Abstract
Microarray analysis with 40 000 cDNA gene chip arrays determined differential gene expression profiles (GEPs) in CD34+ marrow cells from myelodysplastic syndrome (MDS) patients compared with healthy persons. Using focused bioinformatics analyses, we found 1175 genes significantly differentially expressed by MDS versus normal, requiring a minimum of 39 genes to separately classify these patients. Major GEP differences were demonstrated between healthy and MDS patients and between several MDS subgroups: (1) those whose disease remained stable and those who subsequently transformed (tMDS) to acute myeloid leukemia; (2) between del(5q) and other MDS patients. A 6-gene “poor risk” signature was defined, which was associated with acute myeloid leukemia transformation and provided additive prognostic information for International Prognostic Scoring System Intermediate-1 patients. Overexpression of genes generating ribosomal proteins and for other signaling pathways was demonstrated in the tMDS patients. Comparison of del(5q) with the remaining MDS patients showed 1924 differentially expressed genes, with underexpression of 1014 genes, 11 of which were within the 5q31-32 commonly deleted region. These data demonstrated (1) GEPs distinguishing MDS patients from healthy and between those with differing clinical outcomes (tMDS vs those whose disease remained stable) and cytogenetics [eg, del(5q)]; and (2) molecular criteria refining prognostic categorization and associated biologic processes in MDS.
Introduction
The myelodysplastic syndromes (MDS) are a spectrum of clonal myeloid hemopathies with inherent hematopoietic precursor cell (HPC; ie, inclusive of primitive hematopoietic stem cells [HSCs] and committed progenitor cells) anomalies and abnormal hematopoietic regulation.1,2 Heterogeneous subsets of MDS patients have been defined by their clinical (percentage marrow blasts, number of cytopenias) and biologic (specific cytogenetic and molecular lesions) abnormalities.3 Use of these features has provided methods (eg, the International Prognostic Scoring System [IPSS]) to help define the patients' prognoses, including their relative risk of evolving to acute myeloid leukemia (AML) or to have shortened survival.3 However, these approaches are limited in predicting clinical course, and management of patients remains challenging given the uncertainty of the time course of disease progression. Broad-based molecular and cellular analyses are potentially valuable to improve prognostication and the understanding of the mechanisms underlying the defective hematopoietic cell differentiation and abnormal clone expansion in those patients who undergo progression to AML.
Specific gene expression profiles (GEPs) and differentially expressed cellular pathways have been defined and provide insights into the molecular biology of AML and its subtypes.4-6 However, in contrast to the relatively homogeneous marrow population of blasts present in AML for which several well-defined microarray studies have been reported, analysis of MDS marrow is more complex as it contains heterogeneous populations of cells with various degrees of cellular differentiation. Studies evaluating data from enriched marrow HPCs from a variety of MDS patients have been reported,7-10 as have reports for those predominantly with del(5q) MDS.11-14 However, with one exception,11 prior microarray studies in MDS analyzed a limited number of non-del(5q) subjects (10-22 patients). Differing GEPs were described from each study. No association has been reported in these investigations indicating the relationship between GEPs and the long-term outcome of MDS patients. To further evaluate the molecular nature of stable MDS patients as contrasted to those who progressed to AML, using microarray analysis we assessed the GEPs and their functional correlates from CD34+ marrow cells from such patients after prolonged follow-up.
Methods
Patients, bone marrow samples
For microarray analysis of GEPs from patients, CD34+ bone marrow mononuclear cells were obtained by magnetic bead separation (Miltenyi Biotec)15 from 35 MDS patients and 6 age-matched healthy persons. The CD34+ purity was more than 90% on these samples, checked flow cytometrically. CD34+ cells thus obtained were pelleted, frozen in liquid nitrogen, and kept frozen at −80°C until use. MDS patients were categorized by the French-American-British classification, which was the morphologic basis for the IPSS prognostic classification, incorporating refractory anemia with excess blasts in transformation (RAEB-T) patients. AML transformation was thus considered when patients developed morethan 30% marrow blasts. Marrow samples and clinical information were obtained from patients after informed consent in accordance with the Declaration of Helsinki, with the approval of the Stanford Institutional Review Board.
RNA isolation and amplification
RNA was isolated using the RNeasy kit (QIAGEN). We amplified RNA by the method of Wang et al,16 which optimizes amplification of low-abundance RNA samples with high fidelity by combining antisense RNA (aRNA) amplification with a template-switching effect (Clontech). The concentration and quality of aRNA were monitored spectrophotometrically at optical density (OD) 260/280 and 260/230 and with 1% agarose gels. RNA purity and quality were evaluated using the Bioanalyzer 2100 (Agilent Technologies). Cy3-conjugated nucleotide for aRNA from healthy and Cy5-conjugated nucleotide for aRNA from MDS were hybridized to 40 000 gene chip microarrays obtained from the Stanford Functional Genomics Microarray Facility.17 The Gene Expression Omnibus accession number for the deposited microarray data is GSE18366.
