Abstract
Over the past 20 years the molecular bases of almost all the major blood group antigens have been determined. This research has enabled development of DNA-based methods for determining blood group genotype. The most notable application of these DNA-based methods has been for determining fetal blood group in pregnancies when the fetus is at risk for hemolytic disease of the fetus and newborn. The replacement of all conventional serologic methods for pretransfusion testing by molecular methods is not straightforward. For the majority of transfusion recipients matching beyond ABO and D type is unnecessary, and the minority of untransfused patients at risk of alloimmunization who would benefit from more extensively blood group–matched blood cannot be identified reliably. Even if a method to identify persons most likely to make alloantibodies were available, this would not of itself guarantee the provision of extensively phenotype-matched blood for these patients because this is determined by the size and racial composition of blood donations available for transfusion. However, routine use of DNA-based extended phenotyping to provide optimally matched donations for patients with preexisting antibodies or patients with a known predisposition to alloimmunization, such as those with sickle cell disease, is widely used.
Introduction
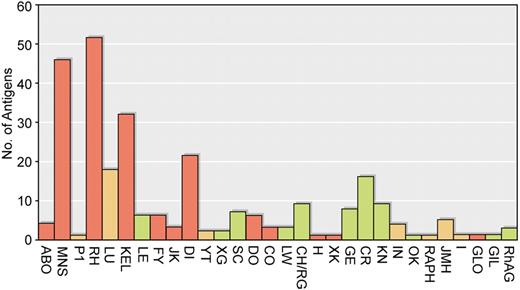
More than 300 inherited blood group antigens have been described on the surface of human red cells.1,2 With rare exceptions, these antigens were first revealed by detection of antigen-specific antibodies present in human serum with the use of the agglutination method. Fewer than 50 of these blood group antigens are known to be polymorphic in any region of the world. That is, to have alternate alleles present in a population at levels greater than can be maintained by recurrent mutation.3 Of those antigens that are polymorphic, few stimulate antibodies of clinical significance by causing transfusion reactions or hemolytic disease of the fetus and the newborn (HDFN)4 (Figure 1). Consequently, safe transfusion for most recipients can be assured by the correct typing of patients and donors with respect to ABO phenotype and screening the patients' serum for the presence of clinically significant antibodies directed against antigens polymorphic in the local population. In populations in which the RhD antigen is polymorphic, it is accepted practice to type all donors and recipients for RhD because anti-D will be found commonly, and it is a major cause of HDFN and transfusion reactions. In the Far East where the D-negative phenotype is uncommon and anti-D is rarely seen, this may not be necessary.5 The “type and screen” procedure has the weakness that antibodies to rare nonpolymorphic antigens will not be detected (approximately 0.06% of cases), but the antibodies missed are rarely of clinical significance.6,7 A further refinement for patients without irregular antibodies is the computer crossmatch, provided appropriate safeguards to prevent issue of ABO-incompatible blood are built into the software.8 Over the past 30 years there has been a trend in blood banking toward reducing the amount of pretransfusion testing and to introducing modified versions of the agglutination test such as the gel test that can be performed safely by laboratory staff without extensive practical training in traditional serologic methods.9 These procedures are perfectly acceptable for most transfusion recipients, but more extensive pretransfusion matching of donor blood for patients with diseases that have a high risk of alloimmunization (sickle cell anemia, thalassemia) is desirable and has become common practice in many laboratories.10,11 In recent years national hemovigilance procedures have confirmed the efficacy of these serologic approaches, and the focus for efforts to increase transfusion safety has moved from laboratory methods to the avoidance of errors occurring after blood has left the blood bank.12,13
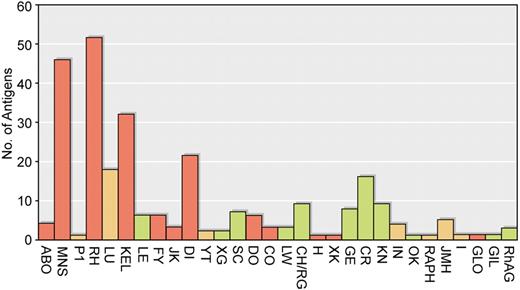
Clinical importance of antibodies to blood group antigens. The blood group systems are given on the abscissa and the number of antigens in each system indicated on the ordinate. Blood group systems containing antigens that often stimulate clinically significant antibodies are shown in red. Blood group systems with antigens that occasionally stimulate clinically significant antibodies are yellow, and blood group systems with antigens that rarely if ever stimulate clinically significant antibodies are green.
Clinical importance of antibodies to blood group antigens. The blood group systems are given on the abscissa and the number of antigens in each system indicated on the ordinate. Blood group systems containing antigens that often stimulate clinically significant antibodies are shown in red. Blood group systems with antigens that occasionally stimulate clinically significant antibodies are yellow, and blood group systems with antigens that rarely if ever stimulate clinically significant antibodies are green.
In parallel with the trend to reduce the amount and complexity of pretransfusion testing, intense research activity has been directed at the molecular characterization of blood group genes and at determining the molecular basis of blood group polymorphisms.14 The knowledge that has been obtained has opened the possibility of a new approach to blood group antigen typing, based on DNA sequence determination rather than on agglutination. DNA-based methods have the fundamental weakness of not detecting directly the presence of an antigen on the surface of a red cell; nevertheless, there are several clinical situations in which this approach has proved to be a valuable addition to the range of blood-grouping methods available. This article reviews the effect such DNA-based methods have already made on the provision of health care in transfusion and neonatal medicine and considers the extent to which they are likely to affect existing routine laboratory procedures for pretransfusion testing.
