Abstract
Nonhomologous end-joining DNA repair factors, including Artemis, are all required for the repair of DNA double-strand breaks, which occur during the assembly of the variable antigen recognition domain of B-cell receptors and T-cell receptors through the V(D)J recombination. Mature B cells further shape their immunoglobulin repertoire on antigen recognition notably through the class switch recombination (CSR) process. To analyze the role of Artemis during CSR, we developed a mature B-cell–specific Artemis conditional knockout mouse to bypass the absence of B cells caused by its early deficit. Although CSR is not overwhelmingly affected in these mice, class switching to certain isotypes is clearly reduced both in vitro on B-cell activation and in vivo after keyhole limpet hemocyanin immunization. The reduced CSR in Artemis-deficient B cells is accompanied by the increase in DNA microhomology usage at CSR junctions, the imprint of an alternative DNA end-joining pathway. Likewise, significant increase in DNA microhomology usage is the signature of CSR junctions obtained from human RS-SCID patients harboring hypomorphic Artemis mutations. Altogether, this indicates that Artemis participates in the repair of a subset of DNA breaks generated during CSR.
Introduction
The large diversity of antigen receptors on T and B cells is provided in lymphoid organs by different mechanisms. The first one, the V(D)J recombination, allows for the somatic rearrangement of V (variable), D (diversity), and J (joining) gene loci into V(D)J segments coding for the variable antigen recognition domain of B-cell receptors and T-cell receptors.1,2 This process is initiated by Rag (recombination acting gene) 1 and 2 endonuclease, which introduces DNA double-strand breaks (DSBs) between V, D, and J coding segments and flanking recombination signal sequences, thereby generating hairpin-sealed DNA coding ends on the chromosome and blunt signal ends excised from the chromosome. The repair of the DSBs is then ensured by the general DNA repair machinery, composed of at least 7 known factors of the nonhomologous end-joining (NHEJ) pathway: Ku70, Ku80, DNA-PKcs, Artemis (Art), XRCC4, Cernunnos/XLF, and DNA ligase IV. The DSB is recognized by the Ku70/Ku80 heterodimer, which recruits DNA-PKcs to form the DNA-PK complex. DNA-PKcs, a DNA-dependent PI3 kinase, recruits and activates Artemis by a phosphorylation process. Artemis is a 5′-3′ exonuclease, which acquires an endonucleolytic activity on 5′ and 3′ overhangs as well as DNA hairpins on activation by DNA-PKcs in vitro whose function during V(D)J recombination is to open the hairpin-sealed coding ends.3 Mutations in genes coding for NHEJ factors are responsible for the onset of various types of severe combined immunodeficiencies (SCID) in humans.4-7 NHEJ represents the major DNA-DSB repair pathway in mammalian cells, and these patients present a cellular hypersensitivity to ionizing radiations (radiosensitive, RS-SCID). Similarly, NHEJ factor gene inactivation in mice results in a profound impaired B- and T-cell development.8-14
After completion of V(D)J recombination, mature immunoglobulin M+ (IgM+) B cells migrate to secondary lymphoid organs, where the immune B-cell repertoire is further shaped by 2 independent processes: somatic hypermutation and class switch recombination (CSR), which enhances the affinity and modifies the effector function of antibodies, respectively, without altering their antigenic specificity. CSR results in the replacement of the IgM constant region (Cμ) with one of the downstream CH gene (γ, ϵ, or α) by a recombination mechanism between large repetitive switch (S) regions located upstream each constant region (except δ), allowing for the fusion of the 2 S regions and excision of intervening DNA.15,16 The B-cell–specific activation-induced cytidine deaminase factor (AID) initiates the CSR reaction by deaminating cytidine residues to uracil within S regions in a transcription-dependent manner, leading to DNA breaks in the 2 opposite strands of DNA.17,18 Much experimental evidence supports the generation of DSBs during CSR,15 although the exact molecular mechanism by which the initial AID-mediated DNA lesion is further processed to DNA-DSB is not yet fully understood.19 As NHEJ represents one of the main pathways for DNA-DSB repair in mammals, it has appeared as an evident candidate for DNA repair during CSR. Analyzing NHEJ during CSR in vivo has been challenging, however, because NHEJ knockout (KO) mice fail to complete V(D)J recombination and are thus devoid of mature B and T lymphocytes, and are embryonic-lethal in the case of XRCC4 and DNA ligase IV inactivation.8-14 To bypass these limitations, we and others have developed conditional XRCC4 KO mice using LoxP/Cre strategy, by which XRCC4 is specifically deleted in mature B cells through the CD21 promoter-driven expression of the Cre recombinase.20-22 These studies, which connect the NHEJ DNA repair pathway in general, and the XRCC4/DNA-ligase IV complex in particular, to the resolution of DSBs required for effective CSR, also demonstrate the existence of an alternative, XRCC4/DNA-ligase IV-independent, DNA end-joining mechanism during CSR. One characteristic of this alternative pathway is the usage of DNA microhomology for the completion of the CSR junction,21 as first noted in various DNA repair deficiency settings in humans.16 Such a microhomology-mediated end-joining pathway during CSR is facilitated by the intrinsic homology that exists between the various S regions.