Data acquisition and analysis
The microarrays were scanned with an Axon GenePix scanner (Axon Instruments) and software. High-resolution scans (10 microns per pixel) were performed to compile a raw dataset for each microarray. Files were submitted to the Stanford Microarray Database,18 and the data were normalized by computer-generated normalization values. From the 40 000 gene chips, 11 000 genes expressed with high quality and intensity levels more than 1.5-fold background were used for further analysis. The gene expression data discussed in this article have been deposited in the NCBI Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/).
GEPs from CD34+ marrow cells from MDS patients were compared with those from age-matched CD34+ healthy marrow cells. aRNA from CD34+ pooled normal marrow cells was used as a reference standard.
SAM
Significance analysis of microarrays (SAM) software was used to measure the strength of the statistical relationship between differentially expressed genes and response variables within our microarray dataset.19 The response variables we used included: unpaired groupings (eg, MDS vs normal, those whose disease subsequently transformed [tMDS] vs normal; those whose disease remained stable [sMDS] vs normal, del(5q) MDS vs normal), multiclass grouping (normal vs sMDS vs tMDS), and censored time to leukemia. A false discovery rate (FDR) was generally set to 10% or less.
Hierarchical cluster dendrograms
Supervised and unsupervised hierarchical clustering methods were used to generate dendrograms from the gene list obtained by SAM analysis.5,17 The graphically ordered tree (dendrogram) indicated the relationships among genes. The cluster program indicating the relationship between genes is represented by a dendrogram tree whose branch lengths indicate the degree of similarity between genes. The computed tree thus groups genes with similar expression patterns to be adjacent and coalesced with arrays from each patient.
PAM
The prediction analysis of microarrays (PAM) methodology is a class predictor for gene expression profiling based on the “nearest shrunken centroid method,” which identified subsets of genes that best characterized each class of samples.20 For example, samples from normal persons and MDS patients as subcategories were compared.
Gene function annotation
Gene functions were assessed using SOURCE, a unified genomic resource provided by the Stanford Microarray Database (http:source.stanford.edu)21 and Gene Ontology (http://www.geneontology.org).22
Gene set enrichment analysis
We subjected our 11 000 gene sets to gene set enrichment analysis (GSEA), a computational supervised analysis methodology that uses aggregated public gene sets (1892 gene sets within a molecular signature database; http://www.broadinstitute.org/gsea/)23 to identify biologic processes present across phenotypes in our microarray dataset. (For a list of biologic processes, see Table 1.) GSEA assigns an enrichment score, which represents the difference between the observed and expected rankings (based on correlation with the chosen phenotype). These enrichment scores are normalized based on the number of genes in the gene set. The gene sets were weighted according to each included gene's correlation with the phenotype.
Hypergeometric analysis
This analysis was performed with OntoExpress software (http://vortex.cs.wayne.edu/projects.htm#Onto-Express),24 which evaluated which of the 250 metabolic and signaling pathways from the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg) database were significantly overrepresented when assessing differentially expressed genes from our database. This method evaluated the statistically significant probability (P < .05) of having the observed number of differentially expressed genes within a given biologic process, using the Fisher exact test to determine these probabilities. Gene Ontology was the basis for Kyoto Encyclopedia of Genes and Genomes pathways.
Kaplan-Meier curves
Kaplan-Meier plots were generated using R software (www.r-project.org). These curves were generated based on either clinical features or gene expression values and the patients' freedom from development of AML transformation.
For assessment of degrees of expression of groups of genes (ie, from gene signatures) associated with AML transformation (ie, the “poor risk” signature as indicated in “Results”), we determined the median value for the combined means of the signature genes for each patient and scored the patients as having overexpression or underexpression of these genes. We then generated Kaplan-Meier curves for the patients based on the combination of the significant genes possessing these dichotomous features.
Real-time polymerase chain reaction
Real-time quantitative polymerase chain reaction (RT-PCR) was used to validate expression data for selected genes.25 The expression level of the aRNA from the CD34+ pooled marrow cell reference standard was used to normalize for differences in input cDNA. Predeveloped TaqMan Assays were used (Assays-on-Demand; Applied Biosystems). Each sample was performed in triplicate, and a reverse-transcriptase negative control was also tested to exclude contaminating DNA amplification. The expression ratio was calculated as 2n, where n is the C(T) value difference for each patient (selected gene minus the reference standard).26
Results
Patient information
GEP analyses were performed on RNA obtained from CD34+ marrow cells from 35 MDS patients and 6 age-matched healthy persons. The clinical and cytogenetic details of the patients classified by French-American-British (but with RAEB patients being subdivided into RAEB-1 and RAEB-2 based on whether they had less than 10% or 10% to 20%, respectively, marrow blasts) and IPSS are described in Table 2. There were 24 IPSS Low/Intermediate-1 (Int-1) [10 with del(5q) cytogenetics] and 11 Int-2/High patients analyzed. The patients had not received disease-specific treatment other than 3 del(5q) patients who had received lenalidomide. Most patients had received recombinant erythropoietin therapy. The patients were monitored clinically, with a median follow-up time of 4.3 years (range, 0.2-8.5 years) from the time the bone marrow sample was obtained. During this follow-up period, 12 of the 35 patients transformed to AML (termed tMDS), all within 14 months, whereas the patients' diseases remained stable in the remaining 23 patients (termed sMDS), at least beyond this time period. The flow cytometric characteristics (forward and side scatter) within the blast gate for the CD34+ cells were similar for the MDS and normal cells.
