Abstract
KLRG1 is an inhibitory receptor expressed on a subset of mature T and NK cells. Recently, E-, N-, and R-cadherin have been identified as ligands for KLRG1. Cadherins are a large family of transmembrane or membrane-associated glycoproteins that were thought to only bind specifically to other cadherins to mediate specific cell-to-cell adhesion in a Ca2+-dependent manner. The consequences of cadherin KLRG1 molecular interactions are not well characterized. Here, we report that the first 2 extracellular domains of cadherin are sufficient to initiate a KLRG1-dependent signaling. We also demonstrate that KLRG1 engagement inhibits cadherin-dependent cellular adhesion and influences dendritic cell secretion of inflammatory cytokines, thereby exerting immunosuppressive effects. Consistent with this, engagement of cadherin by KLRG1 molecule induces cadherin tyrosine phosphorylation. Therefore, KLRG1/cadherin interaction leads to the generation of a bidirectional signal in which both KLRG1 and cadherin activate downstream signaling cascades simultaneously. Taken together, our results provide novel insights on how KLRG1 and E-cadherin interactions are integrated to differentially regulate not only KLRG1+ cells, but also E-cadherin–expressing cells, such as dendritic cells.
Introduction
Epithelial cadherins (E-cadherins), neural cadherins (N-cadherins), and retinal cadherins (R-cadherins) are part of the classical cadherins. These ubiquitously expressed cell adhesion molecules are a large family of transmembrane or membrane-associated glycoproteins comprising an extracellular domain containing 5 cadherin repeats (EC1-5) responsible for cell-to-cell interactions, a transmembrane domain, and a cytoplasmic domain that is linked to the actin cytoskeleton. Typically, cadherins mediate calcium-dependent homophilic adhesion, thereby promoting association of cells expressing the same cadherin family members to form adherens junctions.1,2 The formation of adherens junctions is dependent on the association of cadherin's cytoplasmic tail with β-catenin and its partners.1 Numerous biologic processes, including homeostasis and embryogenesis, rely on the selective adherence of one adhesion molecule to another through precise intermolecular interactions.3 The spatiotemporal regulation of cadherin expression and function are vital to tissue morphogenesis, providing a basis for the formation of epithelial layers of the skin and intestine.4-6
Aside from their homophilic adhesion mode, E-, N-, and R-cadherins have been recently reported to bind in a heterophilic manner with killer cell lectin-like receptor G1 (KLRG1).7-9 KLRG1 is a transmembrane inhibitory receptor belonging to the C-type lectin-like superfamily that contains an immunoreceptor tyrosine-based inhibitory motif (ITIM) in its cytoplasmic domain. The molecule was first identified in the rat basophilic leukemia cell line RBL-2H3 and was originally termed mast cell function-associated Ag (MAFA).10-15
In mice and humans, this well-conserved receptor is found on subsets of T and natural killer (NK) cells.16-24 Cells that express KLRG1 include the most mature and recently activated NK cells as well as effector/memory T cells.16-25 Expression of KLRG1 increases substantially in T and NK cells during viral, bacterial, or parasite infections in mice.20,21,26-28 KLRG1 is also expressed on FoxP3+ regulatory T cells.29,30
Besides its role as a marker to identify lymphocytes in their differentiation stage, KLRG1 has been described to function in multiple roles in a variety of cell types. In both rat and mouse, the ITIM tyrosine residue of KLRG1 is susceptible for phosphorylation leading to the recruitment of phosphatases SH2-containing inositol polyphosphate 5-phosphate (SHIP-1) and SH2-containing protein-tyrosine phosphatase 2 (SHP-2).9,31,32 It has also been shown that engagement of the murine KLRG1 inhibits NK-cell cytotoxicity,7 cytokine production,9,19,21 and Ag-induced T-cell division.8
Although KLRG1 functions are now being uncovered, its physiologic role is still unclear. It is also unknown whether KLRG1 can regulate cadherin functions. Here, we found that upon cell-to-cell contact, cadherin not only sends a signal through the activation of its cognate receptor, but it also rapidly undergoes tyrosine phosphorylation. This cadherin “reverse” signaling, as opposed to the “forward” signaling activated downstream of KLRG1, leads to a disruption of cellular shape and adhesiveness. Notably, KLRG1 inhibits the ability of E-cadherin-expressing dendritic cells (DCs) to release inflammatory cytokines. These data suggest that the interplay of KLRG1 and E-cadherin interaction regulates the reciprocal roles that E-cadherin and KLRG1 play in determining the immune response.
Methods
Mice
C57BL/6J and B10.D2 mice were purchased from Taconic. All mice were maintained at Brown University in accordance with institutional guidelines for animal care and use.
Cells and purification of DCs
Mouse cell lines DO11, A20, L929, and NIH 3T3 were grown in either Dulbecco modified Eagle medium (DMEM) or RPMI 1640 (Invitrogen) with 8% fetal calf serum (FCS; Atlanta Biologicals), 1% penicillin/streptomycin (Gibco), and 50μM 2-mercaptoethanol (Bio-Rad Laboratories). BWZ.36, BWZ.36 H/S64, and BWZ.36 H/S64 YF reporter cells were cultured in 8% RPMI 1640 with hygromycin-B (BD Biosciences) at 200 μg/mL. The retroviral packaging cell line Plat-E was provided by T. Kitamura (University of Tokyo) and was used for retroviral transduction. Mouse bone marrow–derived dendritic cells (BMDCs) were cultured in 8% RPMI 1640 with granulocyte-macrophage colony-stimulating factor (GM-CSF; R&D Systems) at 20 ng/mL. CD11c+ DCs were enriched with anti–CD11c-conjugated beads and columns (Miltenyi Biotec) according to the manufacturer's protocol.