Artemis is absolutely required during V(D)J recombination for opening Rag1/2 generated DNA hairpins but also participates in the repair of a subset of DNA lesions produced by ionizing radiation.23 Given the probable important heterogeneity of DNA-DSB generated during CSR, the possible implication of Artemis in the resolution of some of these can be raised. Rooney et al developed an Artemis-deficient mouse in which the development of mature B cells is rescued by the knock-in of heavy and light chain Ig loci to show that CSR is not impaired in these Artemis-deficient monoclonal B cells.24 On the other hand, conflicting results were obtained on the role of DNA-PK catalytic subunit (DNA-PKcs) on CSR using either DNA-PKcs KO or mutated (scid) mice.25-28 More recently, Franco et al used a sophisticated fluorescence in situ hybridization approach to demonstrate the persistence of AID-dependent DNA-DSB at the IgH locus in the context of either DNA-PKcs or Artemis deficiency, paving the way to a role for Artemis during CSR.29
To directly tackle the role of Artemis during CSR in a physiologic context (that is on polyclonal B cells and in the presence of T cells), we developed a conditional Art-KO mouse model, in which the Artemis gene is specifically inactivated in mature B cells. We show here that, although CSR is not overwhelmingly affected in Artemis-deficient B cells, switching to certain isotypes is clearly diminished. This is the case in vitro for the switch to IgG3 and in vivo to IgA on intragastric keyhole limpet hemocyanin (KLH) immunization. The decreased CSR is accompanied by a modification of CSR junctions in both cases. Likewise, we found that CSR is affected in 2 human patients with hypomorphic Artemis mutations, revealing extensive usage of DNA microhomology at Sμ-Sα junctions.
Methods
Generation of ES cells, Art-KO, and conditional Art-KO mice
The generation of Art-KO embryonic stem (ES) cells and mice was performed at the Mouse Clinical Institute, Institut Clinique de la Souris, Illkirch, France (http://www-mci.u-strasbg.fr). A genomic DNA clone containing a portion of the Artemis (Art) locus (Figure 1A targeting construct) was isolated from a 129/Ola phage genomic library (gift of A. Begue, Institut Pasteur de Lille). The targeting construct was designed to replace exon 12 with floxed exon 12 and floxed PGK-neoR cassette. After transfection of 129 derived P1 ES cells (Institut Clinique de la Souris) and selection under geneticin, homologous recombinants were identified by Southern blot analysis using 5′ (probe B) and 3′ (probe A) external probes (data not shown). One selected homologous recombinant ES clone was further electroporated with a plasmid expressing the Cre recombinase and screened by polymerase chain reaction (PCR) for Cre-mediated deletion events. One ES clone (clone 1) harboring a complete exon12 + Neo deletion and one ES clone (clone 2) containing a floxed exon 12 (with Neo deletion) were injected into C57BL/6J blastocysts to generate chimeric mice. Chimeric mice generated from both ES clones were bred to derive Art−/+ and Artfl/+ heterozygous mouse line, respectively (Figure 1B). Art+/− and Artfl/+ mouse lines were further backcrossed to C57BL/6 mice for at least 6 generations. Homozygous Art−/− mice were generated by intercross of Art+/− mice. Artfl/+ mice were crossed to Art+/− mice and then to CD21-Cre transgenic mice (provided by K. Rajewsky, Harvard Medical School, Boston, MA) to generate conditional Art-deficient (ArtΔ/−) mice. Mice were screened by PCR on tail DNA using the following primers: K5.4F (5′ TTCCTCCTTCCCTTCCCCCACATAG 3′), K5.3R (5′ CACAGCTCTGGCACTCGCATCCCC 3′) and K5.1F (5′ CTTGCCTACTTTTGGAGTTCTCAGGG 3′) for Artemis locus (respectively, primers 1, 2, and 3 in Figure 1B) and CD21-CreF (5′ ACGACCAAGTGACAGCAATG 3′) and CD21-CreR (5′ CTCGACCAGTTTAGTTACCC 3′) for CD21-Cre transgene. All experiments and procedures were performed in compliance with and with the approval of the French Ministry of Agriculture's Regulations for Animal Experimentation (Act no. 87847, October 19, 1987, as modified in May 2001).
Analysis of Artemis deletion by PCR
B-cell–specific Artemis deletion was quantified by PCR on DNA from liver, thymus, and purified splenic mature B cells using primers, respectively, K5.4F, K5.5R (5′ TCCATGACCTTATCCACAGTGAGGC), and K5.3R designed in the genomic sequence upstream, within, and downstream of Artemis exon 12 (primers 1, 5, and 3, respectively, in Figure 1B). The PCR assay was performed on 5 ng of DNA. PCR products were stained with SYBR-Green. The area under the peak corresponding to the Art−Δ and Artfl bands was quantified by ImageJ software Version 1.41 (National Institutes of Health), and the ratio between the 2 alleles was determined after normalization for SYBR-Green incorporation and correction for PCR efficiency.
Analysis of Artemis transcripts by RT-PCR
cDNA was prepared from total RNA extracted from Art+/+ (WT) and Art−/− splenocytes, and Artemis transcripts were amplified using the following primers: Art-exon 1-F (5′-GAAAGCCCGTGCCTACTT-3′) with Art-exon 11-R (5′-GGAGGAGTGAAAAGAGAAGC-3′), Art-exon 12-R (5′-TCTGGCTCTCTTCAATTTTTCCAAGCG-3′) or Art-exon 14-R (5′-GGGTTTCTAGCTCTTCTCCA-3′). Hypoxanthine phosphoribosyl transferase (HPRT)-specific PCR was used as loading control. HPRT-specific primers were: HPRT-F (5′-GTTCTTTGCTGACCTGCTGGATTA-3′) and HPRT-R (5′-GTCAAGGGCATATCCAACAACAAAC-3′).
Flow cytometric analysis of lymphocyte populations and Ig CSR
Cell phenotype on blood, thymus, bone marrow, and splenic lymphoid populations and analysis of CSR were performed by 4-color fluorescence analysis as previously described.20
Purification and activation of splenic mature B cells in vitro
Mature B cells were purified from spleen, labeled with carboxyfluorescein diacetate succinimidyl ester, and stimulated for 4 days with lipopolysaccharide, interleukin-4, anti-mouse CD40 antibody, transforming growth factor-β1, and interferon-γ as previously described.20
In vivo immunization
Mice were immunized via intragastric instillation with 1 mg KLH (Calbiochem) and 10 μg cholera toxin (Sigma-Aldrich) in 0.2 M NaHCO3. Mice were rechallenged in the same conditions 22 days after primary immunization, and serum collected 4 days later to assess KLH-specific serum IgM and IgA by enzyme-linked immunosorbent assay (ELISA).
