to do the complementary experiment with CPSI (data not shown). Taken together, these data support the hypothesis that inhibition of cellular CT-L activity (β5 and LMP7) is sufficient to kill hematologic tumor cells, in particular MM cells, with minimal effects to normal leukocytes.
Inhibition of LMP7 is sufficient to cause an antitumor effect in Δβ5 HS-Sultan cells. (A) HS-Sultan cells transduced with a control shRNA-expressing (ctl) or β5-targeting shRNA vector (Δβ5) were profiled for LMP7 and β5 expression. Control (■) or Δβ5 (□) HS-Sultan cells were treated with a 1-hour pulse dose escalation of IPSI (B) or carfilzomib (C), and cell viability was measured at 24 hours. Mean ± SD are shown (n = 24). For IPSI (B), the dotted line represents the LMP7-selective dose (0.27 μM) described in Figure 3.
Inhibition of LMP7 is sufficient to cause an antitumor effect in Δβ5 HS-Sultan cells. (A) HS-Sultan cells transduced with a control shRNA-expressing (ctl) or β5-targeting shRNA vector (Δβ5) were profiled for LMP7 and β5 expression. Control (■) or Δβ5 (□) HS-Sultan cells were treated with a 1-hour pulse dose escalation of IPSI (B) or carfilzomib (C), and cell viability was measured at 24 hours. Mean ± SD are shown (n = 24). For IPSI (B), the dotted line represents the LMP7-selective dose (0.27 μM) described in Figure 3.
Kinetics of proteasome substrate accumulation
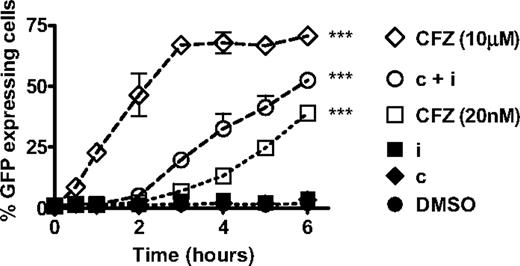
The onset, rate, or extent of proteasome substrate accumulation when treating cells with CT-L-selective versus pan-proteasome inhibitors may be responsible for the difference in the observed antitumor effects. To explore this possibility, we transfected the HS-Sultan cell line with the fluorescent reporter construct (GFPu) that codes for GFP fused to the proteasome degradation signal CL1. We measured the kinetics of GFPu accumulation up to 6 hour after a 1-hour pulsatile exposure to PIs. Monitoring cells past 6 hours was not performed because HS-Sultan cells fully activate the apoptosis pathway by this time point (Figure 4B). In the absence of proteasome inhibition, the GFPu protein is rapidly degraded by the proteasome, and no fluorescence is detected in cells (Figure 6, dimtheyl sulfoxide [DMSO]). Inhibition of either β5 or LMP7 alone with CPSI or IPSI treatment, respectively, had no effect on GFPu accumulation (Figure 6, IPSI, CPSI). In the presence of pan-proteasome inhibition (10 μM carfilzomib), GFPu accumulation was observed as quickly as 30 minutes after treatment and plateaued by 3 hours (Figure 6). Compared with pan-proteasome inhibition, selective CT-L subunit inhibition resulted in a 90-minute delay in the onset of GFPu accumulation. With CT-L subunit inhibition, a slower rate of GFPu accumulation (38%-49%) and a lower total accumulation of GFPu (53%-73%) compared with pan-proteasome inhibition was observed (Figure 6). These data suggest that selective inhibition of CT-L subunits results in proteasome substrate accumulation in the majority of tumor cells preceding the onset of apoptotic pathway activation.
Inhibition of the CT-L subunits leads to rapid accumulation of short-lived proteins. HS-Sultan cells transfected with GFPu were treated with 10 μM carfilzomib (CFZ 10 μM), 0.18 μM CPSI + 0.27 μM IPSI (c + i), 20 nM carfilzomib (CFZ 20 nM), 0.27 μM IPSI (i), 0.18 μM CPSI (c), and DMSO for 1 hour, washed, incubated in compound-free media, and harvested at the indicated times for monitoring fluorescence by flow cytometry. Mean ± SD are shown for 2 independent experiments using 2 separate clones. Two-way analysis of variance was performed and compound treatments resulting in statistically significant differences compared with DMSO treatment group (as determined by Bonferroni post-hoc testing): ***P < .001.