GEPs
MDS versus normal.
Using SAM evaluation, 1175 genes were found to be significantly differentially expressed (FDR = 10%). Of these, 953 genes were overexpressed in MDS and 222 were underexpressed. The median fold change was 2.2 (1.3 to 36) and −2.24 (−1.56 to −33), respectively.
Unsupervised hierarchical clustering using this gene set (Figure 1) clearly separated normal from MDS-derived CD34+ cells and separated MDS patients into 2 major branches, with distinctive signatures derived from their respective GEP clusters. One branch of the MDS patients, highly enriched for those patients who subsequently transformed to AML (tMDS) during follow-up, had a distinctive cluster of overexpressed genes. This patient subgroup, grouped farthest from normal, was composed of 14 patients (top dendrites), within which 10 of the 12 tMDS patients were present. Of interest, only 9 of these patients were classified clinically as having higher risk disease (ie, IPSS Int-2 or High). One of these patients, who did not progress to AML, had subsequently received an allogeneic marrow transplantation 4 months after his marrow sample was obtained. In the other major branch were located the remaining 21 patients, 19 of whose disease remained stable (sMDS), adjacent in the dendrogram, closer to the normals (18 of whom clinically had lower risk IPSS status). Within this patient group, the subgroup of 10 patients with del(5q) abnormalities were present and separable by their distinctive GEP.
GEPs of MDS versus normal CD34+ marrow cells. This unsupervised hierarchical cluster dendrogram depicts differential branches and GEPs from normal and MDS patients (FDR = 10%). Indicated are the clinical and cytogenetic characteristics of these patients as well as whether they subsequently developed AML (purple) or remained stable (blue or brown). Brown dendrites from the patient arrays were from patients with del(5q) MDS.
GEPs of MDS versus normal CD34+ marrow cells. This unsupervised hierarchical cluster dendrogram depicts differential branches and GEPs from normal and MDS patients (FDR = 10%). Indicated are the clinical and cytogenetic characteristics of these patients as well as whether they subsequently developed AML (purple) or remained stable (blue or brown). Brown dendrites from the patient arrays were from patients with del(5q) MDS.
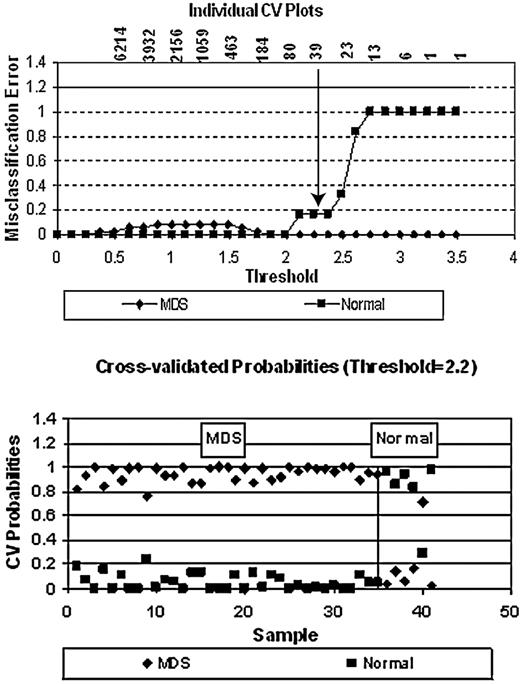
We used PAM (Figure 2) to identify a minimal classifier distinguishing MDS from normal requiring 39 genes, 26 of which were overexpressed and 13 underexpressed in MDS (Table 3). In cross-validation, 5 of 6 healthy persons were classified correctly as were all 35 of the MDS samples. All of the PAM significant classifier genes resided within the most significantly expressed group of SAM significant genes (ie, at FDR = 1%, seen in supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Distinctive classification of MDS from normal using PAM. As indicated (top panel), the classifier distinguishing these groups of persons required a minimum number of 39 genes (the arrow shows the inflection point, below which the misclassification error increases). The specific genes are listed in Table 3. In cross-validation (bottom panel), 5 of 6 healthy persons were classified correctly, as were all 35 MDS samples.
Distinctive classification of MDS from normal using PAM. As indicated (top panel), the classifier distinguishing these groups of persons required a minimum number of 39 genes (the arrow shows the inflection point, below which the misclassification error increases). The specific genes are listed in Table 3. In cross-validation (bottom panel), 5 of 6 healthy persons were classified correctly, as were all 35 MDS samples.
tMDS and sMDS versus normal.
Analysis of GEPs between tMDS and normal and between sMDS and normal demonstrated 1008 and 1052 significantly differentially expressed genes, respectively (FDR = 10%). PAM analysis was used to determine highly differentially expressed gene subset classifiers for tMDS versus normal and sMDS versus normal. This analysis showed distinct segregation between both tMDS and sMDS from normal. The classifier distinguishing tMDS from normal required a minimum of 19 genes and between sMDS and normal required 49 genes. In cross-validation, 5 of 6 normal persons were classified correctly as were all 12 of the tMDS and all 23 of the sMDS samples (6 and 36 genes were unique, respectively, and 13 were concordant). The specific classifier genes for these 2 MDS subgroups are shown in supplemental Tables 2 and 3. SAM analysis depicted 1008 genes differentially expressed at FDR less than 10%, 96 genes at 1%, which also encompassed all of the PAM significant genes. SAM analysis of tMDS versus sMDS revealed 11 highly differentially expressed genes (q value < 10%), overexpressed in tMDS, 5 of which coded for ribosomal proteins (RPs: RPS4X, RPS19, RPS20, RPL6, RPL23, kallikrein-related peptidase 3 [KLK3], tripeptidyl-peptidase II [TPP2], COPB1, SHKBP1, CLID:307029, and CLID:897670).