Blood group genes encoding polymorphic antigens commonly stimulating blood group antibodies of clinical significance
To achieve safe red cell transfusions, 3 major blood group requirements must be satisfied. First, and most important, the red cells to be transfused must be ABO compatible. Second, RhD-positive red cells should not be given to women of phenotype RhD-negative. Third, transfused red cells should lack blood group antigens reactive with any preexisting clinically significant antibodies the recipient may have. Are DNA-based typing methods for ABO, RhD, and other antigens giving rise to clinically significant antibodies sufficiently robust to be considered as replacements for existing serologic methods of pretransfusion testing?
DNA-based typing for ABO antigens
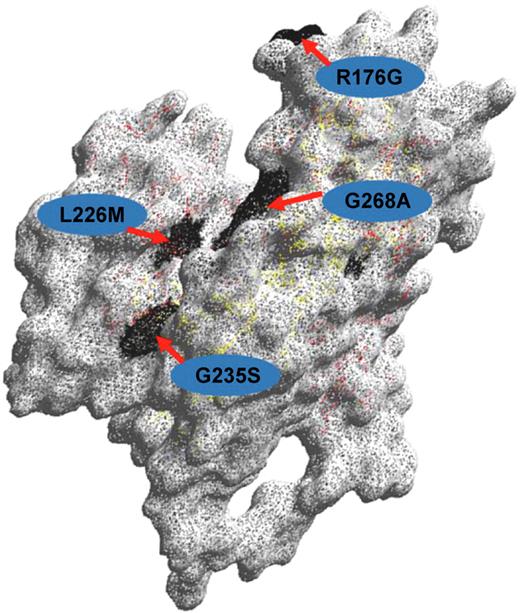
The ABO blood group system is the most clinically important blood group system because antibodies against A or B or both antigens are naturally present in the serum of persons whose red cells express blood group B, A, or O. Every student of transfusion medicine quickly learns that ABO incompatible transfusions are potentially fatal. It follows that universal blood typing with DNA-based methods alone cannot be considered in the absence of a totally robust method for predicting ABO phenotype. The molecular basis of the ABO blood group system was elucidated in 1990.15 The ABO phenotype of a person is determined by the expression of a single glycosyltransferase gene on chromosome 9. The 3-dimensional structure of the B-transferase was elucidated by Patanaude et al16 (Figure 2). The antigens A, B, and their variants result from functional glycosyltransferase genes capable of transferring N-acetyl-D-galactosamine or D-galactose or both to the nonreducing ends of suitable oligosaccharide chains found on red cell membrane glycoproteins and glycolipids. The red cell phenotype denoted O occurs because the glycosyltransferase gene that generates A or B or both antigens is inactive.
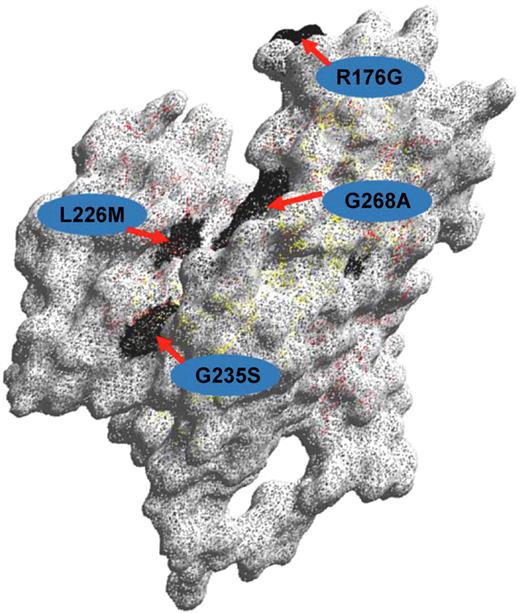
Structure of the ABO glycosyltransferase. Amino acid residues differing between blood group A and B-active transferases, respectively (Arg176Gly;Gly235Ser;Leu266Met;Gly268Ala) are shown with the single-letter code and their positions indicated. The 3-dimensional model was created with the DeepView Swiss Pdb Viewer version 3.7.
Structure of the ABO glycosyltransferase. Amino acid residues differing between blood group A and B-active transferases, respectively (Arg176Gly;Gly235Ser;Leu266Met;Gly268Ala) are shown with the single-letter code and their positions indicated. The 3-dimensional model was created with the DeepView Swiss Pdb Viewer version 3.7.
The fact that group O results from an inactive gene creates a major and fundamental problem for the design of DNA-based methods for ABO typing because inactivating mutations causing a group O phenotype occur in many different places in the coding region of the ABO gene. The most common O allele (O1) is distinguished from the A1 allele because of a G deletion at codon 261 which creates a frameshift that results in a truncated translated protein of 117 amino acids lacking an enzyme active site. In contrast, the O2 allele has 6 SNPs distinguishing it from the A1 allele. The most significant of the SNPs (G802A) results in a Gly268Arg substitution in the active site of the enzyme, rendering it inactive17 (Figure 2). The O3 allele has a G insertion at 804, causing a frameshift that changes the protein sequence at residue 269 within the active site. The O4 allele has a G insertion at nucleotide 88 that results in a frameshift and truncated inactive protein of 56 amino acids. The O5 and O6 alleles have nucleotide substitutions C322T and G542A, respectively, which create stop codons that result in inactive proteins of 107 and 181 residues. These nondeletional alleles all occur on an A1 allele background, so any DNA-based method that is not designed to select all O alleles may erroneously type the sample as A1. Obviously, this is a major problem because of the risk of incorrectly typing a recipient as A when he or she is in fact O and an incompatible transfusion being given. Storry et al described such a case.18 A kidney transplant donor was typed as A with DNA-based methods, but conventional serology showed the donor to be group O with anti-A and anti-B in the serum. Two recent studies provide a comprehensive discussion of the methodologic problems inherent in DNA-based ABO typing particularly with respect to the known O genotypes.19,20
Knowledge of the different molecular bases of the group O phenotype in different world populations permits the development of DNA-based assays capable of detecting all known alleles but cannot allow for novel inactivating mutations not yet observed. Consequently, there will always be the possibility of an occasional ABO incompatible transfusion if such methods are used to replace established procedures. One way of minimizing risk would be additional testing of all samples for the presence of anti-A and anti-B, but even so it is difficult to see how DNA-based methods could be used in the absence of conventional serology to confirm ABO type.