Inhibition of the CT-L subunits leads to rapid accumulation of short-lived proteins. HS-Sultan cells transfected with GFPu were treated with 10 μM carfilzomib (CFZ 10 μM), 0.18 μM CPSI + 0.27 μM IPSI (c + i), 20 nM carfilzomib (CFZ 20 nM), 0.27 μM IPSI (i), 0.18 μM CPSI (c), and DMSO for 1 hour, washed, incubated in compound-free media, and harvested at the indicated times for monitoring fluorescence by flow cytometry. Mean ± SD are shown for 2 independent experiments using 2 separate clones. Two-way analysis of variance was performed and compound treatments resulting in statistically significant differences compared with DMSO treatment group (as determined by Bonferroni post-hoc testing): ***P < .001.
Mechanism of action for selective chymotrypsin-like inhibition
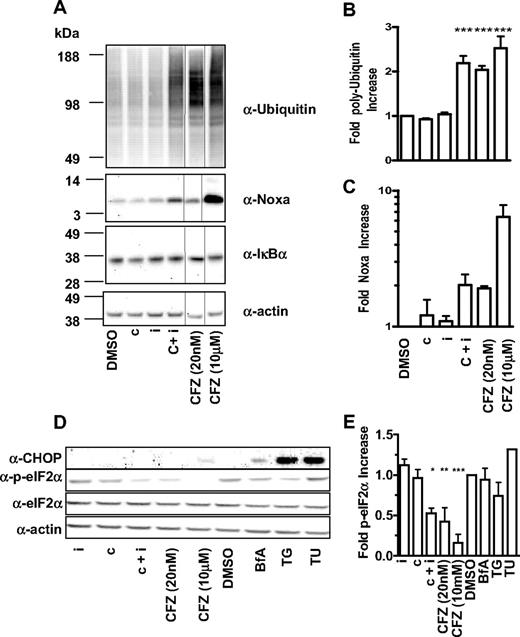
We then explored the molecular sequelae that occur in MM1.S and HS-Sultan cells on selective proteasome subunit inhibition. At various times after pulsatile exposure, cell lysates were prepared and probed with antibodies recognizing polyubiquitin, IkBα, CHOP (CIEBP homologous protein), eIF2α, phospho-eIF2α, and Noxa. Consistent with experiments with GFPu, a 2-fold increase in the accumulation of polyubiquitin chains was observed when both β5 and LMP7 were inhibited in MM1.S (Figure 7A-B) and HS-Sultan cells (data not shown). In MM1.S (Figure 7A-B) and HS-Sultan cells (data not shown), inhibition of either β5 or LMP7 alone did not lead to measurable accumulation of polyubiquitin chains, whereas pan-proteasome inhibition has the most profound effect on polyubiquitin chain accumulation (Figure 7A-B).
Inhibition of CT-L subunits is sufficient to cause modulation of polyubiquitinated proteins, Noxa and phospho-eIF2α. (A) MM1.S cells were 1-hour pulse-treated with DMSO, 0.18 μM CPSI (c), 0.27 μM IPSI (i), 0.18 μM CPSI plus 0.27 μM IPSI (c + i), 20 nM carfilzomib (CFZ 20 nM), or 10 μM carfilzomib (CFZ 10 μΜ), washed and incubated in compound-free media for a total of 4 hours. Cell lysates were electophoresed and transferred for Western blot detection with antibodies targeted against polyubiquitin, actin, Noxa, and IκBα. Quantitation of polyubiquitinated proteins (B) and Noxa (C) in compound-treated MM1.S cells. Mean ± SD for 3 independent experiments are shown. (D) MM1.S cells were 1-hour pulse-treated with 0.27 μM IPSI (i), 0.18 μM CPSI (c), 0.18 μM CPSI plus 0.27 μM IPSI (c + i), 20 nM carfilzomib (CFZ 20 nM), or 10 μM carfilzomib (CFZ 10 μΜ), DMSO, 1 μM brefeldin A (BfA), 1 μM thapsigargin (TG), and 2.5 μg/mL tunicamycin (TU), washed and incubated in compound-free media for a total of 6 hours (brefeldin A, tunicamycin, and thapsigargin were added back to the media). Cell lysates were electophoresed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred for Western blot detection with antibodies targeting CHOP, eIF2α, phospho-eIF2α, and actin. (E) Quantitation of phospho-eIF2α in compound-treated MM1.S. Mean ± SD of 3 independent experiments are shown. One-way analysis of variance was performed and compound treatments resulting in statistically significant differences (as determined by Bonferroni post-hoc testing): *P < .05, **P < .01, ***P < .001.