Reagents and antibodies
Purified anti-cadherin monoclonal antibodies for Western blot were purchased from BD Transduction Laboratories or Sigma-Aldrich. Both anti-Myc (clone 4A6) and anti-phosphotyrosine (clone 4G10) were obtained from Upstate Cell Signaling Solutions. Loading control β-actin antibody was purchased from Abcam. Mouse and rabbit antibodies conjugated to horseradish peroxidase (HRP) were from Jackson ImmunoResearch Laboratories. FACS antibodies TCRβ (clone H57-597), CD11c, and KLRG1 (2F1) were purchased from eBioscience. Rat anti–E-cadherin (mouse) for flow cytometry (clone ECCD-2) was from Zymed Laboratories. KLRG1 tetramer was produced in the laboratory as described in Tessmer et al.9 For some of the in vitro studies, azide-free streptavidin was used to generate KLRG1 tetramer. Recombinant N-cadherin plasmid constructs were a gift from Dr L. Shapiro (Columbia University). Bacteria BL21-CodonPlus (DE3)-RIL Competent Cells (Stratagene) were transformed with the N-cadherin/pET-28b expression vector and induced by the addition of isopropylthiogalactoside to a final concentration of 0.1mM for 18 hours. Bacteria were then lysed and supernatant was used to purify extracellular domain of N-cadherin through Ni2+ affinity chromatography as previously described.33
Flow cytometric analysis
Cells were preincubated with Fc receptor-blocking 2.4G2 mAb, when necessary, and stained for 30 minutes on ice with primary antibody or 20 minutes at room temperature (RT) with KLRG1 tetramer. Washed cells were then stained for 30 minutes on ice with secondary antibody (Jackson ImmunoResearch Laboratories), when necessary, washed, and evaluated on a FACSAria (BD Biosciences) and analyzed with FlowJo software (TreeStar). For N-cadherin blocking experiments, KLRG1 tetramer and TCRβ were preincubated with soluble recombinant extracellular N-cadherin domains 1 and 2 for 30 minutes at RT, followed by addition of DO11 E-cadherin wild-type (WT) cells for 20 minutes at RT.
Reporter cell assay
Soluble recombinant N-cadherin or bovine serum albumin (BSA) in carbonate buffer was coated onto a 96-well plate for 3 hours at 37°C. After washing, N-cadherin or BSA-coated plates were incubated with parental and KLRG1 chimeric receptor reporter cells overnight at 37°C. Fixed cells were washed twice with PBS and treated with X-Gal substrate (Invitrogen) and incubated for 5 hours at 37°C. Cells were imaged at ×10 with an Olympus DP70.
E-cadherin juxtamembrane domain and TD cDNA constructs and retroviral transduction
Using the full-length E-cadherin cDNA as a template, myc-tagged E-cadherin juxtamembrane domain (JMD; lacking the β-catenin binding site) and tail deleted (TD; lacking the whole cytoplasmic tail) were generated. To do so, PCR was performed using 5′ primer 5′-CAGATCTGATGGGAGCCCGGTGCCGCAGC-3′ and 3′ primer (TD) 5′-CGTTAACGCTACAGATCCTCTTCAGAGATGAGTTTCTGCTCCGTTCTCCTCCGTAGAAA3-′; 3′ primer (JMD) 5′-CGTTAACGCTACAGATCCTCTTCAGAGATGAGTTTCTGCTCCCTGTGCAGCTGGCTCAA-3′. The polymerase chain reaction (PCR) product was purified by agarose gel electrophoresis and ligated into the TOPO cloning vector (Invitrogen). The construct was sequenced and inserted into the unique BglII and HpaI sites of the retroviral vector mouse stem cell virus–internal ribosome entry site–green fluorescent protein (MSCV-IRES-GFP). Cells were retrovirally transduced as described.9
Immunoprecipitation
DO11 E-cad WT cells mixed with BWZ.36 cells at a 1:1 ratio and DO11 E-cad WT cells mixed with BWZ.36 H/S64 YF at a 1:1 ratio were incubated 0 minutes (at 4°C) and 5 minutes (at 37°C). After the indicated treatments, cells were then solubilized in ice-cold lysis buffer (1% Triton X-100, 50mM Tris [pH 7.5], 150mM NaCl, 5mM ethylenediaminetetraacetic acid (EDTA), 1mM orthovanadate, and protease inhibitors) at 4°C for 20 minutes. Lysates were centrifuged for 20 minutes at 4°C. For immunoprecipitation, precleared lysates were first incubated for 3 hours at 4°C with 1.5 μg of specific E-cadherin antibodies. Lysates were then incubated with protein G–Sepharose (GE Healthcare) overnight at 4°C. Beads were washed 5 times with ice-cold lysis buffer. Immunoprecipitates were boiled in 3× reducing sample buffer (5% sodium dodecyl sulfate [SDS], 10% glycerol, 3% dithiothreitol, 0.15M Tris/HCl [pH 6.8], 0.012% bromphenol blue) and resolved by 4% to 15% SDS–polyacrylamide gel electrophoresis (PAGE; Bio-Rad Laboratories).
Western blotting and quantitation
After boiling, samples were separated on 4%–15% SDS-PAGE gels and proteins were transferred to nitrocellulose membrane (Bio-Rad Laboratories). Membranes were blocked in either 5% BSA or milk for 1 hour at RT and probed with the indicated antibodies overnight at 4°C. After washing, membranes were incubated with the respective HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) for 1 hour at RT and developed with Super Signal West Pico (Pierce). Membranes were exposed to Biomax MR film (Kodak) and developed. Film images were scanned and analyzed using ImageJ software (National Institutes of Health). Myc signal intensity was compared with β-actin.
