Hedgehog (Hh) ligands bind to the Patched1 (Ptch1) receptor, relieving repression of Smoothened, which leads to activation of the Hh signaling pathway. Using conditional Ptch1 knockout mice, the aim of this study was to determine the effects of activating the Hh signaling pathway in hematopoiesis. Surprisingly, hematopoietic-specific deletion of Ptch1 did not lead to activation of the Hh signaling pathway and, consequently, had no phenotypic effect. In contrast, deletion of Ptch1 in nonhematopoietic cells produced 2 distinct hematopoietic phenotypes. First, activation of Hh signaling in epithelial cells led to apoptosis of lymphoid progenitors associated with markedly elevated levels of circulating thymic stromal lymphopoietin. Second, activation of Hh signaling in the bone marrow cell niche led to increased numbers of lineage-negative c-kit+ Sca-1+ bone marrow cells and mobilization of myeloid progenitors associated with a marked loss of osteoblasts. Thus, deletion of Ptch1 leads to hematopoietic effects by distinct cell-extrinsic mechanisms rather than by direct activation of the Hh signaling pathway in hematopoietic cells. These findings have important implications for therapeutics designed to activate the Hh signaling pathway in hematopoietic cells including hematopoietic stem cells.
Introduction
In vertebrates, the Hedgehog (Hh) signaling pathway is regulated by 3 Hh ligands: Sonic hedgehog (Shh), Indian hedgehog (Ihh), and Desert hedgehog. In the absence of Hh ligands, the transmembrane receptor Patched1 (Ptch1) inhibits the activity of a second receptor known as Smoothened (Smo). Binding of Hh ligands to Ptch1 relieves the repression on Smo, thus allowing pathway activation to proceed by nuclear translocation of activator and repressor forms of the target-gene Gli family of zinc finger transcription factors.1
The Hh pathway is important for the development of numerous cell lineages, including hematopoiesis. In zebrafish, Hh signaling is required for the development of definitive hematopoiesis while in Drosophila, Hh signaling maintains hematopoietic stem cell (HSC) quiescence.2,3 However, effective hematopoietic reconstitution by Smo−/− fetal liver cells suggests that Hh signaling is not required for maintenance of adult HSCs.4 In addition to a role in HSCs, studies of Shh-null and conditional Smo-null mice suggest Hh signaling is required for normal T-cell development.5,6
The effects of increased Hh signaling on hematopoiesis are not well defined. Increased Hh signaling by loss of function mutations of Ptch1 lead to increased proliferation of stem or progenitor cells of the skin and brain, ultimately leading to basal cell carcinomas and medulloblastomas, respectively.7,8 A similar expansion of HSCs or progenitors might also occur by activating the Hh pathway, raising both therapeutic and cancer stem cell implications.9 However, experimental models designed to address the effects of increased Hh signaling in hematopoiesis have been hampered by poor in vitro and in vivo bioavailability of Hh ligands. Incubation of primitive hematopoietic progenitors with Hh ligands, either alone or on stromal feeders, increases the numbers of nonobese diabetic–severe combined immunodeficiency (NOD/SCID) repopulating cells, although this could be an indirect effect.10,11 An alternate method of studying the effects of activating the Hh pathway is through genetic deletion of Ptch1. Whereas Ptch1-null embryos die before the development of definitive hematopoiesis,12 germline Ptch1-heterozygous mice have increased numbers of HSC-enriched, lineage-negative c-kit+Sca-1+ (LKS) bone marrow (BM) cells due to increased cell cycling.13 Noncompetitive transplant assays of Ptch1-heterozgous BM cells into NOD/SCID mice suggested that this aberrant cell cycling improved early (5 weeks) repopulating capacity at the expense of longer-term (8 weeks) repopulating capacity.13 Uhmann et al reported a conditional deletion of Ptch1 in adult hematopoiesis using a ubiquitous Cre under the control of tamoxifen.14 Homozygous deletion of Ptch1 led to increased numbers of LKS, but functional assays were not performed. In this model, deletion of Ptch1 also led to loss of B and T cells, although transplant assays into Rag2−/− γ−/− mice suggested that these lymphoid defects were cell extrinsic.14
Using inducible and tissue-specific deletion of Ptch1 on a congenic C57BL/6J background, the aim of this study was to determine the cell-intrinsic and cell-extrinsic hematopoietic effects of activation of the Hh signaling pathway. Surprisingly, we found that deletion of Ptch1 in hematopoietic cells did not activate the Hh signaling pathway, and consequently had no phenotypic effects. In contrast, activation of the Hh signaling pathway by deletion of Ptch1 in epithelial cells led to apoptosis of lymphoid progenitors, whereas deletion of Ptch1 in the BM cell niche led to increased numbers of LKS and circulating myeloid progenitors. High levels of circulating thymic stromal lymphopoietin (TSLP) and loss of osteoblasts may explain these lymphoid and myeloid progenitor cell defects, respectively. These findings have important implications for therapeutics designed to activate the Hh signaling pathway in hematopoietic cells, including HSCs.