Inhibition of CT-L subunits is sufficient to cause modulation of polyubiquitinated proteins, Noxa and phospho-eIF2α. (A) MM1.S cells were 1-hour pulse-treated with DMSO, 0.18 μM CPSI (c), 0.27 μM IPSI (i), 0.18 μM CPSI plus 0.27 μM IPSI (c + i), 20 nM carfilzomib (CFZ 20 nM), or 10 μM carfilzomib (CFZ 10 μΜ), washed and incubated in compound-free media for a total of 4 hours. Cell lysates were electophoresed and transferred for Western blot detection with antibodies targeted against polyubiquitin, actin, Noxa, and IκBα. Quantitation of polyubiquitinated proteins (B) and Noxa (C) in compound-treated MM1.S cells. Mean ± SD for 3 independent experiments are shown. (D) MM1.S cells were 1-hour pulse-treated with 0.27 μM IPSI (i), 0.18 μM CPSI (c), 0.18 μM CPSI plus 0.27 μM IPSI (c + i), 20 nM carfilzomib (CFZ 20 nM), or 10 μM carfilzomib (CFZ 10 μΜ), DMSO, 1 μM brefeldin A (BfA), 1 μM thapsigargin (TG), and 2.5 μg/mL tunicamycin (TU), washed and incubated in compound-free media for a total of 6 hours (brefeldin A, tunicamycin, and thapsigargin were added back to the media). Cell lysates were electophoresed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred for Western blot detection with antibodies targeting CHOP, eIF2α, phospho-eIF2α, and actin. (E) Quantitation of phospho-eIF2α in compound-treated MM1.S. Mean ± SD of 3 independent experiments are shown. One-way analysis of variance was performed and compound treatments resulting in statistically significant differences (as determined by Bonferroni post-hoc testing): *P < .05, **P < .01, ***P < .001.
Constitutive expression of NF-κB has been implicated in tumor cell survival.22 To explore the effect of CT-L subunit inhibition on NF-κB activity in tumor cells, we examined the levels of the proteasome-dependent NF-κB negative regulators IκBα and p100 in MM1.S and HS-Sultan cells. PI treatment did not lead to stabilization of these factors in tumor cells (Figure 7A; and data not shown). These results are consistent with other reports that have failed to observe the PI suppression of NF-κB in MM23 and melanoma24 cell lines.
Interference with ER stress signals is an alternative explanation for the antitumor effect of PIs in hematologic cancers because MM cells produce high levels of immunoglobulins and, in general, cancer cells have overactive protein synthesis machinery.23,25-27 Induction of the proapoptotic protein CHOP/GADD153 is a hallmark feature of ER stress-induced cell death. We observed that CHOP is induced by pan-proteasome inhibition and ER stressors brefeldin A, thapsigargin, and tunicamycin, but not with inhibition of the CT-L subunits in MM1.S cells (Figure 7D). To further explore the effect of selective CT-L subunit inhibition, we examined the levels of eIF2α, a translation-promoting factor that is modulated by ER stress. Whereas the levels of total eIF2α were unaffected, the levels of the inactive phospho-eIF2α species decreased by a factor of 2 when MM1.S cells were treated with CT-L selective inhibitors (Figure 7D-E). This observation suggests that CT-L inhibition may cause an increase in protein synthesis that may further exacerbate ER stress28 and eventually cause cell death in a CHOP-independent manner.
Lastly, we examined the levels of the proapoptotic factor Noxa, which has been shown to be stabilized on proteasome inhibition in MM29 and melanoma cells.24,29 We observed a reproducible 1.5- to 2-fold increase in the Noxa levels when proteasome CT-L activity was inhibited, but not when either β5 or LMP7 was individually inhibited (Figure 7A-C). Pan-proteasome inhibition induced the highest increase in Noxa levels in both cell types. In sum, these data suggest that inhibition of β5 and LMP7 is sufficient to cause polyubiquitin chain accumulation, phospho-eIF2α suppression, Noxa stabilization, and caspase 3/caspase 7 induction culminating in an antitumor response with minimal cytotoxic effects on nontransformed cells.