Antibody detection by ELISA
Total IgM, IgG1, IgG2b, IgG3, or IgE levels were determined in serum of 9- to 18-week-old mice as previously described.20 ELISA determined KLH-specific IgM and IgA in serum of KLH immunized mice on 2.5 μg/well KLH-coated plates.
CSR junction analyses
Murine Sμ-Sγ1, Sμ-Sγ3, and Sμ-Sα junctions were amplified by PCR from genomic DNA prepared from IgG1+ and IgG3+ splenic B cells after 4 days of stimulation with lipopolysaccharide or purified IgA+ B cells isolated from Peyer patches of KLH-immunized mice, respectively, through a 2-step PCR as described.30 Sμ-Sγ1 junctions were amplified using SμE3: AATGGATACCTCAGTGGTTTTTAATGGTGGGTTTAA; and SγR1: CAATTAGCTCCTGCTCTTCTGTGG. Sμ-Sγ3 junctions were amplified using SμF1: AGAGACCTGCAGTTGAGGCC; and Sγ3R1: GCTAGACCTACTGAGTTCCTGTGCTTG; followed by SμF2: GCAGCTGGCAGGAAG-CAGGTCATG; and Sγ3R2: CTAGCACTCCAGCCTGCCTCAG. Sμ-Sα junctions were amplified Sμ3F1: GCAGGTCGGCTGGACTAACTC; and SαR1: AGCAGTGAGTTTAACAATCC; followed by Sμ3NESTF2: GGCCAGACTCATAAAGCTTGC; and SαNESTR2: GTCCAGTCATGCTAATTCACCC. PCR products were cloned into the PCR2.1 vector (Invitrogen), sequenced, and analyzed using CLUSTALW (http://align.genome.jp) or BLAST in comparison with mouse Sμ (MUSIGCD07), Sγ1 (MUSIGHANB), Sγ3 (MUSIGD18), and Sα (MUSIALPHA) reference sequences. Human Sμ-Sα junctions were amplified by PCR from genomic DNA prepared whole blood and analyzed as described.30 The work on patient material was approved by ethical committee from Necker Hospital for Sick Children and the Commission nationale de l'informatique et des libertés (no. 902096). Patients' families gave their written informed consent for this study in accordance with the Declaration of Helsinki.
Results
Targeted inactivation of the Artemis gene
To explore the role of Artemis in CSR, we generated a conditional allele of the Artemis gene by flanking exon 12 with LoxP sites. The minimal catalytic core of Artemis for V(D)J recombination is composed of the metallo-β-lactamase/β CASP domain (exons 1-13), as previously shown in an in-chromosome V(D)J recombination assay: the level of V(D)J recombination and the quality of the resulting coding joints are identical whether Artemis-deficient fibroblasts are complemented with full-length Artemis protein or with βLact/β CASP domains-only form.31 However, a more truncated form of Artemis (exons 1-12) is unable to complement the V(D)J defect in Artemis-deficient cells (J.-P.d.V., unpublished results, 2004). We therefore reasoned that a truncated form of Artemis composed of exons 1 to 11, which could result from the deletion of exon 12, would not contain the structural elements required for Artemis function. We designed a targeting strategy by which exon 12 was bordered by 2 LoxP sites (Figure 1A). An appropriately targeted ES clone was transfected with a Cre expression plasmid for the further LoxP-mediated deletion of the Neo resistance gene and the exon 12 in vitro. The resulting clones 1 and 2, which have excised neoR and Artemis exon 12 (Art− allele) or neoR only (Artfl allele), respectively, were used to generate the corresponding mouse lines (Figure 1B).
Artemis gene targeting. (A) Schematic representation of the targeted region of murine Artemis locus (WT, Art+), targeting construct, targeted allele, neoR deleted, and exon 12 floxed (Artfl) allele, and neoR deleted and exon 12 deleted (Art−) allele. The PGK-neoR cassette is inserted in the opposite transcriptional orientation. Restriction sites: E, EcoRI. Filled boxes represent exons; filled triangles, LoxP sites. (B) Screening of Art+/− and Artfl/+ mouse line progenies by PCR on tail DNA, using primers 1 (K5.4), 2 (K5.1), 3 (K5.3), and 5 (K5.5); see “Generation of ES cells, Art-KO, and conditional Art-KO mice.” Filled box represents exon 12; filled triangles, LoxP sites. Primers 1 and 3 reveal a 1-kb WT (Art+) and 0.4-kb (Art−) deleted alleles, respectively. Primers 2 and 3 reveal a 0.25-kb WT and 0.34-kb flox (Artfl) alleles, respectively. Vertical lines have been inserted to indicate repositioned gel lanes. (C) RT-PCR analysis of Artemis transcripts in Art−/− mice. RT-PCR was performed on total RNA from spleen of Art−/− (KO) and littermate control Art+/+ (WT) mice. Specific primers were used to detect Artemis transcripts containing exons 1 to 14, 1 to 12, and 1 to 11. HPRT-specific PCR was used as loading control.
Artemis gene targeting. (A) Schematic representation of the targeted region of murine Artemis locus (WT, Art+), targeting construct, targeted allele, neoR deleted, and exon 12 floxed (Artfl) allele, and neoR deleted and exon 12 deleted (Art−) allele. The PGK-neoR cassette is inserted in the opposite transcriptional orientation. Restriction sites: E, EcoRI. Filled boxes represent exons; filled triangles, LoxP sites. (B) Screening of Art+/− and Artfl/+ mouse line progenies by PCR on tail DNA, using primers 1 (K5.4), 2 (K5.1), 3 (K5.3), and 5 (K5.5); see “Generation of ES cells, Art-KO, and conditional Art-KO mice.” Filled box represents exon 12; filled triangles, LoxP sites. Primers 1 and 3 reveal a 1-kb WT (Art+) and 0.4-kb (Art−) deleted alleles, respectively. Primers 2 and 3 reveal a 0.25-kb WT and 0.34-kb flox (Artfl) alleles, respectively. Vertical lines have been inserted to indicate repositioned gel lanes. (C) RT-PCR analysis of Artemis transcripts in Art−/− mice. RT-PCR was performed on total RNA from spleen of Art−/− (KO) and littermate control Art+/+ (WT) mice. Specific primers were used to detect Artemis transcripts containing exons 1 to 14, 1 to 12, and 1 to 11. HPRT-specific PCR was used as loading control.