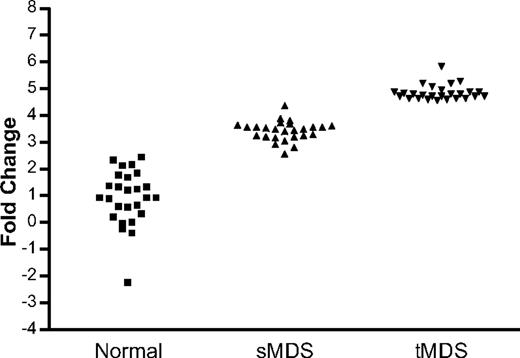
To determine genes potentially involved with disease progression, we performed 2 further statistical analyses. A “time-dependent AML evolution” analysis was performed using SAM to identify the differentially expressed genes that related to the patients' leukemic transformation. This analysis demonstrated 12 significantly differentially expressed genes at FDR less than or equal to 10%, 7 of which coded for RPs (Table 4). In addition, a multiclass progression analysis was performed comparing concordantly expressed genes, which were increased more than or equal to 1.5-fold in sMDS versus normal and a further more than or equal to 1.5-fold increment in tMDS versus sMDS. This analysis demonstrated 26 differentially overexpressed genes (including 8 coding for RPs) to be highly significantly associated (FDR = 1%) with potential for disease progression (Figure 3; Table 5) and 174 genes at less than or equal to 10% FDR.
Multiclass analysis of gene expression in normal persons, sMDS, and tMDS. Comparison of concordantly expressed genes, which were increased more than or equal to 1.5-fold in sMDS versus normal and a further more than or equal to 1.5-fold increment in tMDS versus sMDS demonstrated 26 differentially overexpressed genes to be highly significantly associated with potential for disease progression (FDR = 1%).
Multiclass analysis of gene expression in normal persons, sMDS, and tMDS. Comparison of concordantly expressed genes, which were increased more than or equal to 1.5-fold in sMDS versus normal and a further more than or equal to 1.5-fold increment in tMDS versus sMDS demonstrated 26 differentially overexpressed genes to be highly significantly associated with potential for disease progression (FDR = 1%).
Association of clinical and molecular features with AML transformation
We then determined a “poor risk” gene signature by including genes demonstrated to be highly significantly differentially expressed (FDR < 10%) in all of the 3 following analyses (as described in “tMDS and sMDS versus normal”): time-dependent AML evolution analysis, tMDS versus normal, and multiclass progression analysis (supplemental Table 4, methodologic details). This evaluation demonstrated a group of 6 overexpressed genes (Table 6). Of note, included in this list were 4 coding for RP genes.
Kaplan-Meier curves are shown, which evaluated freedom from AML evolution for patients classified clinically (using IPSS categories; Figure 4A), by their subgrouping in the unsupervised GEP Figure 1 dendrogram (Figure 4B; ie, evaluating the 14 distal patients vs remaining 21 MDS patients in Figure 1), and categorized by their overexpressing (or not) genes comprising the “poor risk” gene signature (Figure 4C; “Methods”). In Figure 4B, analysis of the 14 distal patients in this gene set (group 2) versus the remaining stable (sMDS) patients (group 1) was prognostic, showing increased leukemic transformation in group 2. The GEP was distinct from clinical evaluation (Figure 4A), as only 9 of the 14 GEP high risk subgroup patients (group 2) were clinically higher risk, ie, IPSS Int-2 or High; only 8 were RAEB-2 or RAEB-T.
Freedom from AML evolution for MDS patients classified by clinical and molecular features. Evaluation was performed using (A) clinical features (ie, IPSS categories, P < .001), (B) subgrouping in the unsupervised GEP Figure 1 dendrogram (the 14 distal patients [group 2] vs the remaining 21 MDS patients [group 1]; P = .005), and (C) subgrouping by the overexpression (or not) of genes composing the poor risk gene signature (PRS; P = .01). The Kaplan-Meier curves show significant differences in AML progression using each of these analyses, with significant separation of the IPSS Int-1 subgroup using the poor risk signature (C).
Freedom from AML evolution for MDS patients classified by clinical and molecular features. Evaluation was performed using (A) clinical features (ie, IPSS categories, P < .001), (B) subgrouping in the unsupervised GEP Figure 1 dendrogram (the 14 distal patients [group 2] vs the remaining 21 MDS patients [group 1]; P = .005), and (C) subgrouping by the overexpression (or not) of genes composing the poor risk gene signature (PRS; P = .01). The Kaplan-Meier curves show significant differences in AML progression using each of these analyses, with significant separation of the IPSS Int-1 subgroup using the poor risk signature (C).