Discussion
To our knowledge, this is the first study demonstrating that selective inhibition of the CT-L subunits of the proteasome leads to an antitumor response in hematologically derived tumor cells. In contrast to this finding, concentrations of bortezomib that produce an antitumor response in vitro may inhibit both CT-L and C-L activities30,31 (K.L. and F.P., unpublished results, July 2009). The relationship between CT-L inhibition and tumor cell death was demonstrated by a novel quantitative assay, ProCISE, to characterize tumor cell proteasome composition, novel inhibitors selective for the CT-L subunits of the constitutive proteasome (β5) and the immunoproteasome (LMP7) and a cell line with reduced β5 expression. We found that hematologically derived cells express both CT-L subunits and that inhibition of either subunit alone did not alter cell viability. Inhibition of both β5 and LMP7 is required for an antitumor response despite a range of relative expression levels for these 2 subunits across the different cell lines. The antitumor effect of CT-L inhibition correlated with proteasome substrate accumulation and decreased phospho-eIF2α, resulting in Noxa and caspase activation.
Levels of CT-L subunits, β5 and LMP7, in hematopoietically derived cells
We used the ProCISE assay to determine the amounts of β5 and LMP7 subunits in human primary PBMCs and CD138+ cells as well as cell lines that represent hematopoietically derived and solid tumor-derived cancer cells. Consistent with previously published reports,32,33 the proteasome constitutes 0.5% to 0.8% of total cellular protein in most cell types, including MM cell lines. However, the level of total proteasome in CD138+ cells was consistently lower (0.02%-0.2% of total protein). Several recent publications have demonstrated that proteasome up-regulation may be a possible mechanism of resistance to PI treatment in vitro.34,35 Therefore, it will be important to evaluate whether the level of proteasome expression in CD138+ cells is (1) correlated with sensitivity to proteasome inhibitor therapy and (2) changes as patients progress from diagnosis to relapsed and refractory disease. We have also discovered that PBMCs and CD138+ cells are overwhelmingly LMP7-expressing. Interestingly, the CD138+ cells derived from normal volunteers and newly diagnosed MM patients express a higher percentage of LMP7 compared with CD138+ cells from a patient with relapsed MM (Table 2) and the MM cell lines tested (Table 3). It is intriguing to speculate that progression from early-stage disease to a relapsed and refractory disease may be associated with a change in the CD138+ cells from mostly LMP7-expressing to mostly β5-expressing. However, more patient samples will be necessary to make a definitive correlation between disease state and β5 expression in CD138+ cells.
We also demonstrate that the hematologically derived cancer cell lines HS-Sultan, Molt-4, Arh77, RL, and ANBL6 (Table 3; and data not shown) express high levels of LMP7. On the other hand, solid tumor cell lines HT-29, A549, and MiaPaCa express low amounts of LMP7 (Table 3; and data not shown). Therefore, we conclude that inhibition of the CT-L activity in hematologic cells requires inhibition of both β5 and LMP7.
Antitumor effect of CT-L selective proteasome inhibition
Potent inhibition of the CT-L activity of the proteasome is a shared property of the clinically relevant PIs10 (ie, bortezomib, carfilzomib, NPI-0052, and CEP-18770) and may be a necessary feature of PIs to produce an antitumor response. We set out to test whether selective inhibition of the CT-L subunit is sufficient to produce an antitumor response in hematologically derived tumor cells without causing generalized cytotoxicity in nontransformed cells. In this study, we tested whether inhibition of either β5 or LMP7 subunit alone would lead to an antitumor activity in hematopoietically derived tumor cells. Surprisingly, inhibition of LMP7 had no antitumor effect, including in HS-Sultan cells that express primarily LMP7. Likewise, no antitumor effect was observed in MM1.S or Molt-4 cells treated with CPSI or IPSI. Others have proposed alternative proteasome subunit inhibition profiles as being a preferred strategy for treatment of MM. For example, Kuhn et al36 have suggested that selective LMP2 inhibition may be a viable approach to treatment for MM. One possible explanation for this discrepancy is that Kuhn et al demonstrate a strong antitumor effect when dosing the LMP2 inhibitor at concentrations that also target LMP7 and β536 (F.P. and M.K.B., unpublished results, November 2005), supporting our hypothesis that CT-L subunit inhibition is necessary for an antitumor response in MM cells. Chauhan et al37 have suggested that pan-proteasome inhibition may lead to improved efficacy of PIs in MM. We also observe strong antitumor activity by pan-proteasome inhibition; however, we also found profound cytotoxicity toward normal PBMCs (80% cell death), consistent with previous findings showing a cytotoxic effect in mononuclear cells treated with PIs.38,39 Given the proteasome's central role in numerous cellular pathways, it would be difficult to conceive of a setting where complete proteasome inhibition would not lead to generalized cytotoxicity.