The validation of the exon 12 deletion strategy was performed on Art−/− mice. RT-PCR analysis (Figure 1C) of Artemis expression failed to detect any exons 1 to 12 transcript in Art−/− (KO) splenocytes, thus confirming exon 12 deletion in these mice. Amplification of exons 1 to 14 revealed the expression of a shorter transcript in Art−/− mice compared with Art+/+ (WT) mice. Sequence analysis of this truncated transcript revealed the out-of-frame splicing of exon 11 to 13, generating a premature stop codon immediately downstream exon 11 (data not shown). We conclude that the deletion of Artemis exon 12 produces, if the protein is expressed, a nonfunctional (exons 1-11) truncated form.
Artemis deletion profoundly impairs B- and T-cell development
To verify the loss of function phenotype of Art KO mice, we analyzed B- and T-cell populations in lymphoid organs of Art−/− (KO) and Art+/+ (WT) mice. B- and T-cell development is strongly impaired in Art−/− mice compared with Art+/+ control mice, as shown by a 200-fold reduction in total thymocyte number (1.4 million in Art−/− vs 280 millions in Art+/+), a 30-fold reduction in splenocytes (5.1 million in Art−/− vs 154 millions in Art+/+), and a 6-fold reduction in blood mononuclear cells (650 cells/mm3 in Art−/− vs 3700 cells/mm3 in Art+/+). Flow cytometry analysis confirmed the T-cell development arrest at the CD4− CD8− (DN) progenitor stage in the thymus of Art−/− mice, with almost no emergence of any CD4+ CD8+ (DP) or CD4+ CD8− and CD4− CD8+ (SP) thymocytes (Figure 2A). Although few CD4+ CD8−–positive cells were present in thymus, spleen, and blood, their frequency never reached the level corresponding to the leaky state described in a fraction of Artemis-deficient mice developed by Rooney et al.10 Likewise, B-cell ontogeny was arrested at progenitor stages in Art−/− mice (Figure 2B). The B-cell development block happens before the immature stage in the bone marrow, as shown by the absence of B220+ IgM+ immature and recirculating mature B cells with a residual (albeit reduced) proB-preB B220+ IgM− B-cell population. In addition, no mature B220+ IgM+ B cells are found in the periphery, spleen, and blood (Figure 2B). The block of T- and B-cell development before DP and at proB-preB stages, respectively, is consistent with a profound V(D)J recombination defect. We conclude from these analyses that the deletion of Artemis exon 12 results in a loss of Artemis function, which recapitulates the immunologic features of human RS-SCID patients carrying various deletions or mutations in Artemis βLact/βCASP domain5 as well as the 2 previous Art-KO murine models.10,14
B- and T-cell development in Artemis-deficient mice. The phenotype of B and T cells was analyzed by flow cytometry in lymphoid organs. Staining antibodies are indicated. Plots were gated on viable (propidium iodide–negative) lymphoid cells. Percentages of cells in the quadrants are given. (A) Analysis of T-cell subsets in thymus, spleen, and blood illustrates an absence of mature CD4+ and CD8+ cells in Artemis-deficient mice. (B) Analysis of B-cell subsets in bone marrow (BM), blood, and spleen shows a blockade of B-cell development at pro-B–pre-B (B220+/IgM−) stage in BM (left), leading to an absence of immature and recirculating mature (both B220+/IgM+) B cells in BM, and of B220+/IgM+ mature B cells in spleen and blood.
B- and T-cell development in Artemis-deficient mice. The phenotype of B and T cells was analyzed by flow cytometry in lymphoid organs. Staining antibodies are indicated. Plots were gated on viable (propidium iodide–negative) lymphoid cells. Percentages of cells in the quadrants are given. (A) Analysis of T-cell subsets in thymus, spleen, and blood illustrates an absence of mature CD4+ and CD8+ cells in Artemis-deficient mice. (B) Analysis of B-cell subsets in bone marrow (BM), blood, and spleen shows a blockade of B-cell development at pro-B–pre-B (B220+/IgM−) stage in BM (left), leading to an absence of immature and recirculating mature (both B220+/IgM+) B cells in BM, and of B220+/IgM+ mature B cells in spleen and blood.
Conditional Artemis deletion in mature B cells
To analyze the role of Artemis during CSR, we specifically deleted Artemis in mature B cells through LoxP/Cre technology once a complete immune system has developed. Artfl/− mice (bearing one floxed and one KO allele) were crossed to CD21-Cre transgenic mice, in which the Cre recombinase expression is driven by the CD21 promoter, specifically in mature B cells,32 to create Cre+Artfl/− mice, which will be thereafter named ArtΔ/− (where Δ stands for B-cell–restricted Artemis deletion). Results obtained on ArtΔ/− mice have always been compared with Cre−Artfl/− littermate controls, thereafter named Artfl/−. We evaluated the extent of Artf deletion by PCR analysis on liver, thymus, and purified mature splenic B lymphocyte DNA (Figure 3A) using primers 1, 3, and 5 depicted in Figure 1B. The quantification of Artfl and ArtΔ PCR products (Figure 3B) revealed a 1:1 ratio in liver and thymus of both Cre+ and Cre− mice. In CD43− purified splenic B cells from Cre+ animals, the Artfl allele is reduced by 75%, whereas the ratio between the 2 alleles remain 1:1 in the absence of Cre.