Fibroblast-like cell assay
Parental and E-cadherin transduced cells were mixed with 1 μg/mL or 10 μg/mL dialyzed KLRG1 tetramer or 1× N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) (20mM HEPES, 150mM NaCl, 0.01% sodium azide) and cultured in 24-well plates at 37°C. Cells were imaged at ×20 with an Olympus DP70. Cells obtaining “fibroblast-like” phenotype were counted over total amount of cells, and the percentage of “fibroblast-like” adhesive cells was determined. Imaging and quantitation were performed in a double-blinded manner.
Antigen presentation assay
For in vitro stimulation, parental and E-cadherin transduced A20 cells were pulsed with 10 mg/mL ovalbumin (Sigma-Aldrich) or control for 6 hours at 37°C. Washed A20 cells were incubated with parental or KLRG1 transduced DO11 hybridoma cells for 14-16 hours at 37°C. Supernatants were collected and interleukin-2 (IL-2) was measured by enzyme-linked immunosorbent assay (ELISA). For ex vivo stimulation, antigen presentation assay using BMDCs were performed as above, except BMDCs replace A20s.
Inflammatory cytokine induction assay
E-cadherin–expressing BMDCs were allowed to incubate with mock/parental or KLRG1-transduced cells overnight at 37°C. Supernatants were collected after incubation with 10 μg/mL of LPS (Sigma L43901 E-coli 0111:B4) for 0 (PBS), 6, or 12 hours. IL-6, tumor necrosis factor α (TNF-α), and IL-10 were measured with CBA IL-6, TNF-α, and IL-10 flex sets using a FACSAria and FCAP Array Software (all BD Biosciences).
Statistical analysis
Statistical significance, designated as a P value less than or equal to .05, was determined by paired, 2-tailed Student t test.
Results
Extracellular domains 1 and 2 of N-cadherin binds to KLRG1
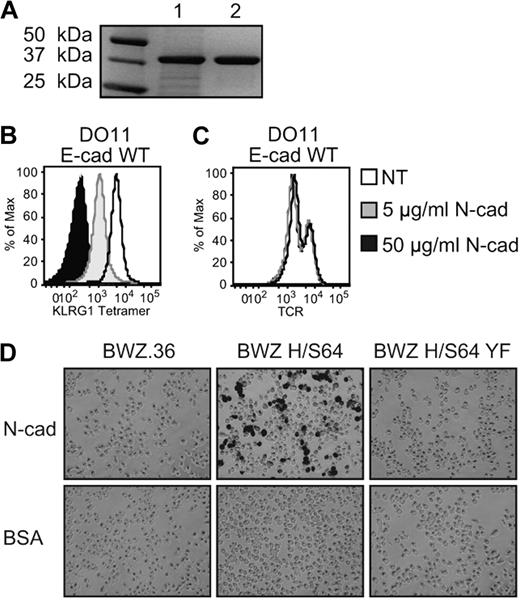
Considering that the first and second extracellular domains of cadherin are critical for homophilic cis and trans interactions,34,35 and that KLRG1 binding to cadherin apparently competes with homophilic cadherin interactions,36 we examined the possibility that extracellular domains 1 and 2 (EC1-2) of N-cadherin could inhibit E-cadherin binding to KLRG1. To determine whether N-cadherin EC1-2 interacts with KLRG1, we produced a purified soluble recombinant N-cadherin EC1-2 (Figure 1A). When E-cadherin–transduced T-cell hybridoma cells were analyzed for KLRG1 tetramer and TCR staining, preincubation of N-cadherin EC1-2 with the staining reagents prevented the binding of KLRG1 tetramer to E-cadherin–transduced cells in a dose-dependent manner (Figure 1B). The specificity of N-cadherin EC1-2 blocking KLRG1 tetramer staining on E-cadherin-transduced DO11 cells was confirmed by the unchanged level of TCR staining regardless of the presence of N-cadherin EC1-2 (Figure 1C). These results cannot be attributed to E-cadherin and N-cadherin interactions, as previous studies indicate that E-cadherin does not interact with N-cadherin.4,37-39 To test for N-cadherin EC1-2 functionality, a reporter cell line9 was used. N-cadherin EC1-2 was able to activate signaling in the KLRG1 chimeric receptor reporter cell (BWZ H/S64), but not the KLRG1 chimeric receptor DAP12 mutant reporter cell (BWZ H/S64 YF; Figure 1D top). Collectively, these data demonstrate that functional N-cadherin EC1-2 not only binds to KLRG1, but also is sufficient to regulate its signaling activities.
Functional recombinant N-cadherin domains 1 and 2 block KLRG1 binding to E-cadherin. (A) SDS-PAGE of recombinant N-cadherin. Extracellular domains 1 and 2 of N-cadherin were produced and purified by Ni2+ affinity chromatography. Lane 1: lysate; lane 2: purified N-cadherin. The protein bands were revealed using Gel Code Blue staining reagent (Coomassie). (B) KLRG1 tetramer and (C) anti-TCRβ mAb were first incubated 30 minutes with N-cadherin before incubation with DO11 E-cadherin WT cells. Cells were washed 3 times and analyzed by FACS. The results are representative of 5 independent experiments. (D) N-cadherin– or BSA-coated plates were incubated with parental or KLRG1 reporter cells. Fixed cells were washed and treated with X-Gal substrate and imaged at ×10 with an Olympus DP70. The results are representative of 3 independent experiments.