DNA-based typing for Rh antigens
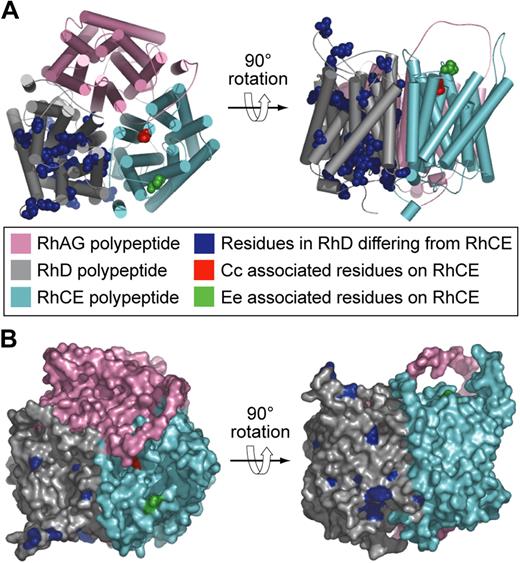
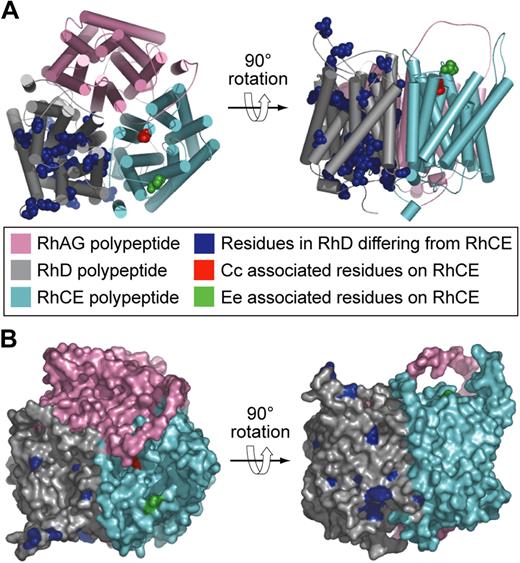
It is almost 20 years since the transcribed products of genes encoding Rh blood group antigens were identified. RHCE was described in 199021,22 and RHD shortly thereafter.23-25 These findings brought to an end 50 years of controversy in which rival groups hypothesized the Rh antigens were either the product of a single RH gene26 or 3 closely linked RH genes, C, D, E.27 It was proposed that the 2 Rh genes give rise to 3 polypeptides with the products of RHCE being separate spliceoforms encoding Cc and Ee antigens, respectively,28 but, when RHCE was expressed in vitro, it was clear that one polypeptide (CE polypeptide) could express both Cc and Ee antigens and that 2 genes RHD and RHCE encode 2 polypeptides denoted D and CE polypeptides, respectively.29 Consequently, the 2 genes/3 polypeptides hypothesis has been abandoned. Recently, the 3D structure of a bacterial analog of the Rh-associated glycoprotein has been reported.30,31 This structure allows more reliable modeling of the D and CE polypeptides than previously possible.32 Figure 3 depicts models of the structure of D and CE polypeptides with the positions of the numerous amino acid differences identified.
Models of the structure of Rh proteins.32 (A) Hypothetical heterotrimer comprising RhAG (pink), RhD polypeptide (gray), and RhCE polypeptide (cyan). Residues in the D polypeptide differing from those in the CE polypeptide are shown as blue spheres. Cc and Ee associated residues on the CE polypeptide are shown as red and green spheres, respectively. (B) Hypothetical heterotrimer comprising RhAG (pink), Rh D polypeptide (gray), and Rh CE polypeptide (cyan) same as in panel A but showing the molecular surface.
Models of the structure of Rh proteins.32 (A) Hypothetical heterotrimer comprising RhAG (pink), RhD polypeptide (gray), and RhCE polypeptide (cyan). Residues in the D polypeptide differing from those in the CE polypeptide are shown as blue spheres. Cc and Ee associated residues on the CE polypeptide are shown as red and green spheres, respectively. (B) Hypothetical heterotrimer comprising RhAG (pink), Rh D polypeptide (gray), and Rh CE polypeptide (cyan) same as in panel A but showing the molecular surface.
The D antigen is often referred to as the most complex of the blood group antigens. The explanation for this apparent complexity is simple. In peoples of European origin a complete deletion of RHD and hence complete absence of the D polypeptide from the red cell surface is very common. Consequently, anti-D has been defined as an antibody failing to react with red cells lacking D polypeptide. D antigen is therefore quite different from other blood group antigens which result from nucleotide transitions changing a single amino acid in the protein sequence of the appropriate blood group–active protein (see “DNA-based typing for other polymorphic antigens commonly giving rise to clinically significant antibodies”). It follows that any person inheriting an RHD with a mutation that changes the protein sequence of the D polypeptide may appear to have an abnormal type of D antigen, particularly if the red cells are tested with a wide variety of monoclonal anti-Ds because those antibodies recognizing the region of the protein containing or affected by the mutation may fail to react with the person's red cells. The situation is further complicated by the occurrence of the adjacent and homologous gene RHCE. Gene conversion events between RHD and RHCE at meiosis can create hybrid alleles encoding proteins with sequence derived from both D and CE polypeptides14 (Figure 4A). After elucidation of the genetic basis of major Rh antigens D, Cc, and Ee (Figure 3), numerous studies of RHD and RHCE in persons having weakly expressed or altered D and other Rh blood group antigens have provided a detailed catalog of genetic variation in these genes in different ethnic groups.14,33 DNA-based methods are ideally suited to the elucidation of variant Rh antigens and provide a much more powerful tool for this purpose than does conventional serology. However, elucidation of the molecular basis of an Rh variant and the frequency with which it occurs in a given population does not show the clinical significance of the variant because that is determined by its immunogenicity.