Methods
Generation of Ptch1-null mice
Ptch1fl mice15 were backcrossed onto a C57BL/6J background for 10 generations. Congenic Ptch1fl mice were bred with transgenic mice expressing Cre recombinase under the control of the interferon-inducible promoter, Mx1.16 Ptch1fl/fl, MxPtch1+/fl, and MxPtch1fl/fl mice were injected with 300 μg polyinosinic-polycytidylic acid (poly(I:C); Sigma-Aldrich) dissolved in normal saline intraperitoneally on alternate days for 3 doses. Deletion of Ptch1 in HSCs was achieved by breeding Ptch1fl mice with transgenic mice expressing the tamoxifen-inducible Cre-ER recombinase under the control of the stem cell leukemia (Scl) stem cell enhancer (HSC-Scl-CreER).17 For inducing Cre-ER recombinase, SclPtch1 mice were treated with 3 intraperitoneal injections of 10 mg tamoxifen (Sigma-Aldrich) diluted in corn oil (50 mg/mL). CD19Cre mice18 and LckCre mice19 were backcrossed onto the C57BL/6 background. Deletion of Ptch1 in epithelial cells was achieved by crossing Ptch1fl mice with Keratin 14 (K14) Cre transgenics.20 Female CD45.1 (B6.SJL(Ptprca[Ly5.1])) mice were purchased from the Walter and Eliza Hall Institute for Medical Research (WEHI) Animal House Facility. Melbourne Health and the University of Melbourne animal ethics committees approved all animal experiments. Deletion of the Ptch1fl allele was confirmed by either Southern blot or polymerase chain reaction (PCR). For Southern blot analysis, genomic DNA was digested by either XbaI or BamHI and then probed with a SacI/NsiI fragment from a 3-kb SpeI fragment of the second intron of Ptch1.15 Primers used for PCR genotyping are shown in supplemental Table 1, available on the Blood website (see the Supplemental Materials link at the top of the online article).
Sample preparation, histology, and immunohistochemistry
All analyses were performed at least 4 weeks after poly(I:C) administration to avoid the effects of the interferon response. For whole blood counts, 250 μL of blood was collected from the retro-orbital plexus into tubes containing potassium EDTA (Sarstedt), and blood counts analyzed using an Advia 2120 automated hematologic analyzer (Bayer). BM cells were harvested by flushing femurs with mouse tonicity (MT)–PBS/2% fetal bovine serum (FBS). Thymi and spleens were filtered through 45-μm nylon screens to generate single-cell suspensions. Histologic analyses were done on paraffin-embedded skin fixed with 10% neutral buffered formalin. All sections were stained with hematoxylin and eosin (H&E). Immunohistochemistry for Keratin 14 (anti–Keratin 14 [AF64]; PRB-155P; Covance Research Products) was performed using standard immunohistochemistry (IHC) techniques. Images were acquired using a Nikon Eclipse 80i microscope equipped with a 40×/0.75 NA objective lens. Images were captured with a Nikon DXM1200C digital camera, and figures were created without manipulation in Adobe Photoshop 10.0.1 (Adobe Systems).