At a concentration selective for the CT-L subunits, carfilzomib induced strong caspase 3/caspase 7 activity and produced an antitumor effect in MM1.S, HS-Sultan, and Molt-4 cells with minimal cytotoxic activity in PBMCs (Figure 4A-B). The superior caspase 3/caspase 7 induction in MMI.S cells compared with Molt-4 and HS-Sultan cells may explain the increased antitumor activity observed in MM1.S cells and is consistent with a previous report.40 We successfully recapitulated these results with a combination of IPSI and CPSI treatment (Figure 4A-B). To address the concern of residual β5 activity after CPSI treatment (25% residual β5 activity, Figure 3), we undertook a genetic approach to knock-down β5 protein expression in HS-Sultan cells (Figure 5A). In a setting where β5 accounted for approximately 5% of the total cellular CT-L activity, treatment with IPSI produced a strong antitumor effect (Figure 5B). In sum, our results support the notion that inhibition of the CT-L activity of the proteasome is sufficient for in vitro antineoplastic activity of PIs.
Effect of chymotrypsin-like selective proteasome inhibition on protein turnover
The effect of selective CT-L subunit inhibition on turnover of long-lived cellular proteins has been previously described.12 In the present study, we examined the turnover rate of a short-lived protein, GFPu, in HS-Sultan cells and monitored polyubiquitin accumulation in MM1.S and HS-Sultan cells. We observed that pan-proteasome inhibition had the most rapid effect on the kinetics of GFPu accumulation and most pronounced effects on total GFPu and polyubiquitin accumulation (Figures 6, 7). On the other hand, selective inhibition of the CT-L subunits had a more moderate effect on the kinetics of GFPu accumulation (38%-49%) and overall accumulation of GFPu (53%-73%) and polyubiquitin (81%-87%) compared with pan-proteasome inhibition (Figures 6–7). Compared with findings by Kisselev et al where inhibition of the CT-L subunit produced a moderate effect on protein turnover of bulk protein (10%-25%),12 we observed a more pronounced effect. This difference in protein turnover caused by CT-L subunit inhibition may reflect the type of proteins or cells studied by Kisselev et al12 (ie, long-lived bulk proteins in HeLa cells) and in this study (ie, short-lived destabilized protein in HS-Sultan cells). Notwithstanding these differences in protein turnover, selective CT-L subunit inhibition translates into a profound antitumor effect in hematologically derived tumor cells, especially in MM1.S cells, that are heavily reliant on the proteasome for protein degradation resulting from overproduction of immunoglobulin molecules. Inhibition of multiple subunits may produce a more significant antitumor effect,37 although generalized cytotoxicity is observed in nontransformed cells (Figure 4A). In sum, selective PI inhibition of CT-L activity may offer the greatest therapeutic index.