Of note, analysis of the impact of the poor risk signature on clinical outcome in the 12 patients having the IPSS Int-1 subtype indicated that, whereas 3 of 6 patients in this clinical group who transformed to AML overexpressed the poor risk signature, all 6 of the patients who lacked this overexpression remained stable (Figure 4C). Those patients in the IPSS Int-2/High risk groups overexpressed the poor risk signature genes, whereas this molecular feature was not present in the low-risk patient group (Figure 4C). These curves all demonstrated significant differences in freedom from AML evolution.
del(5q) MDS versus normal
GEP analysis performed on the dataset comprising del(5q) MDS (n = 10) versus normal (n = 6) demonstrated 540 genes to be significantly differentially expressed (FDR = 10%; supplemental Figure 1). Of these genes, 506 were overexpressed in del(5q) patients and 34 were underexpressed. The median fold change was 3.0 (1.4 to 40) and −3.2 (−2.27 to −71), respectively. The genes that were most significantly overexpressed included GSPT1, ENDOG, ENSA, HCNGP, and SS18L2; those significantly underexpressed were ENG, COG3, COBL, HBA2, and R19275. No clear GEP differences were found between those del(5q) patients before (n = 7) versus after (n = 3) lenalidomide treatment or those with cytogenetic lesions in addition to del(5q) (n = 4). PAM analysis classified del(5q) versus normal well, with the classifier requiring a minimum of 33 genes, 27 overexpressed and 6 underexpressed in del(5q) (Table 7).
del(5q) MDS versus non-del(5q) MDS
GEP analysis was performed comparing the dataset composed of del(5q) MDS (n = 10) versus non-del(5q) MDS patients (n = 25). A total of 1924 genes were found to be significantly differentially expressed (FDR = 10%); 1014 were underexpressed and 901 were overexpressed in del(5q) MDS. The median fold change was 2.28 (1.4 to 54) and −1.89 (−1.11 to −18), respectively. An unsupervised hierarchical clustering dendrogram using these genes showed distinct differences in the GEPs between del(5q) and non-del(5q) MDS patients (supplemental Figure 2). The 10 underexpressed genes within the CDR were AFF4, KIF3A, TGFBI, VDAC1, TCF7, GFRA3, HARSL, ATOX1, FBXO38, and FGFR4.
Functional analyses
The functional categories and biologic processes in which the differentially expressed genes were engaged in MDS versus normal persons (SAM analysis, Figure 1), as determined by Gene Ontology (Table 1) and GSEA, demonstrated a predominance of genes (66%) involved with transcription, cytoskeletal, metabolism, and signaling/transport (at FDR = 10%). Analysis of the most highly differentially expressed genes (ie, at FDR = 1%) demonstrated 96 genes, of which 59% were involved in these same biologic processes (supplemental Table 1). In addition, the genes within these processes were also overrepresented in our dataset compared with the total genes present within the process (using hypergeometric analysis).
GSEA
We subjected our 11 000 gene set to GSEA analysis to identify highly represented differentially expressed genes within our dataset that were common to those in gene sets present within curated public databases. We compared our rank-ordered list of MDS versus normal genes to 412 gene sets obtained from Molecular Signature Database, a database detailing which genes were involved in specific biologic processes. These gene sets were associated with the 12 distinct cellular processes relevant to our MDS dataset (Table 1). Significantly increased numbers of genes involved with RP biosynthesis, Myc and Wnt signaling pathways were present in tMDS patients compared with normal (Table 8; supplemental Figure 3). This contrasted with increased levels of apoptosis-related genes present in sMDS compared with normal persons (supplemental Figure 3). Further, in contrast to the relative overexpression of the ribosomal, Myc and Wnt target genes in tMDS versus normal, these genes were relatively underexpressed in del(5q)MDS versus other MDS patients (Table 8). Table 9 shows the representative gene sets within the public databases related to our tMDS versus sMDS dataset, also demonstrating the predominantly enriched translational (including ribosomal)-, Myc-, and Wnt-related gene sets in tMDS versus sMDS.
Biologic processes
To further clarify the differential expression of specific groups of genes within patient subgroups, we analyzed the genes that were represented in the “poor risk” signature. Because RPs were overrepresented within the signature and in GSEA, we assessed the representation of the entire group of RPs (70 total) and found 37 of them to be differentially expressed. Of interest, these ribosomal genes were all overexpressed in comparisons of MDS versus normal and tMDS versus sMDS (Figure 5A-B), whereas they were underexpressed in the del(5q) group versus the remainder of MDS (Figure 5C). The relative expression of 14 RPs concordantly expressed in the 3 compared subgroups is shown in Figure 5D, including 3 of the 4 RPs within the poor risk signature (RPS4X, RPS25, and RPL23). These genes were also overrepresented as determined by hypergeometric analysis.
Differentially expressed RP expression in MDS subsets. (A) MDS versus normal. (B) tMDS versus sMDS. (C) del(5q) versus non del(5q) MDS. (D) Comparative expression of 14 RPs in these MDS subsets. These data demonstrated increased RP expression in MDS versus normal and in tMDS versus sMDS in contrast to their underexpression in del(5q) MDS versus other MDS patients.