Functional recombinant N-cadherin domains 1 and 2 block KLRG1 binding to E-cadherin. (A) SDS-PAGE of recombinant N-cadherin. Extracellular domains 1 and 2 of N-cadherin were produced and purified by Ni2+ affinity chromatography. Lane 1: lysate; lane 2: purified N-cadherin. The protein bands were revealed using Gel Code Blue staining reagent (Coomassie). (B) KLRG1 tetramer and (C) anti-TCRβ mAb were first incubated 30 minutes with N-cadherin before incubation with DO11 E-cadherin WT cells. Cells were washed 3 times and analyzed by FACS. The results are representative of 5 independent experiments. (D) N-cadherin– or BSA-coated plates were incubated with parental or KLRG1 reporter cells. Fixed cells were washed and treated with X-Gal substrate and imaged at ×10 with an Olympus DP70. The results are representative of 3 independent experiments.
KLRG1 binding to E-cadherin inhibits E-cadherin–dependent cell adhesion
E-cadherin contains a large cytoplasmic tail consisting of the JMD and a β-catenin binding site (Figure 2A). To investigate the function of E-cadherin when bound to KLRG1, several constructs were generated, including WT, JMD (lacks the β-catenin binding site), and TD (lacks the entire cytoplasmic tail) mutant E-cadherin constructs (Figure 2A). Because E-cadherin antibodies used for Western blots recognize only the cytoplasmic tail, we N-terminally Myc tagged WT and mutant E-cadherin constructs. The retroviral vector contains an IRES-GFP cassette, allowing E-cadherin and GFP to be linked by a single transcript. Although cells transduced with WT, JMD, or TD E-cadherin exhibited comparable levels of GFP proteins, E-cadherin cell surface expression was not proportional to the GFP level. We found, in 2 different cell lines, that TD E-cadherin cell surface expression level was higher than WT E-cadherin (Figure 2B and supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). In contrast, JMD E-cadherin is poorly expressed at the cell surface compared with WT E-cadherin (Figure 2B). Notably, immunoblot analysis revealed that comparable amounts of protein were produced (Figure 2C), suggesting that these proteins used distinct trafficking routes.
E-cadherin cell surface expression is modulated by its cytoplasmic domain. (A) Schematic representative of E-cadherin constructs. (B) Parental or the various E-cadherin transfectants as indicated were stained with KLRG1 tetramer or E-cadherin antibody and analyzed by FACS. Data are representative of at least 5 independent experiments. (C) Cell lysates were blotted for Myc and reprobed with β-actin. Data are representative of at least 4 independent experiments. Quantitation was performed as described in “Western blotting and quantitation.”
E-cadherin cell surface expression is modulated by its cytoplasmic domain. (A) Schematic representative of E-cadherin constructs. (B) Parental or the various E-cadherin transfectants as indicated were stained with KLRG1 tetramer or E-cadherin antibody and analyzed by FACS. Data are representative of at least 5 independent experiments. (C) Cell lysates were blotted for Myc and reprobed with β-actin. Data are representative of at least 4 independent experiments. Quantitation was performed as described in “Western blotting and quantitation.”
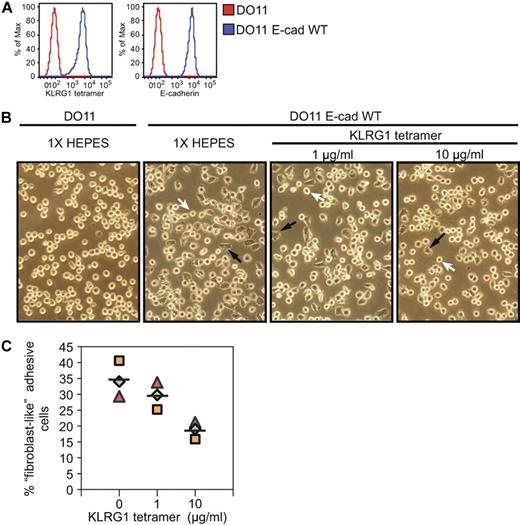
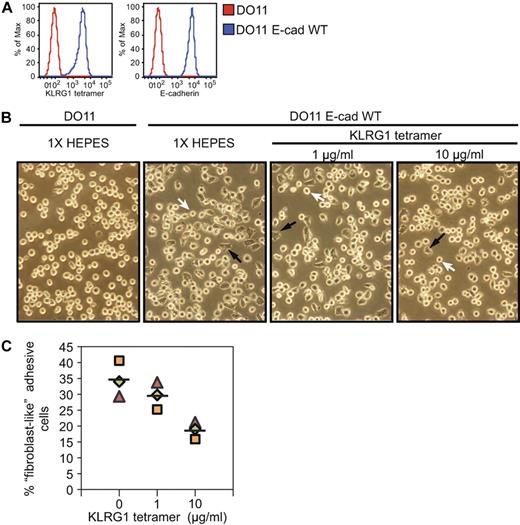
Interestingly, we observed that DO11-E cadherin WT cells lose their round shape morphology, acquire a “fibroblast-like” morphology, and become strongly adherent (Figure 3B black arrows). The change in morphology appears to be largely dependent on the E-cadherin cytoplasmic tail, because JMD and TD E-cadherin–transfected cells did not display this phenotype, suggesting that a functional cadherin-catenin complex is needed for the switch in morphology (data not shown). We hypothesized that KLRG1 might interfere with cadherin-dependent adhesion properties. To test this possibility, KLRG1 tetramer was incubated with parental and WT E-cadherin–transduced cells. We found that KLRG1 binding to cadherin induces a reduction of the number of adhesive fibroblast-like cells in a dose-dependent manner but has no noticeable effect on parental cell lines (Figure 3B,C). We conclude that KLRG1 interaction with E-cadherin can inhibit cadherin-dependent cell adhesion.