Flow cytometric analysis and sorting
Antibodies used for analyses were obtained from BD Pharmingen: CD8a (clone 53-6.7), CD45.2 (104), annexin V, B220 (RA3-6B2), IgM (AF6-78), and Sca-1 (E13-161-7) as fluoroscein isothiocyanate (FITC) conjugates; CD4 (GK1.5), Mac-1a (M1/70), CD19 (1D3), CD21 (7G6), CD45.1 (A20), and c-kit (2B8) as phycoerythrin (PE) conjugates; B220 and c-kit as allophycocyanin (APC) conjugates; and biotinylated CD3e (145-2C11), CD4, CD8, CD23 (B3B4), CD11b, B220, Gr-1 (RB6-8C5), and TER-119. Second-stage fluorescent reagents were either SAv-APC or SAv-PerCP-Cy5.5. The appropriate conjugated rat anti–mouse isotypes were used as negative controls. Viability was determined by exclusion of propidium iodide (Sigma-Aldrich). For cell-cycle analysis, sorted cells were fixed in 70% ethanol, washed with PBS, and stained with ice-cold 2 mg/mL Hoechst 33342 (Molecular Probes) and 4 mg/mL Pyronin Y (Polysciences) for 20 minutes before analysis with a BD LSR Benchtop flow cytometer (BD Biosciences). For isolation of LKS and lineage-negative c-Kit+Sca-1− (LK) cell fractions, cell sorting of immature BM cells after immunomagnetic bead depletion of lineage marker–positive BM cells was performed before sorting on a FACSVantage SE system (BD Biosciences). The purity of sorted cells was determined by reanalyzing a small sample of the collected cells and was 80% to 90%. All data were analyzed using a WEHI in-house program, Weasel version 2.3.
Transplant assays
Competitive repopulation assays were performed as previously described.21 Female CD45.1 mice were used as recipients and as a source of competitor BM cells (2 × 106). Donor BM cells (2 × 106) were obtained from MxPtch1Δ/Δ, MxPtch1Δ/+, or Ptch1fl/fl mice 6 to 8 weeks after poly(I:C). Each transplant inoculum was injected into a minimum of 3 recipients. Secondary recipients received 107 BM cells from primary recipients.
Real-time PCR
RNA from sorted cells was isolated using TRIZol (Invitrogen), and genomic DNA was removed using Turbo DNA-free kit (Ambion). Reverse transcription of 2 μg DNA-free RNA into cDNA was carried out with the Omniscript Reverse Transcription Kit (QIAGEN). Real-time quantitative (Q)–PCR 20-μL reactions each contained 1 μL cDNA, the fluorescent DNA binding dye SYBR green (Molecular Probes), and specific primers (supplemental Table 1) were used for all hematopoietic tissues. Amplification was performed in a RotorGene 2000 PCR instrument (Corbett Research) at 95°C for 5 minutes followed by 40 cycles of 15 seconds at 94°C, 30 seconds at 60°C, and 30 seconds at 72°C, and 30 seconds at 82°C. SYBR Green dye intensity was analyzed using the RotorGene 200 software, and all Ct values were normalized to HPRT. For expression of TSLP and Gli1 in skin and whole-bone samples, Taqman MGB probes and FAM dye-labeled Expression Systems were used (Applied Biosystems). Preincubation was performed in a Roche LightCycler 480 II (Roche) at 50°C for 2 minutes followed by 20 seconds at 95°C. Amplification was performed at 95°C for 3 seconds followed by 30 seconds at 60°C for 45 cycles before cooling at 40°C for 30 seconds.