Conditional Artemis deletion in mature B cells. (A) PCR analysis of Artemis deletion. Artfl and ArtΔ alleles were amplified from genomic DNA from liver, purified splenic mature B cells, and thymocytes from Artfl/− expressing Cre (+) or not (−) using a combination of primers 1, 3, and 5 (depicted in Figure 1B). (B) Quantification of the Artemis deletion in mature B cells. PCR products from panel A were quantified using ImageJ to calculate the ratio between the Artfl and the ArtΔ alleles. B cells expressing the Cre recombinase show a 74% deletion of the Artfl allele. In all the other situations, the ratio between the 2 alleles is approximately 1:1. (C) Analysis of splenic B-cell subpopulations on Artemis deletion. The phenotype of B-cell subpopulations was analyzed by flow cytometry in spleen. Staining antibodies are indicated. Plots were gated on viable (propidium iodide–negative) IgM+ lymphoid cells. Percentages of cells in the quadrants are given. T1 indicates transitional type 1 B cells (IgM+/CD21−/CD23−); T2, transitional type 2 B cells (IgM+/CD21+/CD23+); MZ, marginal zone B cells (IgM+/CD21+/CD23−); and M, mature follicular B cells (IgM+/CD21low/CD23+).
Conditional Artemis deletion in mature B cells. (A) PCR analysis of Artemis deletion. Artfl and ArtΔ alleles were amplified from genomic DNA from liver, purified splenic mature B cells, and thymocytes from Artfl/− expressing Cre (+) or not (−) using a combination of primers 1, 3, and 5 (depicted in Figure 1B). (B) Quantification of the Artemis deletion in mature B cells. PCR products from panel A were quantified using ImageJ to calculate the ratio between the Artfl and the ArtΔ alleles. B cells expressing the Cre recombinase show a 74% deletion of the Artfl allele. In all the other situations, the ratio between the 2 alleles is approximately 1:1. (C) Analysis of splenic B-cell subpopulations on Artemis deletion. The phenotype of B-cell subpopulations was analyzed by flow cytometry in spleen. Staining antibodies are indicated. Plots were gated on viable (propidium iodide–negative) IgM+ lymphoid cells. Percentages of cells in the quadrants are given. T1 indicates transitional type 1 B cells (IgM+/CD21−/CD23−); T2, transitional type 2 B cells (IgM+/CD21+/CD23+); MZ, marginal zone B cells (IgM+/CD21+/CD23−); and M, mature follicular B cells (IgM+/CD21low/CD23+).
The overall development of B cells from CD21− to CD21+ mature B cells (in which CSR takes place) was not affected in ArtΔ/− mice as revealed by the normal distribution of the various splenic B-cell subpopulations (transitional T1: IgM+/CD21−/CD23−, T2: IgM+/CD21+/CD23+, marginal zone: IgM+/CD21+/CD23−, and follicular mature: IgM+/CD21low/CD23+) (Figure 3C). Furthermore, T-cell subpopulations in thymus, spleen, and peripheral blood were not different in frequency and absolute numbers between ArtΔ/− and Artfl/− mice as expected (data not shown). These observations guarantee that mature B cells from ArtΔ/− mice will receive T-cell costimulation signals required for an efficient CSR process.
Normal Ig isotype representation in serum from Artemis KO mice
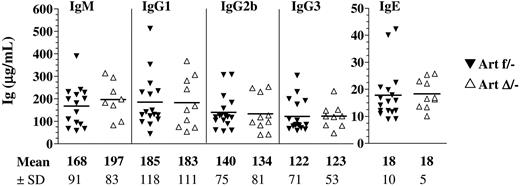
As a first indication of a possible role of Artemis in CSR in vivo, we quantified the basal Ig levels of different isotypes in the serum by ELISA. IgM and the various switched Ig titers were equivalent between ArtΔ/− and Artfl/− mice (Figure 4), indicating that Artemis is not critically required during CSR as first suggested by Rooney et al using Artemis-deficient IgH/L KI mice.24
Seric Ig production in ArteΔ/− mice. Sera from 9- to 18-week-old Artfl/− and ArtΔ/− mice were collected, and total IgM, IgG1, IgG2b, IgG3, and IgE (μg/mL) were determined by ELISA. None of the differences between groups reached statistical significance (.46 < P < .97).
Seric Ig production in ArteΔ/− mice. Sera from 9- to 18-week-old Artfl/− and ArtΔ/− mice were collected, and total IgM, IgG1, IgG2b, IgG3, and IgE (μg/mL) were determined by ELISA. None of the differences between groups reached statistical significance (.46 < P < .97).
Decreased switch to IgG3 in vitro in ArtΔ/− B cells
The accumulation of seric Ig can mask a slight CSR defect given the long half-life of Ig (notably IgG) and of plasma cells. To evaluate the CSR in Artemis-deficient mice in more details, we analyzed purified splenic mature B cells from ArtΔ/− and Artfl/− mice for their ability to undergo CSR in vitro on appropriate stimulation. Surface Ig expression was analyzed by flow cytometry after stimulation of splenic CD43− mature B cells with various polyclonal B-cell activators (Figure 5). Because B-cell capacity to undergo CSR is related to their proliferation potential,33 we linked the analysis of CSR to that of B-cell proliferation through labeling with carboxyfluorescein succinimidyl ester (CFSE). The frequency of switched Ig+ ArtΔ/− B cells was not different from controls for IgG1, IgE, and IgA, and slightly reduced for IgG2b (10% decrease, P = .08). We also noticed a significant (P > .003) 23% decrease in the frequency of IgG3+ ArtΔ/− B cells (3.6%) compared with WT controls (4.7%). CFSE labeling coupled to Ig isotype determination demonstrated that the overall B-cell proliferation was not affected by the Artemis deletion and that the altered switching to IgG2b and IgG3 CSR in Artemis-deficient mature B cells was not attributable to a cell proliferation defect (Figure 5B-D). Altogether, these results show that Artemis deletion in polyclonal mature B cells has a slight effect on the ability of B cells to undergo CSR in vitro.