KLRG1 inhibits adhesive properties on E-cadherin-expressing cells. (A) Parental or WT E-cadherin cells were stained with KLRG1 tetramer or E-cadherin antibody and analyzed by FACS. (B) Approximately 7.5 × 104 parental or WT E-cadherin–transduced cells were either treated with KLRG1 tetramer at the indicated concentrations or left untreated (1× HEPES) for 24 hours at 37°C. Fibroblast-like adhesive properties were imaged at ×20 with an Olympus DP70. Black arrowheads show fibroblast-like cells, and white arrowheads depict round circular cells. Data are representative of 3 independent experiments performed in a double-blinded manner. (C) Quantitative analysis of the experiments described in panel B. Three fields were analyzed, and between 50 and 200 cells were counted per field.
KLRG1 inhibits adhesive properties on E-cadherin-expressing cells. (A) Parental or WT E-cadherin cells were stained with KLRG1 tetramer or E-cadherin antibody and analyzed by FACS. (B) Approximately 7.5 × 104 parental or WT E-cadherin–transduced cells were either treated with KLRG1 tetramer at the indicated concentrations or left untreated (1× HEPES) for 24 hours at 37°C. Fibroblast-like adhesive properties were imaged at ×20 with an Olympus DP70. Black arrowheads show fibroblast-like cells, and white arrowheads depict round circular cells. Data are representative of 3 independent experiments performed in a double-blinded manner. (C) Quantitative analysis of the experiments described in panel B. Three fields were analyzed, and between 50 and 200 cells were counted per field.
KLRG1/E-cadherin interaction leads to tyrosine phosphorylation of E-cadherin
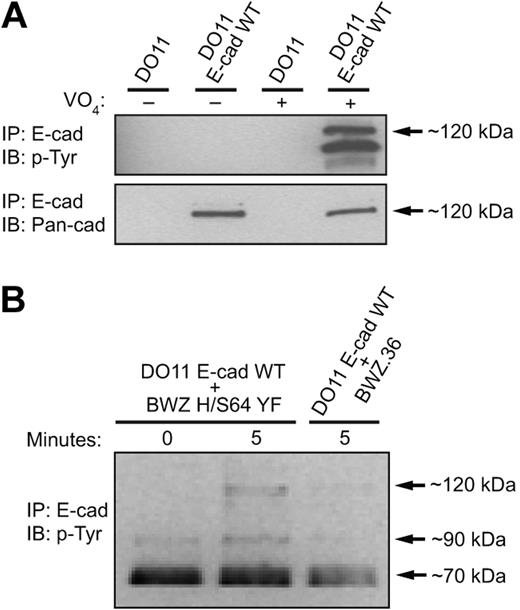
The adhesive function of E-cadherin has been known to be dynamically regulated. Activation of tyrosine kinases in epithelial cells results in the disassembly of E-cadherin–mediated cell-cell adhesions.40,41 We therefore hypothesized that the KLRG1-mediated inhibition of E-cadherin–dependent cell adhesion observed in Figure 3 must result from E-cadherin phosphorylation. To test this hypothesis, we first investigated whether E-cadherin was capable of being tyrosine phosphorylated. E-cadherin was immunoprecipitated from pervanadate-treated cells, and its phosphorylation status was examined by Western blot. Using these conditions, we found that E-cadherin as well as associated proteins are tyrosine phosphorylated (Figure 4A). Next, we examined whether KLRG1 interaction with E-cadherin induces E-cadherin phosphorylation. E-cadherin–transfected cells and parental cell lines were therefore incubated with inactive KLRG1 transfectants (BWZ H/S64 YF) or parental cell lines (BWZ.36) at different time points. We found that after incubation of WT E-cadherin–transduced cells with KLRG1+ target cells for 5 minutes at 37°C, a tyrosine phosphorylated band at approximately 120 kDa was reproducibly seen in E-cadherin immunoprecipates (Figure 4B). This was not observed when the mixture of cells were kept at 4°C or when the parental cell line was incubated with E-cadherin–transduced cells (Figure 4B).
E-cadherin is tyrosine phosphorylated when bound to KLRG1.(A) Parental or WT E-cadherin–transduced cells were treated with pervanadate or left untreated and E-cadherin was immunoprecipitated from lysates. Immunoprecipitated proteins were analyzed by Western blot using the indicated mAbs. (B) E-cadherin transfectants were incubated at different time points with parental or KLRG1 chimeric receptor reporter cells. E-cadherin immunoprecipitated lysates were resolved by SDS-PAGE and transferred to nitrocellulose membrane for Western blot analysis with anti-pY mAb 4G10. The results are representative of at least 5 independent experiments.
E-cadherin is tyrosine phosphorylated when bound to KLRG1.(A) Parental or WT E-cadherin–transduced cells were treated with pervanadate or left untreated and E-cadherin was immunoprecipitated from lysates. Immunoprecipitated proteins were analyzed by Western blot using the indicated mAbs. (B) E-cadherin transfectants were incubated at different time points with parental or KLRG1 chimeric receptor reporter cells. E-cadherin immunoprecipitated lysates were resolved by SDS-PAGE and transferred to nitrocellulose membrane for Western blot analysis with anti-pY mAb 4G10. The results are representative of at least 5 independent experiments.