Progenitor assays
BM, spleen, and peripheral blood cells were cultured in 0.3% agar cultures and analyzed as previously described.22 Recombinant cytokines were used at the following concentrations: 10 ng/mL mIL-3, 50 ng/mL mIL-6, and 50 ng/mL rSCF (R&D Systems).
Cell culture
Mouse mesodermal cells derived from 14- to 17-day-old embryos, C3H/10T1/2 (C3H), were obtained from ATCC. Cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% (vol/vol) FBS and 100 U/mL penicillin/streptomycin and were cultured in 5% CO2 and 95% air. For experiments, cells were seeded into 12-well plates at 70 000 cells/well and allowed to attach overnight. The next day, Shh-conditioned medium was added at a 1:5 dilution with 5% FCS DMEM and incubated on the cells for 3 days. Cells were then harvested for RNA using TRIZol.
TSLP immunoassay
Serum samples were collected from kill bleeds of mice. Serum TSLP levels were measured using the Quantikine mouse TSLP immunoassay (catalog no. MTLP00; R&D Systems).
Results
Deletion of Ptch1 in adult mice leads to death of B- and T-cell progenitors
To delete Ptch1 in adult hematopoiesis, we intercrossed MxCre transgenic mice with animals carrying a conditional “floxed” (fl) allele of Ptch1 (Ptch1fl).15 Deletion of the Ptch1fl allele (Ptch1Δ) early in development is embryonic lethal and phenocopies the germline Ptch1-null mice.12,15 Ptch1-null (MxPtch1Δ/Δ) hematopoiesis was generated by treating MxPtch1fl/fl mice with poly(I:C). Control animals, also treated with poly(I:C), included Ptch1-wild-type mice (Ptch1fl/fl) and Ptch1-heterozygous mice (MxPtch1+/Δ). Deletion of one Ptch1 allele had no effect on hematopoiesis and therefore, in some experiments, MxPtch1+/Δ mice were used as controls. Southern blot analysis of hematopoietic organs confirmed more than 90% deletion of the targeted Ptch1fl allele (Figure 1A).
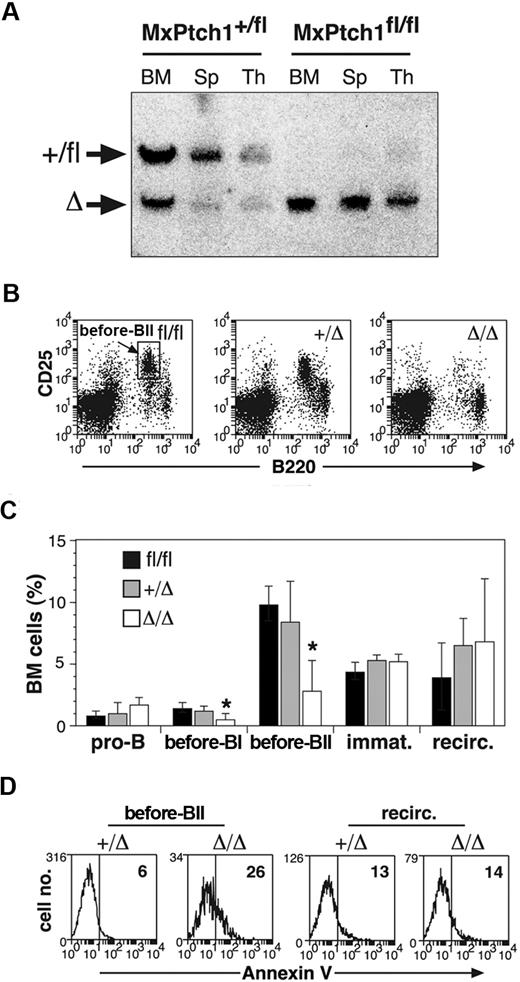
B-cell defects in the absence of Ptch1. (A) Southern blot analysis for Ptch1 deletion status. Bone marrow (BM), spleen (Sp), and thymus (Thy) from MxPtch1+/fl and MxPtch1fl/fl mice harvested 4 weeks after administration of poly(I:C). Wild-type Ptch1 allele (+) and loxP-flanked Ptch1 allele (fl) have the same size band. Deleted Ptch1fl allele (Δ). (B) Dot plots of BM cells stained with B220 and CD25. The MxPtch1Δ/Δ dot-plot is an example of a severely affected animal. (C) B-cell subsets in BM from Ptch1-wild-type (Ptch1fl/fl), heterozygous (MxPtch1+/Δ) and null (MxPtch1Δ/Δ) mice: see supplemental Figure 1 for definitions of B-cell subsets. Mean plus SD calculated from 5 mice of each genotype. *P < .05 by one-way ANOVA. (D) Annexin V expression on pre-BII and recirculating B cells within the BM. The mean percentage of annexin V+ cells calculated from 3 mice for each genotype is shown.