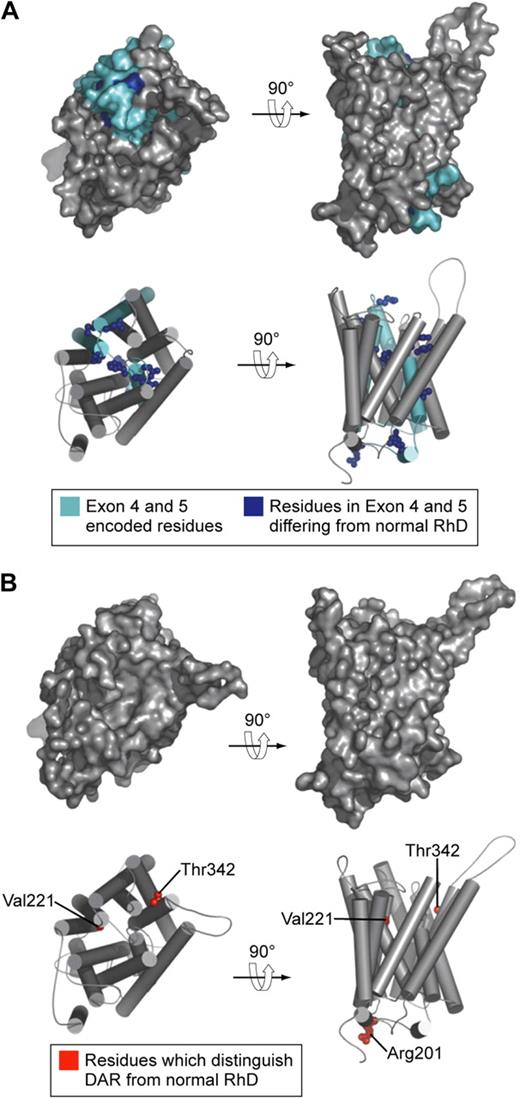
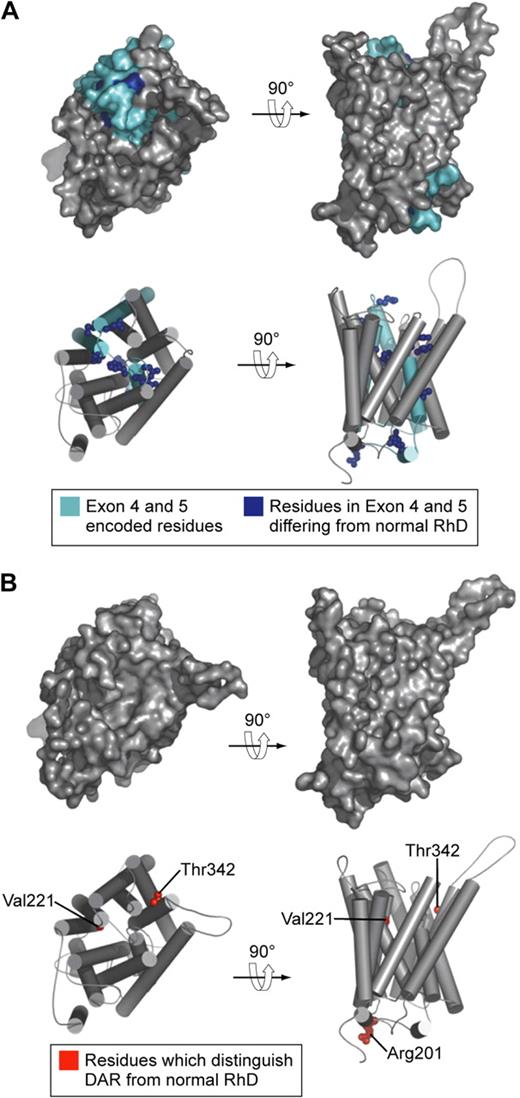
Models of the structure of D antigen variants DVI type I and DAR. (A) DVI type I polypeptide. Residues encoded by exons 4 and 5 (which are replaced by CE sequence in DVI type I) are shown in cyan. Residues in exons 4 and 5 that differ from those in normal D polypeptide are shown as blue spheres. (B) DAR polypeptide. Residues that distinguish DAR from normal D polypeptide are shown as red spheres. In this model DAR-associated residues are not surface exposed and not visible on the molecular surface.
Models of the structure of D antigen variants DVI type I and DAR. (A) DVI type I polypeptide. Residues encoded by exons 4 and 5 (which are replaced by CE sequence in DVI type I) are shown in cyan. Residues in exons 4 and 5 that differ from those in normal D polypeptide are shown as blue spheres. (B) DAR polypeptide. Residues that distinguish DAR from normal D polypeptide are shown as red spheres. In this model DAR-associated residues are not surface exposed and not visible on the molecular surface.