KLRG1/cadherin interactions regulate both KLRG1 and E-cadherin–expressing cells
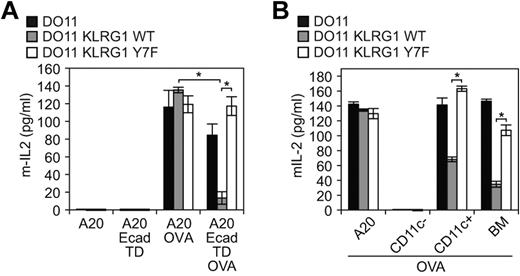
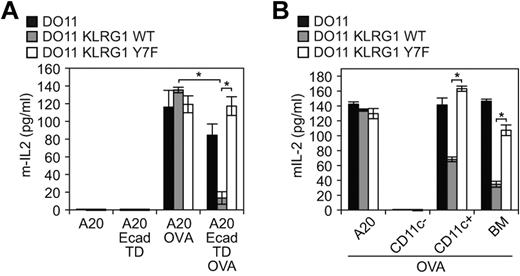
The results described above suggest that cadherin signaling may have had an impact on previous studies that aimed at understanding cadherin effects on the inhibitory properties of KLRG1. We therefore took advantage of the TD E-cadherin that cannot signal to revisit these findings. To elucidate the function of KLRG1 when ligated with cadherins, TD E-cadherin–transduced A20 cells (antigen-presenting cells [APCs]), as well as WT and Y7F mutant KLRG1-transduced T-cell hybridomas, were used. The T-cell hybridoma DO11 used for this analysis is able to recognize peptides processed from ovalbumin (OVA) protein presented by major histocompatibility complex (MHC) class II molecule Iad. When pulsed with OVA, the Iad-positive B cells, A20, present OVA peptide to DO11 cells. The TD E-cadherin–transduced A20 cells simultaneously express Iad and an inactive E-cadherin, allowing us to define specifically the inhibitory properties of KLRG1. Using this system, we found that WT KLRG1-transduced T-cell hybridomas had a significant decrease in IL-2 levels, whereas parental and Y7F KLRG1-transduced T cells exhibited similar levels of IL-2 when incubated with OVA-pulsed E-cadherin–transduced APCs (Figure 5A). Although TD E-cadherin is expressed at a much higher level than WT E-cadherin in A20 cells, it is expressed at a level comparable with endogenous E-cadherin in DCs (supplemental Figures 1-2). To directly study the role of endogenous E-cadherin, we used BMDCs. B6 BMDCs express E-cadherin42 but cannot present OVA to DO11 cells due to MHC restriction. We therefore used B10.D2 (d haplotype) that can present OVA to DO11 cells. We first determined and compared the levels of cadherin expression on BMDCs from B6 and B10.D2 mice. Flow cytometric analysis revealed that CD11c+ BMDCs from B6 and B10.D2 strains of mice express high levels of E-cadherin (supplemental Figure 2A). Because KLRG1 can also bind N-cadherin, we also tested whether BMDCs express N-cadherin. A Western blot analysis demonstrates that BMDCs express E-cadherin but not N-cadherin (supplemental Figure 2B-C). The N-cadherin mAb is functional because N-cadherin was detected by immunoblot on lysates from N-cadherin–transfected cells (supplemental Figure 2B). Notably, R-, P-, and M-cadherin were not detected on BMDCs by Western blot using specific mAbs (data not shown). We then tested the role of E-cadherin expressing BMDCs on regulating the function of KLRG1 on DO11 hybridomas. Consistent with the TD E-cadherin–transduced APCs, BMDC (> 90% CD11c+, supplemental Figure 2A) presentation of OVA to DO11 cells is inhibited only if DO11 cells express WT KLRG1 (Figure 5B). Taken together, these data reveal an important function for E-cadherin on DCs and the predominant role of the ITIM tyrosine in the inhibitory properties of KLRG1.
E-cadherin binding to KLRG1 inhibits T-cell activation. (A) In vitro or (B) ex vivo approach using E-cadherin-expressing cells pulsed with or without ovalbumin and washed before incubation with parental cells or KLRG1 transfectants. Levels of mIL-2 were measured by ELISA. For the in vitro approach, results are representative of at least 5 independent experiments (*P < .001). For the ex vivo approach, results are representative of 4 independent experiments (*P < .005).
E-cadherin binding to KLRG1 inhibits T-cell activation. (A) In vitro or (B) ex vivo approach using E-cadherin-expressing cells pulsed with or without ovalbumin and washed before incubation with parental cells or KLRG1 transfectants. Levels of mIL-2 were measured by ELISA. For the in vitro approach, results are representative of at least 5 independent experiments (*P < .001). For the ex vivo approach, results are representative of 4 independent experiments (*P < .005).