In vitro CSR analysis in ArteΔ/− B cells. Surface expression of switched isotypes. Purified B cells from Artfl/− and ArtΔ/− mice were labeled with CFSE and stimulated for 4 days with various polyclonal B-cell activators for switching to IgG1, IgE, IgG2b, IgG3, and IgA (“Purification and activation of splenic mature B cells in vitro”), and stained with anti-B220 and specific anti-Ig antibodies for flow cytometric analysis. All cells were B220+, assessing for the efficiency of B-cell purification. (A) Percentage of total B cells expressing IgG1, IgE, IgG2b, IgG3, and IgA, as determined by flow cytometric analysis on B cells from Artfl/− (▴) and ArtΔ/− mice (▵), after 4 days of in vitro stimulation. **Statistically significant difference (P > .003, 2-tailed Mann-Whitney test). (B) Analysis of CSR in the various subsets of proliferative B cells according to CFSE intensity. Plots show IgG1, IgE, IgG2b, IgG3, and IgA staining together with CFSE, on viable (propidium iodide–negative) lymphoid cells. The percentages of switched cells are indicated. (C-D) Percentages of total (C) and IgG1-, IgE-, IgG2b-, IgG3-, or IgA-switched (D) B cells that have undergone the indicated numbers of cell division, for Artfl/− and ArtΔ/− mice. Columns and bars represent mean and SD of independent animals depicted in panel A.
In vitro CSR analysis in ArteΔ/− B cells. Surface expression of switched isotypes. Purified B cells from Artfl/− and ArtΔ/− mice were labeled with CFSE and stimulated for 4 days with various polyclonal B-cell activators for switching to IgG1, IgE, IgG2b, IgG3, and IgA (“Purification and activation of splenic mature B cells in vitro”), and stained with anti-B220 and specific anti-Ig antibodies for flow cytometric analysis. All cells were B220+, assessing for the efficiency of B-cell purification. (A) Percentage of total B cells expressing IgG1, IgE, IgG2b, IgG3, and IgA, as determined by flow cytometric analysis on B cells from Artfl/− (▴) and ArtΔ/− mice (▵), after 4 days of in vitro stimulation. **Statistically significant difference (P > .003, 2-tailed Mann-Whitney test). (B) Analysis of CSR in the various subsets of proliferative B cells according to CFSE intensity. Plots show IgG1, IgE, IgG2b, IgG3, and IgA staining together with CFSE, on viable (propidium iodide–negative) lymphoid cells. The percentages of switched cells are indicated. (C-D) Percentages of total (C) and IgG1-, IgE-, IgG2b-, IgG3-, or IgA-switched (D) B cells that have undergone the indicated numbers of cell division, for Artfl/− and ArtΔ/− mice. Columns and bars represent mean and SD of independent animals depicted in panel A.
Reduced in vivo Ig switch to IgA on KLH immunization
To extend the notion of a possible role of Artemis in CSR, we set up to analyze the consequence of Artemis deletion on a specific immune response. For this, 4 different series of mice were immunized mice twice with KLH via intragastric instillation and analyzed for anti-KLH IgA specific response both in serum and in Peyer patches. As shown in Figure 6A, the frequency of IgA+ B cells in the Peyer patches after immunization was constantly lower (22.7%-61.4% inhibition) in the Artemis-deleted mice compared with littermate controls. This inhibition reached statistical significance in the 2 series with several animals analyzed (2-tailed Student t test, P = .048 in series 2 and P = .024 in series 4). Considering the 8 immunized mice together, Artemis deficiency caused a significant 52% decrease in IgA+ B cells after immunization (2-tailed Mann-Whitney test, P < .005). The important reduction (77.2%-83.9% inhibition) of anti-KLH specific IgA in the serum of immunized deficient mice (Figure 6B) further attested for the significant CSR defect (2-tailed Mann-Whitney test, P < .038 when considering the 8 mice) caused by Artemis deletion in these mice.
Impaired IgA production in ArtΔ/− mice on KLH immunization. Four series of mice were immunized with KLH orally for a total of 8 ArtΔ/− and Artfl/− animals. (A) Percentage of Peyer patch B220+ cells expressing IgA, as determined by flow cytometric analysis. The decrease in IgA+ cells is statistically significant in series 2 and 4 (2-tailed Student t test, P = .048 and P = .024, respectively). (B) Serum levels of KLH-specific IgA and IgM were measured by ELISA (8 ArtΔ/− and 8 Artfl). The variations in immunization efficiency were normalized by considering the ratio (anti-KLH IgA/anti-KLH IgM) for each mouse.
Impaired IgA production in ArtΔ/− mice on KLH immunization. Four series of mice were immunized with KLH orally for a total of 8 ArtΔ/− and Artfl/− animals. (A) Percentage of Peyer patch B220+ cells expressing IgA, as determined by flow cytometric analysis. The decrease in IgA+ cells is statistically significant in series 2 and 4 (2-tailed Student t test, P = .048 and P = .024, respectively). (B) Serum levels of KLH-specific IgA and IgM were measured by ELISA (8 ArtΔ/− and 8 Artfl). The variations in immunization efficiency were normalized by considering the ratio (anti-KLH IgA/anti-KLH IgM) for each mouse.