The major clinical problem associated with Rh proteins is HDFN caused by anti-D (for review see Klein and Anstee34 ). In Western Europe the D-negative phenotype results from a deletion of RHD.35 The cause of the D-negative phenotype in the various populations of Central Asia has not been formally determined. However, methods designed to detect the deletion of RHD in fetal DNA give reliable results for D-negative Asian Indians residing in the United Kingdom, suggesting a similar deletion phenotype to that found in Western Europe is present in Central Asia (Kirstin Finning, Bristol Institute for Transfusion Sciences, written communication, 2007). A significant proportion of D-negative persons of African origin do not have a complete deletion of RHD.36 In the indigenous peoples of South East Asia the D-negative phenotype is rarely encountered, and consequently HDFN resulting from anti-D is not a common clinical problem. Yan et al tested 305 572 Chinese of whom 99.53% were D positive.37 Because of the very high incidence of D positive in Taiwan, routine D typing of donors and patients was discontinued in 1988, and no increase in the incidence of anti-D resulted.5
Polyclonal anti-D immunoglobulin (synonym Rh immune globulin) is a well-established prophylactic treatment for HDFN.38 This product has an excellent safety record, although a recent report describing a patient with factor XI deficiency who developed an inhibitor antibody to factor XI after receiving Rh immune globulin shows that the use of such a product is not entirely risk free.39 Human monoclonal anti-D would avoid this risk and bring the added value of homogeneity and security of supply, but the very high costs of bringing such a product to market have so far delayed its introduction. An alternative approach currently under investigation is the use of immunodominant peptides derived from the D polypeptide sequence to induce tolerance to D antigen in D-negative women. Early studies with the use of immunodominant D peptides in humanized HLA-DR15 transgenic mice suggest such an approach is feasible.40
The most valuable contribution to health care arising from characterization of the genes encoding Rh polypeptides has been the introduction of diagnostic testing to determine whether a pregnant D-negative woman carries a fetus of phenotype D positive or D negative. Initially, testing was carried out on fetal DNA obtained by amniocentesis from D-negative mothers who had already given birth to a baby affected by HDFN. Amniocentesis is not without risk to the fetus, and so wider application of fetal DNA testing was not a practical option until it was shown that sufficient fetal DNA for testing can be derived from a maternal blood sample.41 Determination of D phenotype with fetal DNA from maternal plasma of D-negative mothers with preexisting anti-D is now a routine diagnostic procedure in many countries.42 A logical extension of this work would be to test fetal DNA from maternal blood samples of all pregnant D-negative women so that Rh immune globulin is given only to those women carrying a D-positive fetus. In this way a significant number of women (∼ 40% of pregnancies in the United Kingdom) would avoid unnecessary exposure to Rh immune globulin. The first steps toward achieving this goal have now been taken with 2 studies reporting successful application of automated testing systems capable of typing large numbers of antenatal samples.43,44 HDFN resulting from Rh antibodies other than anti-D is far less common, but fetal DNA typing is no less valuable in these cases. Suitable tests for c and E are available.45
The case for application of DNA-based methods in typing Rh antigens to aid the management of HDFN is clear. Less clear is the proposed routine use of DNA-based methods for determination of the many variants of D found in blood donors and patients. There is no doubt the technology to do this is available.46,47 What is yet to be resolved is whether sufficient clinical benefit would be derived from such testing to justify the cost of implementing the methodology.48,49 The importance of determining whether a patient or donor has a D variant is governed by the likelihood of anti-D stimulation and the probability the anti-D produced will be clinically significant. D-variant antigens are often subdivided into partial D and weak D, although the distinction between these types of D antigen is so vague as to be meaningless.50 A more useful subdivision would be to list D-variant antigens known to have stimulated clinically significant antibodies. Few D-variant antigens occur with frequencies greater than 1 or 2 per 1000 anywhere in the world. Taken together with the paucity of reports of alloimmunization or HDFN resulting from D-variant antigens, a more selective introduction of DNA-based methods for patient testing may be appropriate. One approach would be the use of DNA-based methods to identify only those D variants whereby the variant is known to stimulate antibody in recipients, in populations in which the D variant is polymorphic. In people of European origin the DVI variant falls into this category; however, it can be detected with conventional serology with 2 monoclonal anti-D reagents, one of which reacts with DVI-positive red cells and the other which does not (Figure 4A). D-variant antigens are more common in people of African origin and comprise 3 types denoted as DIVa, DAU, and weak D type 4 (includes DIIIa and DAR51 ; Figure 4B). In this context it is interesting to note that the amino acid substitutions associated with DAR are not predicted to be accessible on the outer surface of the red cell (Figure 4B), yet persons homozygous for DAR can make allo anti-D,52 leading one to conclude that subtle changes in the relative positions of external contact residues in DAR-positive persons are sufficient to allow stimulation of anti-D on exposure to wild-type D antigen. Such a conclusion is consistent with the proposal of Chang and Siegel to account for the fact that the footprints of anti-Ds with different epitope specificities are essentially identical.53 Castilho et al report the occurrence of D-variant antigens DIIIa or DAR or both in 8 of 130 patients with sickle cell disease, 3 of whom had made anti-D, and they suggest there may be a case for including DNA-based assays for these D variants in selected patient groups.54 However, the clinical significance of anti-D produced by these and the majority of D-variant antigens is unknown.
If gene chips are introduced for donor testing in blood centers, inclusion of the additional reactions necessary to detect D variants can easily be accommodated. If gene chips are used for routine testing of patients for genetic susceptibility to disease and a full blood group genotype determination is incorporated into the same chips (discussed further under “Assimilation of red cell genotyping into routine practice”), the whole debate about the cost effectiveness and clinical value of D-variant testing will cease because the information will be obtained en passant.