B-cell defects in the absence of Ptch1. (A) Southern blot analysis for Ptch1 deletion status. Bone marrow (BM), spleen (Sp), and thymus (Thy) from MxPtch1+/fl and MxPtch1fl/fl mice harvested 4 weeks after administration of poly(I:C). Wild-type Ptch1 allele (+) and loxP-flanked Ptch1 allele (fl) have the same size band. Deleted Ptch1fl allele (Δ). (B) Dot plots of BM cells stained with B220 and CD25. The MxPtch1Δ/Δ dot-plot is an example of a severely affected animal. (C) B-cell subsets in BM from Ptch1-wild-type (Ptch1fl/fl), heterozygous (MxPtch1+/Δ) and null (MxPtch1Δ/Δ) mice: see supplemental Figure 1 for definitions of B-cell subsets. Mean plus SD calculated from 5 mice of each genotype. *P < .05 by one-way ANOVA. (D) Annexin V expression on pre-BII and recirculating B cells within the BM. The mean percentage of annexin V+ cells calculated from 3 mice for each genotype is shown.
Hematopoiesis of MxPtch1Δ/Δ mice was examined 4 to 6 weeks after poly(I:C), a time-point after resolution of the interferon effects from poly(I:C). Analysis of peripheral blood counts of MxPtch1Δ/Δ mice revealed a significant increase in neutrophils but no effect on hemoglobin, lymphocyte count, or platelet count (Table 1). The cell surface markers on B220+ B cells of the BM and spleen used to enumerate B-cell subsets are shown in supplemental Figure 1A. Although numbers of pro-B cells (B220loCD19−) were normal, MxPtch1Δ/Δ mice had a 4-fold loss of pre-BI cells (B220loc-kit+) and pre-BII cells (B220loCD25+) in the BM (Figure 1B-C). Despite loss of pre-B cells, deletion of Ptch1 did not significantly affect the numbers of immature (B220loIgM+) and recirculating (B220hiIgD+) B cells within the BM. B-cell subset analysis of the spleen revealed a 3-fold reduction in the proportion of follicular B cells (supplemental Figure 1B). However, absolute numbers of follicular B cells were normal because the spleen size of MxPtch1Δ/Δ mice was increased 3-fold due to increased numbers of myeloid cells (supplemental Figure 1C). The loss of BM pre-B cells was due to increased cell death, as this population, but not recirculating B cells, displayed increased expression of annexin V (Figure 1D). In contrast, Ptch1 was not required for cell-cycle control or maturation of pre-B cells, as determined by Hoechst staining (supplemental Figure 1D) and in vivo BrdU labeling (supplemental Figure 1E), respectively. Thus, deletion of Ptch1 by the MxCre transgene leads to death of BM pre-B cells.