Altered CSR junction in ArtΔ/− B cells
In mice with B-cell–specific inactivation of either XRCC4 or DNA-ligase IV, not only is the CSR activity reduced,20,21 but sequencing the CSR junctions revealed an increased usage of DNA microhomology,21 suggesting the existence of an alternative, XRCC4 and DNA-ligase IV–independent, DNA end-joining pathway in these situations. The same observation was made on CSR junctions obtained from human SCID patients with hypomorphic DNA-ligase IV mutations34 (J.P.d.V., unpublished observations, 2007). To gain further insights in the role of Artemis within these 2 end-joining pathways, we sequenced Sμ-Sγ1 and Sμ-Sγ3 junctions (Figure 7A-B) from in vitro activated ArtΔ/− B cells and Sμ-Sα junctions (Figure 7C) from freshly isolated IgA+ B cells in Peyer patches from KLH-immunized ArtΔ/− mice and Sμ-Sγ3 compared them with CSR junctions (Figure 7C-D) from in vitro activated ArtΔ/− B from Artemis-proficient littermate controls (Figure 7D). Whereas the median length of DNA microhomology (with the tolerance of one mismatch in the stretch of microhomology) was 2.5 bp and 3.0 bp for Sμ-Sγ3 and Sμ-Sα, respectively, in the WT situations, these values raised to 3.0 bp and 4.0 bp in Artemis-deficient B cells. Although these differences in microhomology usage do not reach statistical significance (2-tailed Mann-Whitney test, P = .532 and P = .119, respectively), they are in the range with what has been observed in XRCC4 KO mice.21 Interestingly, there was no variation in microhomology usage (2.0 bp) within Sμ-Sγ1 CSR junctions between WT and Art-deficient mice, an isotype the switch to which is not affected by Artemis deletion (Figure 5A).
Analysis of CSR junctions in ArtΔ/− B cells. (A-C) Samples of Sμ-Sγ1, Sμ-Sγ3, and Sμ-Sα CSR junction sequences in ArtΔ/− and Artfl/− mice. Microhomology is identified by the longest region of homology (1-bp mismatch accepted, underlined nucleotide). Annotated Sμ sequence (top row); CSR junctions (middle row); Sγ1, Sγ3, and Sα sequence (bottom row). Vertical line represents no sequence identity; gray highlight, microhomology; and underlined text, mismatch. Nucleotide overlaps are shown on the right of each sequence. (D) Distribution of microhomology length of Sμ-Sγ1, Sμ-Sγ3, and Sμ-Sαand junctions in ArtΔ/− and Artfl/− mice. Each triangle represents a unique sequence junction of a given length. Vertical lines and numbers indicate the median microhomology length.
Analysis of CSR junctions in ArtΔ/− B cells. (A-C) Samples of Sμ-Sγ1, Sμ-Sγ3, and Sμ-Sα CSR junction sequences in ArtΔ/− and Artfl/− mice. Microhomology is identified by the longest region of homology (1-bp mismatch accepted, underlined nucleotide). Annotated Sμ sequence (top row); CSR junctions (middle row); Sγ1, Sγ3, and Sα sequence (bottom row). Vertical line represents no sequence identity; gray highlight, microhomology; and underlined text, mismatch. Nucleotide overlaps are shown on the right of each sequence. (D) Distribution of microhomology length of Sμ-Sγ1, Sμ-Sγ3, and Sμ-Sαand junctions in ArtΔ/− and Artfl/− mice. Each triangle represents a unique sequence junction of a given length. Vertical lines and numbers indicate the median microhomology length.
Increased microhomology at CSR junctions in Artemis-deficient human SCIDs
We identified in our series of Artemis-defective radiosensitive SCID patients 2 siblings harboring a homozygous (T425Fs) mutation in the Artemis exon 14 gene, leading to a hypomorphic phenotype compatible with the development of few B and T lymphocytes as previously noted35 (J.P.d.V., unpublished results, 2007). We analyzed the sequence of Sμ-Sα junctions by PCR from B lymphocytes of these 2 patients (Figure 8). Sequences obtained from healthy 6 and 8 healthy adult and pediatric controls indicated a median length of microhomology of 4.0 bp and 6.0 bp, respectively (with the tolerance of one mismatch in the stretch of microhomology). In contrast, the 2 Artemis-deficient patients presented 13.5 and 14.5 bp of microhomology usage at Sμ-Sα junctions, respectively (Figure 8B), which were statistically significantly higher than control values (2-tailed Mann-Whitney test, P < .001 and P < .001, respectively), thus reinforcing the impact of Artemis on the resolution of a subset of CSR-generated DNA-DSB, as noted elsewhere.36 Altogether, these results suggest that CSR may proceed through the previously proposed alternative end-joining pathway(s) in the absence of Artemis. However, in contrast to the XRCC4 and DNA-ligase IV–deficient situations, one cannot formally exclude that another, less specialized, protein may take over the absence of Artemis in the classic NHEJ pathway in our mice.
Analysis of CSR Sμ-Sα junctions in Artemis-deficient patients. (A) Samples of Sμ-Sα CSR junction sequences amplified from whole blood obtained from 14 healthy controls (8 children and 6 adults) and 2 RS-SCID patients (Art1 and Art2) with hypomorphic Artemis mutations. Microhomology at the junction (middle row) is identified by the longest region of sequence homology (1-bp mismatch accepted, underlined nucleotides, gray box) between the Sμ donor sequence (top row) and the Sα acceptor sequence (bottom row). (B) Distribution of microhomology length at Sμ-Sα junctions. Each dot represents a unique Sμ-Sα junction sequence. Vertical lines and numbers indicate the median value of microhomology length in base pairs. The difference in the median length of microhomology between the 2 Artemis patients (13.5 bp and 14.5 bp) and the pool of 8 pediatric controls (6.0 bp) is highly significant (2-tailed Mann-Whitney test, P < .001 for each patient).
Analysis of CSR Sμ-Sα junctions in Artemis-deficient patients. (A) Samples of Sμ-Sα CSR junction sequences amplified from whole blood obtained from 14 healthy controls (8 children and 6 adults) and 2 RS-SCID patients (Art1 and Art2) with hypomorphic Artemis mutations. Microhomology at the junction (middle row) is identified by the longest region of sequence homology (1-bp mismatch accepted, underlined nucleotides, gray box) between the Sμ donor sequence (top row) and the Sα acceptor sequence (bottom row). (B) Distribution of microhomology length at Sμ-Sα junctions. Each dot represents a unique Sμ-Sα junction sequence. Vertical lines and numbers indicate the median value of microhomology length in base pairs. The difference in the median length of microhomology between the 2 Artemis patients (13.5 bp and 14.5 bp) and the pool of 8 pediatric controls (6.0 bp) is highly significant (2-tailed Mann-Whitney test, P < .001 for each patient).