DNA-based typing for other polymorphic antigens commonly giving rise to clinically significant antibodies
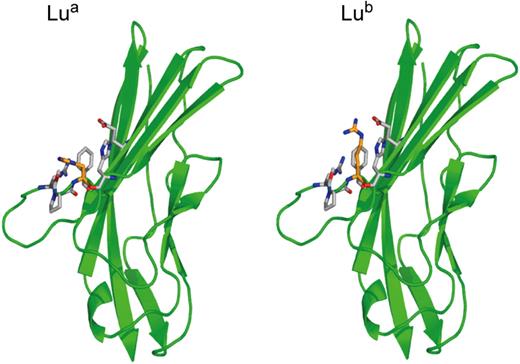
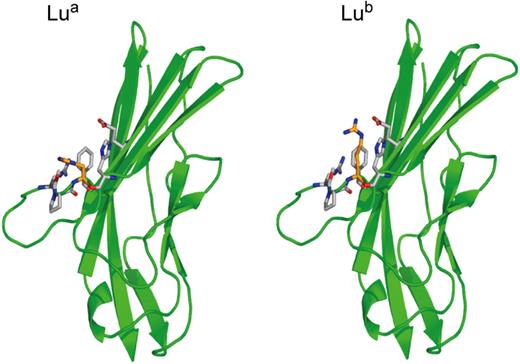
The inherent problems associated with attempting to develop robust DNA-based methodologies for ABO and Rh typing do not apply to polymorphic antigens in other blood group systems. Here, allelic antigens generally result from single nucleotide transitions changing one amino acid in the blood group–active protein (synonym single nucleotide polymorphisms [SNPs]). Almost all the genes encoding polymorphic blood group antigens in blood group systems have been cloned, and the SNPs responsible for the polymorphic antigens associated with clinically significant antibodies have been identified. Computer modeled 3D structures are available for many of these blood group–active proteins based on the crystal structures of structural homologues. Actual structures are available for Aquaporin 1, which expresses the Colton blood group antigens,55 and for the amino terminal domains 1 and 2 of Lutheran glycoprotein.56 The actual structures of Lua and Lub antigens are shown in Figure 5 to show the structure of a blood group antigen defined by a single amino acid substitution and to emphasize the difference between this type of antigen and D antigen (compare Figure 3).
Structure of the Lua and Lub blood group antigens. Crystal structure of the amino-terminal domain of the Lutheran glycoprotein from Mankelow et al,56 depicting the nucleotide transition (SNP) giving rise to His77(Lua) or Arg77(Lub).
Structure of the Lua and Lub blood group antigens. Crystal structure of the amino-terminal domain of the Lutheran glycoprotein from Mankelow et al,56 depicting the nucleotide transition (SNP) giving rise to His77(Lua) or Arg77(Lub).
K/k; Kpa/Kpb; Jsa/Jsb
Antibodies to the Kell antigen (K1) are the most frequently encountered clinically significant alloantibodies after ABO and Rh antibodies in peoples originating from Europe.34 The gene encoding Kell glycoprotein was cloned in 1991.54 It is a type II membrane protein with homology to zinc endopeptidases (M13 family). It occurs in the red cell membrane disulfide bonded to the XK protein. A structural model based on the crystal structure of NEP and depicting amino acid changes that are the loci for antigens within the Kell blood group system is provided by Lee et al.57 Several methods for DNA-based typing of polymorphic antigens in the Kell blood group system have been described (reviewed in Lee58 ). However, it cannot be assumed that typing for these SNPs alone will always give a correct interpretation of Kell phenotype. Poole et al described a family in which the red cells have weak K1 expression yet type as homozygous K2 at nucleotide 587.59 In this case a 577>T change converts codon 193 to serine.
Anti-K1 is responsible for severe neonatal anemia in approximately 40% of K1-positive babies of women with anti-K1.60 The Kell glycoprotein is expressed very early in erythropoiesis.61 The antibody appears to inhibit erythropoiesis,62 suggesting a functional role for Kell glycoprotein additional to endopeptidase activity. A method for determining the presence of DNA encoding K1 in fetal DNA from maternal plasma has been described.45
Fya/Fyb
The gene encoding Fy antigens was cloned in 199363 and fully characterized in 1996.64 It encodes a 7-membrane spanning protein with the Fya/Fyb polymorphism defined by a nucleotide transition giving Gly(Fya) or Asp(Fyb) at position 42 in the extracellular amino-terminal domain.65 The widespread occurrence of the red cell phenotype Fy(a-b-) in populations deriving from areas where the malarial parasite Plasmodium vivax is endemic necessitates the concomitant assay of DNA for a nucleotide substitution in the GATA 1 binding site upstream of FY66 when molecular Fy typing is undertaken. Other SNPs give rise to weak expression of Fy antigens (Fyx reviewed in Castilho67 ).
Jka/Jkb
The gene encoding Jk antigens was cloned in 1994,68 and the molecular basis of the Jka/Jkb polymorphism was elucidated in 1997.69 The gene encodes a urea transporter (HUT 11) predicted to have 10-membrane spanning domains. The Jka/Jkb polymorphism results from a SNP (838G>A) that changes Asp280(Jka) to Asn(Jkb) in the fourth predicted extracellular loop. Some alleles giving rise to the Jk(a-b-) phenotype may be polymorphic in Asia (particularly Polynesia) and Finland, necessitating the determination of additional SNPs for accurate phenotype prediction.70
Dia/Dib
The gene encoding Di antigens is the anion exchange protein gene SLC4AE1 (synonym band 3). A nucleotide transition (C2561T) changes Leu854(Dia) to Pro854(Dib).71 Dia is rarely found in peoples of European or African origin, but it is polymorphic in many of the indigenous peoples of Asia and of North, Central, and Southern America.
