In this issue of Blood, Chung and colleagues reported that undifferentiated hematopoietic cells are characterized by a genome-wide undermethylation dip around the transcription start site and a hierarchical epigenetic plasticity.
Every somatic cell with the exception of B and T lymphocytes, which during differentiation rearrange immunoglobulin or T-cell receptor genes, respectively, contains the same repertoire of genes. However, these genes, because of epigenetic modifications during cell maturation, are differently expressed in various cell types. Thus, had George Orwell been a geneticist, he could have come to the conclusion that “all cells are equal, but some cells are more equal than others.” Would he have been right in this particular case?
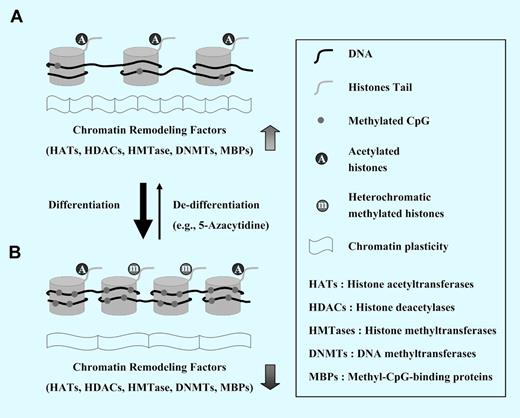
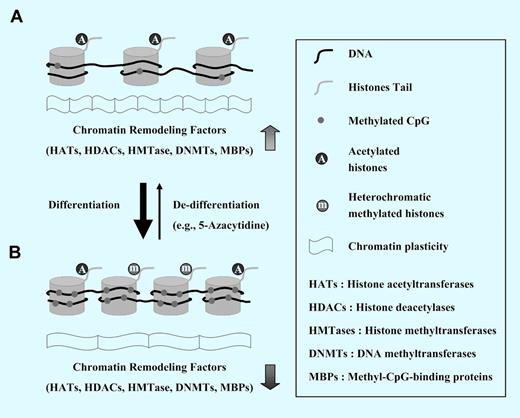
The role of epigenetics in chromatin plasticity. (A) Chromatin in most primitive pluripotent stem cells (eg, embryonic stem cells) is in an open/active state (euchromatin) and several genes are transcribed. This state is due to wide undermethylation of DNA in and its association with acetylated histones that are a mark of euchromatin. (B) The cell differentiation is linked to gradual methylation of cytosine at cytosine-phosphate-guanine (CpG) dinucleotides that reside within DNA regulatory sequences affecting the gene expression. These CpG dinucleotides are gathered in the DNA strand in repetitive sequences present usually around transcription start sites (TSS) of promoters of some genes; they are called CpG islands (CGI). Whereas CGI+ genes encode developmental regulators or housekeeping genes, CGI− genes are highly enriched for lineage-specific genes. The methylation of CpGs usually negatively affects transcription of neighboring genes. Transcription of particular genes is also affected by another epigenetic mechanism: recruitment of transcription repressive complexes by CpG-enriched DNA and posttranslational modification of histone tails that leads to a decrease in their acetylation. Thus, during the differentiation process, the open type of euchromatin changes to the more condense and genetically silent heterochromatin.
The role of epigenetics in chromatin plasticity. (A) Chromatin in most primitive pluripotent stem cells (eg, embryonic stem cells) is in an open/active state (euchromatin) and several genes are transcribed. This state is due to wide undermethylation of DNA in and its association with acetylated histones that are a mark of euchromatin. (B) The cell differentiation is linked to gradual methylation of cytosine at cytosine-phosphate-guanine (CpG) dinucleotides that reside within DNA regulatory sequences affecting the gene expression. These CpG dinucleotides are gathered in the DNA strand in repetitive sequences present usually around transcription start sites (TSS) of promoters of some genes; they are called CpG islands (CGI). Whereas CGI+ genes encode developmental regulators or housekeeping genes, CGI− genes are highly enriched for lineage-specific genes. The methylation of CpGs usually negatively affects transcription of neighboring genes. Transcription of particular genes is also affected by another epigenetic mechanism: recruitment of transcription repressive complexes by CpG-enriched DNA and posttranslational modification of histone tails that leads to a decrease in their acetylation. Thus, during the differentiation process, the open type of euchromatin changes to the more condense and genetically silent heterochromatin.