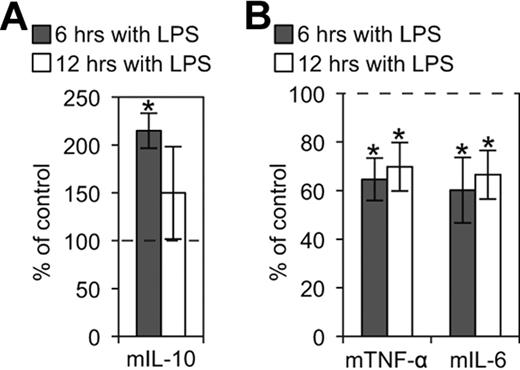
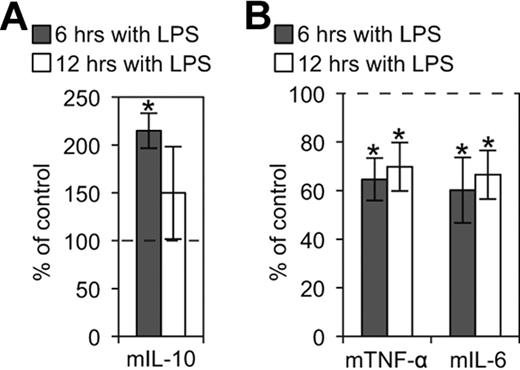
It has been demonstrated that ligation of E-cadherin on Langerhans cells inhibits their maturation,43 whereas disruption of E-cadherin–mediated adhesion induces a functionally distinct DC maturation.42 To determine whether KLRG1 binding to E-cadherin affects DC function, CD11c+ and E-cadherin+ BMDCs were treated with LPS in the presence of KLRG1-expressing cells or controls. We found that KLRG1 and cadherin interaction enhanced release of the anti-inflammatory cytokine IL-10 (Figure 6A), but suppressed the secretion of proinflammatory cytokines IL-6 and TNF-α on DCs at 6 and 12 hours postincubation (Figure 6B). This effect was observed using DO11 KLRG1 transfectants (Figure 6B) and 3T3 KLRG1 transfectants (data not shown). Thus, KLRG1 binding to E-cadherin promotes the up-regulation of anti-inflammatory cytokines and the down-regulation of proinflammatory cytokines.
KLRG1 modulates inflammatory cytokines produced by LPS-matured BMDCs. (A) CBA flex set for the anti-inflammatory cytokine mIL-10 was performed on supernatants from LPS-matured BMDCs incubated with or without the presence of KLRG1-expressing cells. The cytokines produced from these BMDCs in the presence of KLRG1 were then normalized to the cytokines produced from BMDCs in the presence of parental cells (parental cell average of 93 and 357 pg/mL for 6 and 12 hours, respectively). (B) CBA flex sets for the proinflammatory cytokines mTNF-α (parental cell average of 20 000 and 20 708 pg/mL for 6 and 12 hours, respectively) and mIL-6 (parental cell average of 19 874 and 25 834 pg/mL for 6 and 12 hours, respectively) were performed as described in panel A. The results are representative of at least 3 independent experiments (*, P < .05).
KLRG1 modulates inflammatory cytokines produced by LPS-matured BMDCs. (A) CBA flex set for the anti-inflammatory cytokine mIL-10 was performed on supernatants from LPS-matured BMDCs incubated with or without the presence of KLRG1-expressing cells. The cytokines produced from these BMDCs in the presence of KLRG1 were then normalized to the cytokines produced from BMDCs in the presence of parental cells (parental cell average of 93 and 357 pg/mL for 6 and 12 hours, respectively). (B) CBA flex sets for the proinflammatory cytokines mTNF-α (parental cell average of 20 000 and 20 708 pg/mL for 6 and 12 hours, respectively) and mIL-6 (parental cell average of 19 874 and 25 834 pg/mL for 6 and 12 hours, respectively) were performed as described in panel A. The results are representative of at least 3 independent experiments (*, P < .05).
Discussion
Although not known to be a major player to the immune system, E-cadherin has recently been implicated to have an important role on the ability of DCs to control the immune response.42,43 A recent study by Jiang et al demonstrated that maturation of BMDCs was in part due to the loss of E-cadherin contacts,42 yet how E-cadherin contacts could be disrupted in a physiologic setting was not explored. We therefore hypothesized that KLRG1/cadherin interactions must affect both KLRG1 and cadherin-expressing cells. In this report, we found several unexpected consequences of KLRG1/cadherin interactions. We show that the first 2 extracellular domains of cadherin interact with KLRG1 and are sufficient to induce KLRG1 signaling. We also demonstrate that KLRG1 engagement induces cadherin phosphorylation, inhibits cadherin-dependent adhesion, and influences DC secretion of inflammatory cytokines, thereby exerting immunosuppressive effects.
The KLRG1 binding site on cadherin (EC1-2) is also critical for binding to another cadherin ligand the integrin αE(CD103)β7.44-47 This raises the question of how these molecules interact with cadherin if expressed on the same cells and what are the consequences of coexpression of these molecules. Although it is unknown whether KLRG1 and integrin αE(CD103)β7 are coexpressed on the same cell, there is indirect evidence that they may be on some T-cell subsets. KLRG1 is expressed on short-lived effector mouse CD8+ T cells25 and on FoxP3+ regulatory T cells,29,30 as well as on some human central memory CD4+ T lymphocytes,48 whereas integrin αE(CD103)β7 appears to be expressed on some CD8+ T cells,49-52 as well as FoxP3+ regulatory cells.29,53,54 Therefore, these 2 cadherin ligands are likely to be coexpressed on some CD8+ T cells and Treg cells. Note, however, that E-cadherin homophilic interactions do not exclude simultaneous interaction with integrin αE(CD103)β7.46 In contrast, KLRG1 binding to E-cadherin apparently competes with homophilic E-cadherin interactions,36 suggesting that KLRG1 and integrin αE(CD103)β7 do not bind to the same binding site. The function of integrin αE(CD103)β7 is not clear but it can act as an accessory molecule that strengthens positive T-cell signaling.52 Since the T-cell activation pathway regulates the strength of interaction between E-cadherin and integrin αE(CD103)β7, it is possible that KLRG1 inhibits integrin αE(CD103)β7 activation. In this situation, KLRG1 would control integrin αE(CD103)β7-expressing lymphocyte migration and functions. Further analysis will be required to determine the exact binding affinity and binding site of KLRG1 and integrin αE(CD103)β7 to cadherin to define the molecular basis for their interaction.