Differentially expressed RP expression in MDS subsets. (A) MDS versus normal. (B) tMDS versus sMDS. (C) del(5q) versus non del(5q) MDS. (D) Comparative expression of 14 RPs in these MDS subsets. These data demonstrated increased RP expression in MDS versus normal and in tMDS versus sMDS in contrast to their underexpression in del(5q) MDS versus other MDS patients.
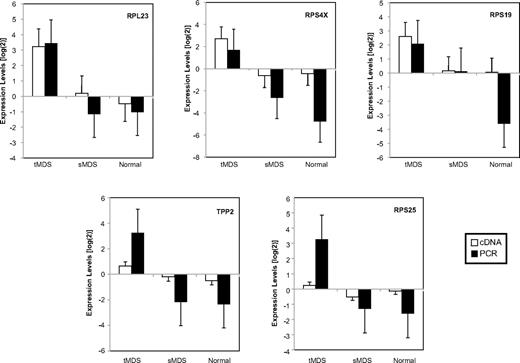
Quantitative RT-PCR
RT-PCR analysis of 7 representative genes, including 5 from the “poor risk” signature (Figure 6), showed similar relative levels of altered gene expression compared with the data generated by the microarray determinations obtained from 9 patients (5 tMDS, 4 sMDS) and 4 healthy persons for which there was adequate remaining material. Noteworthy are the relatively differing expression levels of relevant genes between tMDS and sMDS patients, as also demonstrated by microarray analysis. For example, increased expression was noted from tMDS versus sMDS patients for those genes in the poor risk signature (RPL23, RPS4X, RPS19, RPS25, and TPP2). Combined tMDS and sMDS patients had higher and similar expression levels than healthy persons for GARS and GSPT1 (supplemental Table 5).
Expression of representative genes assessed by quantitative RT-PCR. Comparison of the relative expression levels obtained from RT-PCR and cDNA microarray experiments for 5 genes present in the “poor risk” signature from CD34+ marrow cells from patients with MDS and healthy persons. Demonstrated are the similar degrees of expression for these genes using both analytic methods, as related to the reference standard (mean ± SEM in log2 scale). Also shown are the differing levels of expression of these genes in tMDS (n = 5, increased) versus sMDS (n = 4, decreased) patients, which are further decreased in healthy persons (n = 4).
Expression of representative genes assessed by quantitative RT-PCR. Comparison of the relative expression levels obtained from RT-PCR and cDNA microarray experiments for 5 genes present in the “poor risk” signature from CD34+ marrow cells from patients with MDS and healthy persons. Demonstrated are the similar degrees of expression for these genes using both analytic methods, as related to the reference standard (mean ± SEM in log2 scale). Also shown are the differing levels of expression of these genes in tMDS (n = 5, increased) versus sMDS (n = 4, decreased) patients, which are further decreased in healthy persons (n = 4).
Discussion
Our study provides the initial paper evaluating GEPs from CD34+ marrow cells of MDS patients with prolonged clinical follow-up. SAM was used to differentiate MDS from normal, and then unsupervised hierarchical clustering using this gene set demonstrated 2 major MDS subgroups: those with a high potential to develop AML (tMDS) within 14 months and those whose disease remained stable (sMDS) (Figure 1). These 2 unsupervised GEP subgroups were prognostic and distinct from the clinical evaluation of the patients (Figure 4A-B). This finding led to our subsequent supervised analyses of the tMDS and sMDS patients.
Using a variety of bioinformatic methods and comparative analyses, we demonstrated GEPs valuable for classifying these 2 subgroups of patients and defined a “poor risk” signature of6 genes, which correlated with their subsequent development of leukemia within 14 months (Table 6). This signature also correlated with GEP differences between tMDS and sMDS and showed progressive alterations of expression with more advanced disease status. We demonstrated that patients with overexpression of the genes within the poor risk signature had adverse clinical outcomes (ie, AML transformation). As “controls,” those patients in the IPSS Low and Int-2/High categories had gene signature findings consistent with outcomes generally associated with these features (Figure 4C). Further, of particular note, this association was also evident within the IPSS Int-1 patient group (ie, 3 of 6 such patients overexpressing the signature genes developed AML, whereas all 6 Int-1 patients lacking such expression remained stable). Because clinical determination of prognosis in Int-1 patients remains somewhat problematic, these molecular findings may provide a useful approach to aid evaluation of prognostic features for these patients.