All these epigenetic modifications, however, are reversible as shown in nuclear transfer experiments when the nucleus of a differentiated fibroblast after transfer into ovum may acquire the full developmental potential to create an embryo. Thus, with the advancement of regenerative medicine, much effort has been dedicated to elucidate mechanisms that regulate gene expression in primitive versus more differentiated cells. Hematopoietic stem cells (HSCs) at different levels of maturation are a great model for such studies.
A DNA helix that contains genetic information is wrapped around histone proteins and together makes up nuclear chromatin. The structure of chromatin at specific loci differs between cells and is determined by heritable mechanisms known as epigenetic modifications that lead to changes in gene expression without altering the sequence of DNA. The most important epigenetic mechanisms include DNA methylation and histone tail modification (see figure).
To maintain undifferentiated status, primitive stem cells like embryonic stem cells (ESCs) possess an open/active state chromatin (euchromatin) and display more dynamic epigenetic status, due to the high expression level of chromatin-modifying factors. Moreover, according to extensive genome-wide studies, important developmental regulators including homeodomain-containing transcription factors (eg, Dlx, Irx, Lhx, Pou, Pax, Six) are regulated in ESCs by specific epigenetic marks called “bivalent domains,” in which 2 transcriptionally active—and repressive—histone marks are physically linked together.1,2 These plastic epigenetic marks prevent undifferentiated ESC differentiation, but in response to environmental cues, they may turn on the specialized transcriptomes to regulate development of more differentiated cells. Although similar epigenetic mechanisms involving “bivalent domains” can occur in adult stem cells,3 the results of genome-wide approaches to better characterize such domains in undifferentiated HSCs are still missing.
In this issue of Blood, Chung et al used a genome-wide comparison of DNA methylation to study epigenetic marks in human umbilical cord (UCB) CD34+ (undifferentiated) and CD34− (differentiated) cells.4 They found in CD34+ cells a presence of characteristic hypomethylation dip around transcription start sites (TSS) of promoters and hypermethylation in flanking regions. The undermethylated DNA methylation pattern near TSS is different in CpG islands (CGI+) and non-CGI (CGI−) genes and seems to be related to an active gene transcription and dynamic chromatin status in a population of primitive HSCs. Furthermore, undifferentiated HSCs exhibited dynamic open-type chromatin associated with transcriptionally active acetylated histones more than terminally differentiated ones. In the next step, authors inhibited chromatin condensation (heterochromatin) by preventing methylation of DNA and histones by employing 5-azacytidine or trichostatin A and noticed enhanced self-renewal of murine bone marrow (BM)–derived HSCs that were transplanted into lethally irradiated recipients but not of “steady-state” BM-isolated HSCs. Interestingly, similar treatment of more mature cells leads to partial phenotypic de-differentiation and apoptosis that correlated with the level of their hematopoietic maturation. The authors conclude that the undifferentiated state of hematopoietic cells is characterized by unique epigenetic signature that includes both (1) dynamic chromatin structure and (2) an epigenetic plasticity of gene expression that correlates with the level of differentiation.
This paper is important because it addresses several issues related to developmental biology and potential “plasticity” of HSCs. First, the authors clearly demonstrate the dynamic structure of chromatin and its epigenetic modification during differentiation of hematopoietic cells. Second, data presented explain the efficacy of 5-azacytidine and trichostatin A for ex vivo expansion of undifferentiated HSCs at a molecular level. This may be relevant during generation of inducible pluripotent stem cells using the so-called small molecular chromatin modifying agents (eg, BIX-01294 and BayK8644).5 However, because aberrant epigenetic regulation can lead to tumorigenesis and premature stem cell aging,6 full understanding of epigenetic processes that are a kind of “2-edged sword” mechanism is a timely challenge. Finally, additional studies on purified HSCs to elucidate biological significance of unique DNA methylation patterns found in current genomewide analysis will help to better understand epigenetic mechanisms that govern hematopoiesis.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■