In addition to their role in cell adhesion, cadherins can function as signal-transducing molecules.55 Cadherin cytoplasmic tail contains many serine, threonine, and tyrosine residues that are putative phosphorylation sites. We report herein that KLRG1 binding to cadherin induces cadherin tyrosine phosphorylation. The relatively weak signal obtained could be explained by the experimental approach, which entails biochemical analysis after engagement of natural ligands and receptors in the absence of antibodies. Natural ligands to receptors are weaker in transmitting signals compared with artificial cross-linking of receptors with antibodies because of the technical challenges arising from the lack of synchronization between the population of cells engaged in synapse formation.56 As a likely consequence of cadherin tyrosine phosphorylation, we observed a decreased number of adhesive cadherin-expressing cells when they are incubated with KLRG1. In addition, it has been shown that E-cadherin tyrosine phosphorylation leads to cadherin ubiquitination and subsequent degradation in lysosomes.57,58 It is therefore tempting to speculate that KLRG1-induced cadherin phosphorylation will result in cadherin down-regulation. In support of this idea, we noticed that KLRG1 induces a slight reduction of cadherin cell surface expression when KLRG1+ cells and E-cadherin+ cells are incubated for 12 hours (supplemental Figure 3), providing a possible explanation for the observed cell adhesion decrease. Future studies will shed light on the molecular and cellular regulation of cadherin during its interaction with KLRG1.
We initially developed a system that would allow us to have cadherin and an antigen-presenting molecule expressed on the same cells. While we were studying this pathway, we uncovered an interesting phenotype of cadherin cell surface expression. We found that cadherin cell surface expression is dependent on its cytoplasmic tail (TD > WT > JMD), demonstrating the importance of cellular trafficking for cadherin cell surface expression. Importantly, WT E-cadherin cell surface expression is variable and depends on which cell subset it is expressed. For instance, E-cadherin cell surface expression is much higher on NIH 3T3 cells than on A20 cells (despite comparable level of the GFP reporter), most likely reflecting cell line–specific trafficking properties. Expression variability is not unique to KLRG1 ligands. Recently, Raulet and colleagues reported that expression of Mult1 protein, a ligand for the activating receptor NKG2D, is controlled by a mechanism dependent on cytoplasmic lysine residues that is associated with polyubiquitination of the protein.59 Mult1 degradation and ubiquitination is reduced in response to stress resulting in increased cell surface expression.59 Importantly, the Mult1 di-lysine motif is located at the same position within the juxtamembrane region as a di-leucine motif on E-cadherin, which is required for E-cadherin sorting and trafficking60,61 (and data not shown). It is therefore tempting to speculate that under certain conditions such as stress or viral infections E-cadherin expression may also be affected, resulting in increased NK and/or CD8+ T-cell responses. In support of this possibility, repressed E-cadherin expression has been observed in tumorigenesis and metastasis as well as in viral infections.62-64
Previous work from others and from us has revealed some inhibitory functions and signaling pathways of KLRG1.7-9,20,31,65 Upon activation, KLRG1 recruits SHIP-1 and SHP-2 phosphatases to presumably block T- and NK-cell activation. Although the ITIM tyrosine is essential for the recruitment of these phosphatases, there have been some discrepancies regarding the exact contribution of the KLRG1 tyrosine when KLRG1 was cross-linked with an anti-KLRG1 mAb.9,65 Using a well-controlled system, which minimizes the role of cadherin “reverse” signaling, we revisited these findings. Although we cannot exclude a potential role for serine, threonine, or other residues that are localized in the KLRG1 cytoplasmic tail, our data demonstrate the predominant role of the ITIM tyrosine in the inhibitory properties of KLRG1 when engaged by its ligands. In addition, using DCs expressing physiologic cadherin expression levels, we confirmed the ability of endogenous cadherin to inhibit T-cell functions. Importantly, we found that the KLRG1/cadherin interaction is bidirectional, as it cannot only inhibit T-cell cytokine production in a KLRG1 ITIM-dependent manner but also exerts immunosuppressive effects by modulating the cytokine production of DCs in a cadherin-dependent fashion. These data reveal a previously unappreciated role for KLRG1/cadherin interaction and demonstrate that KLRG1 regulates DC function.
Bidirectional communication that allows pairs of coreceptors on adjacent cells to engage in a crosstalk by reciprocally acting as ligands and receptors has been described in embryonic development66 but has been relatively overlooked for immune receptors besides SIRP-α67 and T cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT).68 Interestingly, TIGIT, an immune receptor found on T and NK cells, has been shown to enhance the production of IL-10 and diminish the production of IL-12p40 when it engages its ligand on DCs.68 It is likely that other inhibitory receptors besides KLRG1 and TIGIT will have similar functions. Altogether this illustrates the complexity of the immune system and emphasizes the role of NK-cell/DC crosstalk in the coordination of innate and adaptive immune responses.69 Further insights on KLRG1 and cadherins are needed to fully understand the interplay between the immune response against developing tumors and viral infections as well as tissue organization and integrity. Therefore, the heterophilic interaction of E-cadherin with KLRG1 warrants future investigation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr L. Shapiro for reagents, S. Terrizzi for technical support and cell sorting, and Dr M. Tessmer for helpful discussions. We also thank Vinh Nguyen and Dr Robbins for critical reading of the manuscript.
This work was supported by a National Institutes of Health (NIH) research grant (AI58181) to L.B. and by an NIH National Research Service Award F31 grant (AI080230) to C.B.
National Institutes of Health
Authorship
Contribution: C.B. and L.B. designed and performed experiments, analyzed the data, and wrote the paper; and C.F. performed and analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laurent Brossay, Department of Molecular Microbiology and Immunology, Division of Biology and Medicine, Box G-B597, Brown University, Providence, RI 02912; e-mail: Laurent_brossay@brown.edu