Discussion
Given the absolute prerequisite of NHEJ factors in general, and Artemis in particular, for the completion of V(D)J recombination and, hence, the development of both B and T lymphocytes, the analysis of NHEJ requirement in the mature immune system, downstream of V(D)J recombination, has been quite challenging. Several models have been developed in which the reconstitution of the B-cell compartment was achieved by the introduction of rearranged IgH and IgL chains knock-in alleles (HL mice) in mice deficient for the NHEJ factors Ku70, Ku80, Artemis, and DNA-PKcs, as well as in scid mice (which carry mutation in the DNA-PKcs gene). Ku70- and Ku80-deficient B cells had severely impaired CSR, but this could be the consequence of proliferative defect or increased apoptosis of B cells, although Ku-deficient Bcl2-transgenic B cells that divide still undergo CSR.37-39 Conflicting results were reported concerning the role of DNA-PKcs in CSR, both in DNA-PKcs KO and in scid mice. In 129/C57Bl6 DNA-PKcs KO mice reconstituted with H/L transgenes, Manis et al demonstrated a defect of class switching to all isotypes but IgG1.25 On the contrary, Kiefer et al26 showed an absence of CSR defect in balb/c DNA-PKcs KO mice reconstituted with H/L transgenes coding for an antigen receptor with a different specificity, and engrafted with T cells from B-cell–deficient donors (JH−/− mice). In addition, they could detect postswitch transcripts in HL B cells deficient for DNA-PKcs after induction of CSR in vitro.26 The specificity of the transgenic B-cell receptor, the genetic background of mice, and the presence of T cells could explain the differences in the results obtained in these 2 models. HL scid mice display partial and variable impairment in CSR, depending on the isotypes, on the H and L transgenic chains used for B-cell repertoire reconstitution and on control mice (wild-type or Rag1 KO) analyzed.27,28
We developed a conditional Artemis KO mouse model resulting in the deletion of Artemis DNA repair factor specifically in mature B cells to more directly analyze the role of Artemis during CSR in an otherwise physiologic immune system (polyclonal B cells and the presence of T lymphocytes). We conclude from our analyses that, although Artemis is not essential for an efficient CSR to occur, as previously noted by Rooney et al,24 it most probably intervenes in the resolution of a subset of DNA breaks generated during CSR, which is in accord with the recent observation of persistent DNA breaks at IgH loci in Artemis KO mice carrying IgH/L transgenes.29 Artemis is a DNA endonuclease/exonuclease whose function is critical to resolve DNA hairpin structures at coding ends during V(D)J recombination and process a subset of DNA modifications introduced by ionizing radiations or radiomimetic drugs, such as bleomycin.3,5,23 During CSR, DNA-DSB occurs as a result of the processing of the uracil introduced through cytidine deamination by AID. However, neither the exact nature/structure of the DNA-DSB nor the molecular pathway(s) that leads to the formation of this break starting from the uracil is known with certainty. These DNA-DSBs exist either as blunt or staggered DNA extremities.19 Schrader et al proposed that, depending on the proximity of the DNA single strand breaks resulting from the uracil deamination by AID, the nature of the ultimate DNA-DSB would be different.40 Moreover, the increase in DSB detection within S regions on prior treatment of the DNA with T4 DNA polymerase further accredits the view of the heterogeneous nature of these breaks, some of which require subsequent processing before end joining. To add to the complexity, Zan and Casali reported recently on the AID-dependent modification of preexisting (AID-independent) DNA breaks during CSR.41 The nature of these breaks is unknown. In this context, the finding that Artemis, a DNA processing enzyme, participates in CSR to some extent not only strengthens this hypothesis of the heterogeneity of DNA-DSB during CSR but also induced processing before religation.
With Artemis being implicated in the resolution of DNA breaks during CSR, the question of the consequence of an Artemis defect in mature B lymphocytes undergoing CSR becomes an important question with regard to the possible onset on B-cell malignancies in such situations. Indeed, several studies have shown that CSR is a highly mutagenic process in essence that is prone to chromosomal translocations.42-45 Likewise, several types of mature B-cell lymphomas in humans are thought to arise from chromosomal translocation secondary to defective CSR reaction.46 Murine Artemis-deficient B cells undergoing CSR display increased chromosomal instability, which is directly caused by the unrepaired, AID-generated DNA breaks.29 Lastly, we have reported several cases of immunodeficiency and predisposition to lymphoma in patients with hypomorphic Artemis mutations similar to those harbored by the 2 patients studied here.35 The survey of patients with Artemis hypomorphic mutations and the specific design of murine Artemis B-cell–deficient models on the P53−/− tumor–accelerating background should help clarify this important issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr K. Rajewky for providing the CD21-Cre transgenic mice and A. Rossard and M. Gaillard for excellent animal care.
This work was supported by Inserm, the Ligue Nationale contre le Cancer (Equipe labellisée LA LIGUE), Commissariat à l'Energie Atomique (LRC-CEA 40V), and the INCa/Cancéroplole IdF. P.R.-M. was supported by INCa/Cancéropole Idf and the Association de Recherche contre le Cancer. P.S.-S. was supported by Inserm, Electricité de France, and the Fondation Singer Polignac. G.L.G. was supported by the Ministère de la Recherche et de la Technologie and Association de Recherche contre le Cancer.
Authorship
Contribution: P.R.-M., P.S.-S., J.-P.d.V. designed, performed, and analyzed research and wrote the paper; G.L.G. and V.A. performed research; S.B. and F.P. contributed essential reagents; and A.F. analyzed research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean-Pierre de Villartay, Inserm U768, Hôpital Necker Enfants Malades, 149 rue de Sèvres, F-75015 Paris, France; e-mail: jean-pierre.de-villartay@inserm.fr.
References
Author notes
P.R.-M. and P.S.-S. contributed equally to this study.