Doa/Dob
The Dombrock blood group glycoprotein is a member of the ADP-ribosyltransferase gene family. The polymorphic antigens Doa and Dob result from a nucleotide transition (A793G) which converts Asn265(Doa) to Asp(Dob).72
Coa/Cob
The Colton blood group antigens are located on the water transporter, aquaporin 1 (AQP1). The polymorphic antigens Coa and Cob result from a nucleotide transition (C134T) which converts Ala45(Coa) to Val(Cob) on the first extracellular loop.55
S/s
Applications of red cell genotyping in transfusion practice
There are several well-established and useful applications of DNA-based typing in transfusion and neonatal medicine (reviewed in Westhoff76 and Hillyer et al77 ; summarized in Table 1). Fetal DNA typing has been used widely for more than a decade and is of proven utility in the management of HDFN (discussed under “DNA-based typing for Rh antigens”).
Other applications of DNA-based methods include determining the red cell genotype of patients who have received a large transfusion or have autoimmune hemolytic anemia whereby conventional serology is not a reliable method of determining the patients blood group.
Extensive screening to identify donors with rare blood groups or to establish the frequency of blood group polymorphisms in a given population are useful applications which may be achieved with a high-throughput automated system.74,78
However, significant problems preclude any thought of replacing all serologic procedures for blood typing with DNA-based methods (see “DNA-based typing for ABO antigens”). In addition to the fundamental problem posed by determination of the group O phenotype, the genes encoding some common nonpolymorphic antigens (Vel;Jra;Ata;MAM;AnWj), which occasionally give rise to clinically significant antibodies, have not been cloned and characterized. Therefore, it is not possible to use molecular methods to detect the absence of these antigens and so identify persons who may have clinically significant antibodies. These antibodies are very rare but can cause severe transfusion reactions and HDFN. Therefore, the introduction of DNA-based blood group typing would have to take place alongside conventional ABO typing and antibody screening and not in isolation. Given the simplicity of conventional methods for D typing, there would have to be substantial advantages apart from the benefit accruing from identification of some potentially immunogenic D-variant antigens, to introducing more costly DNA-based methods into routine procedure.
High throughput automated DNA-based typing would, in principle, allow all transfusion recipients to receive the most blood group–compatible blood available from blood centers. However, the most blood group–compatible blood available is unlikely to match the genotype of the patients at more than 3 or 4 blood group loci because of inventory limitations even in countries with blood services collecting millions of donations a year. In any case it makes no sense to attempt such a total matching service, given that less than 10% of transfusion recipients make alloantibodies with current matching procedures.79,80 Available evidence supports the view that the ability to make alloantibodies to blood group antigens is genetically determined.80 Clearly, there is a need for DNA-based methods capable of identifying those patients most likely to make antibodies in response to transfusion. If such methods were available, then it might be feasible to target this small subset of patients (“responders”) so that they receive the most blood group–compatible blood available, whereas nonresponders receive ABO- and D-compatible blood in the conventional way. Higgins and Sloan80 argued that, because patients who have already made an alloantibody are responders and very likely to make further antibodies, it would be sensible to make more fully typed units available to these groups, which would include patients with sickle cell disease and women of childbearing age.
Extensive prophylactic red cell genotyping to select donors for patients who will receive repeated transfusions over a long period is an attractive application of DNA-based blood grouping. This is particularly relevant to patients with sickle cell disease in whom the rate of alloimmunization is high.81 A recent survey of 1182 North American laboratories showed that most laboratories (n = 743) typed patients with sickle cell for ABO and D only. Most laboratories that performed more extensive phenotyping (n = 330) typed for C, E, and K.82 Prophylactic matching for patients with sickle cell disease with the use of DNA-based methods does not necessarily require the use of extensive DNA arrays (gene chips). In principle, it could be carried out in the blood bank with the use of a combination of conventional serology (for ABO and Rh typing) and a DNA-based method that used beads or microspheres for other blood group antigens (Karpasitou et al83 ; Figure 6). However, this application of red cell genotyping also has limitations. Castro et al in a study of 137 alloimmunized patients with sickle cell noted that, when using limited phenotype matching (for Cc, Ee, K in addition to ABO and D), all alloantibodies would have been prevented for more than one-half (53.3%) of the patients.84 If the phenotyping had been extended to include S, Fya, and Jkb, all antibodies would have been prevented in 70.8% of patients, but, whereas 13.6% of random white donors of blood cells would match the limited phenotype, only 0.6% would match the extended phenotype. By combining recruitment of African American donors and screening for those with the Fy(a-b-) phenotype with the use of monoclonal anti-Fy3, 12% of donors were extended phenotype matches for 41 patients with sickle cell.85 Olujohungbe et al reported a much higher incidence of alloimmunization in patents with sickle cell in the United Kingdom (76% of those transfused) compared with a comparable group in Jamaica (2.6% of those transfused), further emphasizing the consequences of disparity between donor and recipient populations.86 Clearly, the usefulness of DNA-based methods of blood typing to reduce alloimmunization in sickle cell donors is limited by the availability of compatible blood donations.