Our data using hierarchical clustering algorithms and dendrograms, obtained from a heterogeneous group of MDS patients, also demonstrated substantial differences in GEPs from their marrow CD34+ cells compared with those from normal persons. These data confirm and extend those from prior studies, which generally had smaller numbers of patients and used a different molecular platform (oligonucleotide arrays rather than the cDNA arrays we used).7-14 These prior studies showed various identity, numbers, and functional correlates of differentially expressed genes in marrow cells from MDS patients, generally with different genes being demonstrated from each study.7-14
A primary biologic observation in our study was the consistent differential expression of ribosomal transcripts in MDS. We demonstrated that a substantial number of RP genes were overexpressed in MDS and more prominently in tMDS versus normal (Figure 5), in distinction to the decreased ribosomal expression in del(5q) MDS (later in “Discussion”). In addition, 4 of the overexpressed genes within the 6-gene poor risk signature were those generating RPs (RPL23, S4X, S19, and S25; Table 6). These findings reflect much prior information in several other neoplastic conditions in which RP overexpression is associated with disease progression and aggressiveness.27-30 Data are accumulating regarding the extraribosomal functions of RPs, with reports showing relationships between overexpression of genes encoding RPs and cancer.31,32 RPs play a direct role in growth regulation. Of note, RPL23, a negative regulator of a Myc antagonist, promotes cell proliferation.33
In addition to the overexpressed RPs represented in the poor risk signature are 2 other genes, which generate known proteolytic enzymes: KLK3 and TPP2. Human tissue kallikreins (KLKs or kallikrein-related peptidases) are a family of extracellular serine proteases that act on a wide variety of physiologic substrates, display aberrant expression patterns in several neoplasms, and have been reported as potential cancer biomarkers.34,35 Prostate-specific antigen (also known as KLK3) is the most widely recognized member of this family.34,35 TPP2 is a protease involved in intracellular proteolysis that is up-regulated in irradiated glioblastoma cells, enhancing tumor cell survival and radio-resistance.36
Confirmation of the relative levels of increased gene expression of specific genes, including those in the poor risk signature assessed by microarray was demonstrated using PCR analysis. Noteworthy are the relatively differing expression levels of relevant genes between tMDS and sMDS patients, as shown by the 2 independent methods.
Using analytic methods accessing public databases (GSEA), we determined functional correlates of these genes and found a high degree of enrichment in MDS (particularly tMDS) compared with normal of the Myc-, Wnt-, and translational-related genes (including RPs) with gene sets in previously curated datasets (Tables 8–9). Further, using additional methods (hypergeometric analyses) to define the relative overrepresentation of the specific genes, we demonstrated these intracellular hematopoietic signaling pathways known to be associated with leukemic progression or AML (Myc and Wnt)37-41 were also overexpressed in the tMDS patients relative to those with sMDS. The Wnt pathway plays important roles in hematopoiesis and stem cell biology.42 Myc is linked to the Wnt pathway by being an important transcriptional target of β-catenin43 and mediates the hyperproliferative effects of Wnt activation.44 In contrast, genes involved with apoptosis-related pathways were more prominent in the sMDS patient subgroup compared with tMDS. These findings are consistent with prior studies indicating enhanced apoptosis and Myc:Bcl2 oncoprotein expression within the CD34 cell compartment early in MDS, with a switch to a decrease in this process concomitant with disease progression.45 Consistent with these findings, recent data have indicated that deregulation of protein translation (including sustained translation of the Myc oncogene) is critical for leukemic cell survival in AML.46
For the more global analysis of the functional correlates of the differentially expressed genes from evaluation of MDS versus normal marrow CD34+ cells, 12 general processes were overrepresented, with the main functions (66%) being those involved with transcription, cytoskeletal metabolism, and signaling/transport (Table 1). We also demonstrated groups of novel genes associated with potential for disease progression (ie, tMDS). Prior MDS microarray studies evaluating marrow cells indicated differentially expressed genes to be associated with stress-related protectors, immune processes, signaling, or the cell cycle or apoptotic inhibitors.7-14
Patients with del(5q) chromosomal abnormalities are a separable MDS subgroup with anemia who have selective beneficial responses to the drug lenalidomide compared with the remainder of MDS patients.47 We found that these patients' marrow CD34+ GEPs were also distinctive; but in contrast to the tMDS patients, these persons generally had decreased RP expression. Prior studies have shown decreased levels of other genes (SPARC)14 and a specific RP (RPS14)48 in this disorder. In addition, consistent with our findings, Pellagatti et al have also shown ribosomal- and translation-related probe sets to be significantly differentially expressed, with approximately 90% of these showing lower expression levels in the 5q− syndrome patient group compared with normal and other MDS patients.12 Several congenital types of anemias (eg, Diamond-Blackfan anemia, dyskeratosis congenita), which have a propensity to develop leukemia, also have mutations or decreases in RP synthesis or expression, including RPS19, S20, L5, and L11.49,50 Of note, the del(5q) patients in our study had decreased expression of genes coding for RPs, including RPS20, L5, and L11 (Figure 5). These are potentially relevant ribosomal relationships between congenital types of anemias (which are not only not clonal hemopathies initially but have highly vulnerable and noncompetitive stem cells) and MDS patients (which are clonal and the clones have out-competed the nonclonal stem cells). A possible explanation for these findings is that initially the del(5q) patients may be comparable with the pre-MDS DBA or dyskeratosis patients, whereas the del(5q) MDS patients have evolved clonally in a way that alters the defect in a maladaptive way. This is consistent with the differentially expressed genes in the del(5q)MDS versus normal and the non-del(5q) MDS comparisons.