Molecular sequelae of chymotrypsin-like selective proteasome inhibition
Because PI treatment leads to pleiotropic effects on protein homeostasis, numerous mechanisms of action of PIs in hematologic malignancies have been uncovered in an effort to explain the observed antitumor effects. A favored PI mechanism of action postulates that MM cells constitutively express the active form of NF-κB by down-regulating IκBα and p100.39,41-43 In our study, the levels of IκBα and p100 were unaffected by either selective CT-L inhibition or pan-proteasome inhibition in MM1.S, HS-Sultan, and Molt-4 cells (Figure 7A; and data not shown). This difference in observations may be explained by the fact that these previous studies were performed in the presence of continuous (24-72 hours) proteasome inhibition, a situation that does not reflect the in vivo experience with either bortezomib44 or carfilzomib13 and will result in greater inhibition of proteasome activities.10,31 Moreover, other investigators have failed to observe the PI suppression of NF-κB activity in their systems.23,24 It is doubtful that NF-κB inhibition is the sole determinant for the antitumor activity of PIs because selective inhibition of the NF-κB pathway with PS-1145 does not produce an equivalent antitumor response in MM cells compared with PI treatment.22
ER stress is an alternative pathway that has been implicated in proteasome inhibitor-mediated cell death.28,40,45,46 To demonstrate whether inhibition of the CT-L activity of the proteasome modulates ER stress, we examined the ER stress marker CHOP and found its levels increased on PI treatment with pan-proteasome inhibition but not CT-L subunit inhibition (Figure 7). Unexpectedly, we also found that levels of phospho-eIF2α were decreased in the presence of both CT-L selective and pan-proteasome inhibition before the onset of cell death. A decrease in phospho-eIF2α has been reported in PI-treated pancreatic cancer cells28 and MM cells40 at 4 to 12 hours after PI treatment. On the other hand, an increase in phospho-eIF2α has been observed in nontransformed cells46 and tumor cells at 24 hours after PI treatment.40,45 In this study, phospho-eIF2α was monitored 4 to 6 hours after PI treatment (Figure 7D; and data not shown) to uncover the initial cellular effects of CT-L inhibition. The consequence of CT-L inhibition is an initial decrease28,40 in the ratio of phospho-eIF2α to eIF2α (Figure 7E), which may lead to an increase in protein synthesis.28 A possible downstream consequence of an initial increased protein synthesis is protein accumulation in the ER further exacerbating cellular stress, leading to protein synthesis shut-down,40,45 CHOP induction,28,40,45 and eventual up-regulation of the proapoptotic markers.14,24,29,37 Lastly, we observed the up-regulation of the Bcl2/Bax family member Noxa in cells where CT-L activity was inhibited. Noxa up-regulation has been consistently observed in MM29 and other cell types24 treated with PIs.
How can one explain the therapeutic window for PIs in the hematologic cancer setting given the role of the proteasome in the turnover of both bulk and regulated proteins in both nontransformed and cancer cells? It is well established that aneuploidy is a feature of most cancer cells. There is emerging evidence that cells with aberrant chromosome number have an imbalanced transcription and translation machinery leading to the expression of misfolded proteins and protein complexes.47-49 Compared with nontransformed cells, this feature of cancer cells puts a burden on the cancer cell's degradation machinery to dispose of these misfolded proteins.25-27 MM is a paramount example of a tumor type that has a strong need for the proteasome to accommodate the heavy protein burden, resulting from overexpression of immunoglobulin molecules.23 In light of this evidence, it is safe to assume that specific interventions in MM and other cancer cells with PI treatment regimens require accurate and specific modulation of proteasome activity to take advantage of the overtaxed need for protein degradation in these tumor cells without causing cytotoxicity in nontransformed cells. Our results are consistent with a model in which selective inhibition of the CT-L activity of the proteasome (both β5 and LMP7) results in short-lived protein buildup in cells (as measured by polyubiquitin and GFPu accumulation) and may lead to an ER stress response (decrease in phospho-eIF2α). These events lead to an increase in the proapoptotic marker Noxa and caspase3/caspase 7 activation resulting in cancer cell death with minimal cytotoxicity in nontransformed cells. Our results are consistent with models proposed by other groups that strongly suggest that the principal mode of action of PI is via disruption of the unfolded protein response/ER stress pathway, leading to apoptosis and cell death.28,46,50
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Christopher Kirk (Proteolix) and Dr Raymond Deshaies (California Institute of Technology) for their insightful suggestions in the writing of this manuscript.
Authorship
Contribution: F.P. designed, performed, and interpreted many of the experiments and wrote the manuscript; S.J.L., M.A., E.S., and K.L. performed experiments; J.B.L., D.R.M., and P.R. generated the Δβ5 HS-Sultan cells; C.S., Y.L., and K.D.S. designed and synthesized the CPSI, IPSI, and PABP compounds; and M.K.B. supervised the progress of the work.
Conflict-of-interest disclosure: F.P., S.J.L., M.A., E.S., K.L., C.S., Y.L., K.D.S., and M.K.B. receive(d) stock options as part of their employment with Proteolix. The remaining authors declare no competing financial interests.
The current address of Dr Micklem is BerGenBio A/S, Bergen, Norway.
Correspondence: Francesco Parlati, Proteolix Inc, 333 Allerton Ave, South San Francisco, CA 94080; e-mail: francescoparlati@gmail.com or fparlati@proteolix.com.