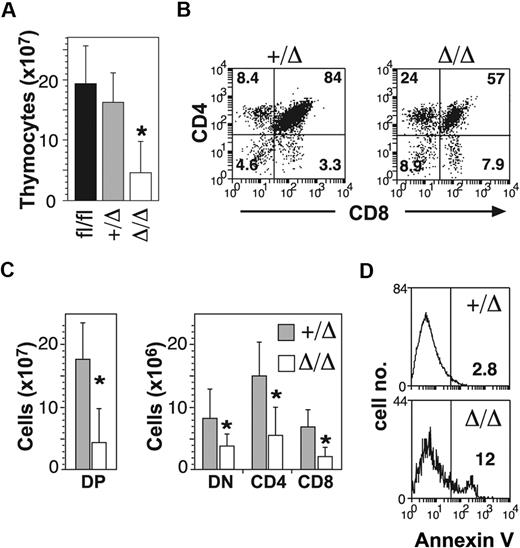
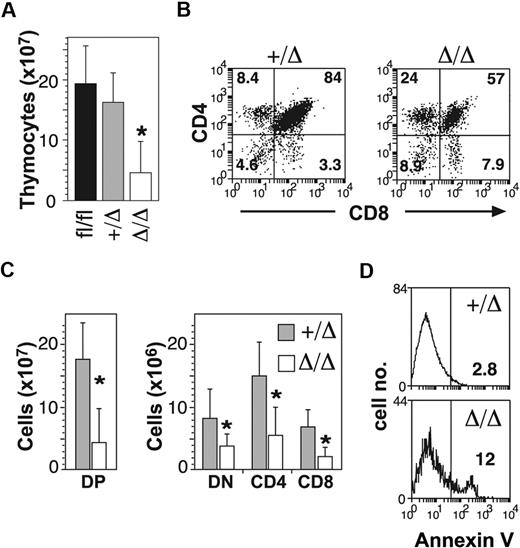
In addition to B-cell defects, MxPtch1Δ/Δ mice developed thymic atrophy (Figure 2A). Flow cytometry of thymic T-cell subsets demonstrated significant loss of the CD4+CD8+ double-positive (DP) cells (Figure 2B). Although the proportions of the other subsets (double negative [DN] and single positive) were relatively increased, the absolute number of all T-cell subsets was significantly reduced in the MxPtch1Δ/Δ thymi (Figure 2C). The proportions of DN subsets were unaffected by deletion of Ptch1 (supplemental Figure 2). Similar to the B-cell phenotype, increased expression of annexinV+ on DP cells indicated increased apoptosis (Figure 2D). Thus, deletion of Ptch1 by the MxCre transgene leads to death of thymocytes, especially DP cells.
T-cell defects in the absence of Ptch1. (A) Total numbers of thymocytes (mean plus SD calculated from 8 mice of each genotype). *P < .05 by one-way ANOVA. (B) Representative dot-plots of T-cell subsets. The mean percentage shown for each subset was calculated from 8 mice of each genotype. (C) Total numbers of T-cell subsets: CD4−CD8− double negative (DN); CD4+CD8+ double positive (DP); CD4+ single positive (CD4); CD8+ single positive (CD8). Mean plus SD calculated from 8 mice of each genotype. *P < .05 by 2-tailed Mann-Whitney test. (D) Annexin V expression on DP thymocytes. The mean percentage of annexin V+ calculated from 3 mice for each genotype is shown.
T-cell defects in the absence of Ptch1. (A) Total numbers of thymocytes (mean plus SD calculated from 8 mice of each genotype). *P < .05 by one-way ANOVA. (B) Representative dot-plots of T-cell subsets. The mean percentage shown for each subset was calculated from 8 mice of each genotype. (C) Total numbers of T-cell subsets: CD4−CD8− double negative (DN); CD4+CD8+ double positive (DP); CD4+ single positive (CD4); CD8+ single positive (CD8). Mean plus SD calculated from 8 mice of each genotype. *P < .05 by 2-tailed Mann-Whitney test. (D) Annexin V expression on DP thymocytes. The mean percentage of annexin V+ calculated from 3 mice for each genotype is shown.