Our findings of distinctive marrow CD34+ cell GEPs in MDS patient subsets provide molecular insight into mechanisms underlying the disease and its propensity to progress to a more aggressive stage. Of particular importance in our study was the definition of a “poor risk” signature associated with the propensity of MDS patients to undergo AML transformation. Assessing the impact of multiple molecular abnormalities on disease phenotype, particularly of RPs, by this gene array technology supplements those studies evaluating single-gene analyses and clinical features. These findings, if verified, should prove to be valuable in the future for diagnostically and prognostically classifying such patients. It will be important to expand the number of patients analyzed by these methods and to validate the poor risk signature and GEPs found in this study against an additional cohort of MDS patients. Such studies are ongoing in our laboratory and those of others.
The online version of this article contains a data supplement.
Presented in part at the American Society of Hematology 47th Annual Meeting, Atlanta, GA, December 12, 2005.51
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Note added in proof:
After completion of our study and just before submission of our manuscript, we noted the article by Mills K, Kohlmann A, Williams PM, et al. Microarray-based classifiers and prognosis models identify subgroups with distinct clinical outcomes and high risk of AML transformation of myelodysplastic syndrome. Blood. 2009;114(5):1063-1072. These authors used nonenriched mononuclear cells for their analyses rather than CD34+ cells as used in our study.
Acknowledgments
This study was supported by the Muriel and Ira Coleman Leukemia Research Fund, the William E. Walsh Leukemia Research Fund, the Eugene, Elizabeth and Christina Cronkite Fund for Hematology, California Cancer Research Program (grant 99-00520V-10144), the Leukemia & Lymphoma Society (SCOR grant), Veterans Administration Palo Alto Health Care System (resources and use of facilities), and the National Institutes of Health (R01 grant LM009719; A.J.B.).
National Institutes of Health
Authorship
Contribution: K.S. performed and designed the research and assisted in writing the manuscript and analyzing the data; D.T.R. analyzed the data and reviewed the manuscript; R.T. provided statistical analysis; A.J.B. provided bioinformatics and statistical analyses; and P.L.G. designed the research, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current address of D.T.R. is Applied Genomics Inc, Burlingame, CA.
Correspondence: Peter L. Greenberg, Hematology Division, Stanford University Medical Center, 875 Blake Wilbur Dr, Rm 2335, Stanford, CA 94305; e-mail: peterg@stanford.edu.



![Figure 4. Freedom from AML evolution for MDS patients classified by clinical and molecular features. Evaluation was performed using (A) clinical features (ie, IPSS categories, P < .001), (B) subgrouping in the unsupervised GEP Figure 1 dendrogram (the 14 distal patients [group 2] vs the remaining 21 MDS patients [group 1]; P = .005), and (C) subgrouping by the overexpression (or not) of genes composing the poor risk gene signature (PRS; P = .01). The Kaplan-Meier curves show significant differences in AML progression using each of these analyses, with significant separation of the IPSS Int-1 subgroup using the poor risk signature (C).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/23/10.1182_blood-2009-08-236422/4/m_zh89990945180004.jpeg?Expires=1768477718&Signature=sc4Gf5xxkUJdekGjUUZGPrAzhCjXTJY9HJk0Yk5P6YOGt0g3C-pjmTP2gxKXgj16DbTo~Rc3JVQEoS7fTIB48~KXiE9S8jrgNKLJa~~XSAXz34H3-swkOWKAMvz64Y3HzhAwgtiMG4DblKISq38jE8SOR9tuh39e3W44Kpss9PurjwUlmGvZFXWrVqoGGrzDxJuQ8yOznY94jq-nuJpm8s0fzsEua29mg11HMQ5J4IuNmrMnaHPYE2uhR5vlzz33YSuIAC1ESa1NaB1IiPiTA2jrPmT785hjYW8~pqfKnwP5UAB~xv2DIe14PvenMIxMjhBYX75mbMgXa7wM-YTfqw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





![Figure 4. Freedom from AML evolution for MDS patients classified by clinical and molecular features. Evaluation was performed using (A) clinical features (ie, IPSS categories, P < .001), (B) subgrouping in the unsupervised GEP Figure 1 dendrogram (the 14 distal patients [group 2] vs the remaining 21 MDS patients [group 1]; P = .005), and (C) subgrouping by the overexpression (or not) of genes composing the poor risk gene signature (PRS; P = .01). The Kaplan-Meier curves show significant differences in AML progression using each of these analyses, with significant separation of the IPSS Int-1 subgroup using the poor risk signature (C).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/23/10.1182_blood-2009-08-236422/4/m_zh89990945180004.jpeg?Expires=1768477719&Signature=NRSr2y6FwN31Kpc2kOA44QsJqm4hiSyNiGcTguP1j6AYxCCAFGP3kZD5FN9urpYtT0xOcxWszP8YmZ8YFGnCmY-ueis6UEM8wu~zTv3xaDRY9AmVv54OFas-XC946SgE5rm~i3GJA5IzrXOLe8pkMU6zcvLE7oQPACgSLsW8efhfXnPEom3QuUfkuHVVnSskj~Uja6a9ujHn-RFgWDfWP98bjdIQ~5W4nu-0Y8RSSC3~8kuIqJg78TayAVSFu8TXI2h1yx34IjjrX2E2jsG9bJVN~7uGeSwlg3x2zA-2ieEcvrxOsSWmHXzbt25Khamw9KqYy03l67~C6YKhyE2FxA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)