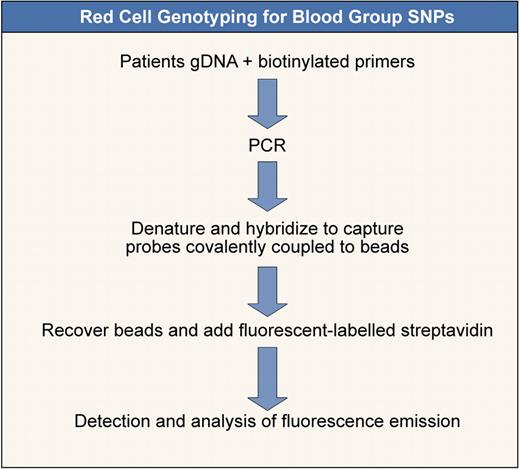
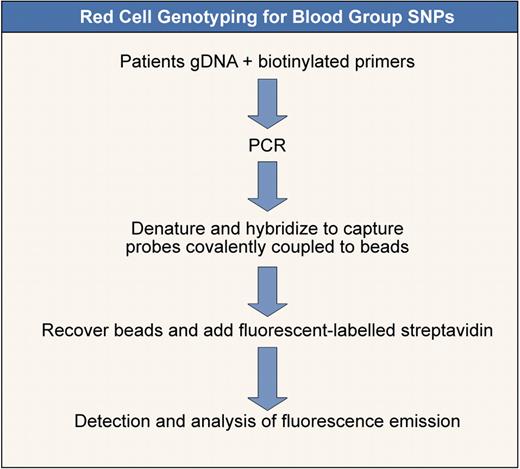
Red cell genotyping for blood group SNPs. Outline of a method to determine blood group SNPs based on that of Karpasitou et al.83 Many other methods are available (see also Hashmi et al,74 Hillyer et al,77 and Avent et al78 ). This one is illustrated because it is suitable for “in-house” development.
Red cell genotyping for blood group SNPs. Outline of a method to determine blood group SNPs based on that of Karpasitou et al.83 Many other methods are available (see also Hashmi et al,74 Hillyer et al,77 and Avent et al78 ). This one is illustrated because it is suitable for “in-house” development.
Assimilation of red cell genotyping into routine practice
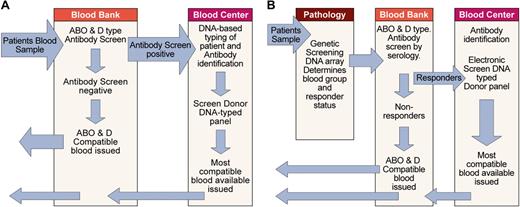
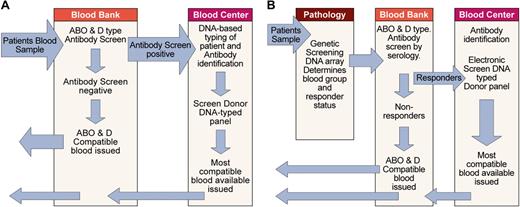
As previously discussed, optimal use of red cell genotyping in routine practice requires that those patients likely to make alloantibodies can be distinguished from those that are unlikely to make antibodies. At the present time this distinction cannot be made, and so the best available option is to use DNA-typed donors for patients who have preexisting antibodies and are therefore known responders and those patients most likely to make alloantibodies such as patients with sickle cell disease. One possible scenario for the assimilation of DNA-based typing into pretransfusion testing protocols is depicted in Figure 7A. In this scenario the patient sample is ABO and D typed and screened for antibodies with conventional methods. If antibody is detected and identified, patients' DNA is typed by blood bank or blood center personnel for other blood groups with the use of a method similar to that depicted in Figure 6. The blood center database of available DNA-typed donors is then searched electronically to find the best match. At blood centers with large numbers of donations, automated typing of donated blood for multiple blood group loci would most likely be carried out with DNA arrays or gene chips.
Assimilation of red cell genotyping into routine practice. (A) Depicts one scenario for incorporating red cell genotyping into routine practice based on technology currently available. It is also possible DNA-based testing could be undertaken at the blood bank or another laboratory. (B) Depicts one possible future scenario, assuming genetic screening with DNA arrays to identify disease susceptibility will become routine and a genetic test capable of identifying those patients (responders) likely to make alloantibodies to blood group antigens will be available.
Assimilation of red cell genotyping into routine practice. (A) Depicts one scenario for incorporating red cell genotyping into routine practice based on technology currently available. It is also possible DNA-based testing could be undertaken at the blood bank or another laboratory. (B) Depicts one possible future scenario, assuming genetic screening with DNA arrays to identify disease susceptibility will become routine and a genetic test capable of identifying those patients (responders) likely to make alloantibodies to blood group antigens will be available.
Another scenario is possible if the use of DNA arrays for genetic screening to establish susceptibility to common diseases becomes part of routine practice in pathology laboratories. In this case it would be feasible to include the necessary additional reactions of the patient to blood group on the same gene chip. Blood bank personnel would confirm the patient's ABO and D type and perform an antibody screen. If the antibody screen was positive and the antibodies identified or responder status determined, interrogation of the blood center database of available donors could proceed directly because DNA-based typing has already been performed (Figure 7B).
Conclusions
DNA-based blood group typing provides a valuable adjunct not an alternative to traditional methods of pretransfusion testing. Traditional methods for ABO and D typing are likely to continue, and methods for antibody detection and identification will still be required. Theoretically, it is possible to match patient and donor blood group genotypes electronically with the use of gene chip technology. However, this assumes DNA-based methods are totally robust, which they are not because novel mutations arise continually. It also assumes the blood available for transfusion at any given time will match all patients' blood group phenotypes for all possible polymorphic antigens capable of stimulating clinically significant antibodies, which is unrealistic. Tandem application of DNA-based methodology and existing methods will however provide improvements in the provision of extensively blood group–phenotyped red cells for patients with alloantibodies.
Acknowledgments
I thank Nick Burton for preparing Figures 3, 4, and 5; Ed Anstee for preparing Figure 1; Martin Olsson for Figure 2; and Geoff Daniels and Joyce Poole for critical reading of the manuscript.
This work was supported by the Department of Health (England).
Authorship
Contribution: D.J.A. was the single author of this paper.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: David J. Anstee, Bristol Institute for Transfusion Sciences, Southmead Rd, NHS Blood and Transplant, Bristol, BS10 5ND, United Kingdom; e-mail: david.anstee@nhsbt.nhs.uk.