Deletion of Ptch1 promotes cell cycle and mobilization of immature myeloid progenitors
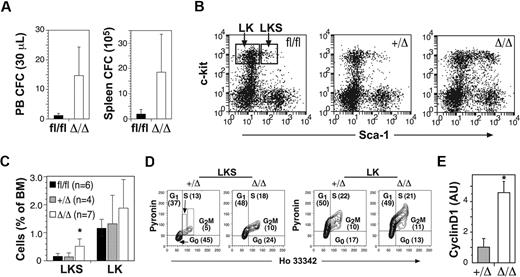
To examine the effects of Ptch1 deletion on myelopoiesis, we measured myeloid progenitors within the BM, spleen, and peripheral blood of MxPtch1Δ/Δ mice. There was no change in either the numbers or types of myeloid progenitors within the BM (Table 2). However, the spleen and peripheral blood of MxPtch1Δ/Δ mice contained at least 10-fold more granulocyte-macrophage progenitors (Figure 3A). To assess the consequences of Ptch1 deletion on HSCs, we enumerated early progenitor populations by flow cytometry. MxPtch1Δ/Δ mice had a 3-fold increase in numbers of LKS cells, a fraction enriched for HSCs and multipotent myeloid progenitors (Figure 3B-C). However, the numbers of LK cells, a cell fraction containing more mature myeloid-restricted progenitors, was not significantly increased by deletion of Ptch1, consistent with normal numbers of BM myeloid progenitors formed in 7-day culture assays.
Increased cell cycle and mobilization of multipotent progenitors in the absence of Ptch1. (A) Numbers of circulating peripheral blood and spleen myeloid colony-forming cells in Ptch1 wild-type (Ptch1fl/fl) and null (MxPtch1Δ/Δ) mice 4 to 6 weeks after poly(I:C). Mean plus SD calculated from 3 mice of each genotype. (B) Representative dot-plots for c-kit and Sca-1 expression on lineage-negative BM cells. Gates used to calculate proportions of LKS and LK cells are shown. (C) Proportions of LKS and LK cells expressed as a percentage of nucleated BM cells. The numbers of mice used to calculate the mean plus SD are shown in parentheses. *P < .05 by one-way ANOVA. (D) Cell-cycle analysis of FACS-isolated LKS and LK cells stained with Hoechst 33342 and Pyronin Y. The mean percentage of each stage of cell cycle calculated from 4 mice is shown. (E) Quantitative RT-PCR for cyclin D1 mRNA expression in LKS. The control samples have been normalized to 1, and the mean plus SD from 3 mice of each genotype are shown. *P < .05 by 2-tailed Mann-Whitney test.
Increased cell cycle and mobilization of multipotent progenitors in the absence of Ptch1. (A) Numbers of circulating peripheral blood and spleen myeloid colony-forming cells in Ptch1 wild-type (Ptch1fl/fl) and null (MxPtch1Δ/Δ) mice 4 to 6 weeks after poly(I:C). Mean plus SD calculated from 3 mice of each genotype. (B) Representative dot-plots for c-kit and Sca-1 expression on lineage-negative BM cells. Gates used to calculate proportions of LKS and LK cells are shown. (C) Proportions of LKS and LK cells expressed as a percentage of nucleated BM cells. The numbers of mice used to calculate the mean plus SD are shown in parentheses. *P < .05 by one-way ANOVA. (D) Cell-cycle analysis of FACS-isolated LKS and LK cells stained with Hoechst 33342 and Pyronin Y. The mean percentage of each stage of cell cycle calculated from 4 mice is shown. (E) Quantitative RT-PCR for cyclin D1 mRNA expression in LKS. The control samples have been normalized to 1, and the mean plus SD from 3 mice of each genotype are shown. *P < .05 by 2-tailed Mann-Whitney test.
As deletion of Ptch1 promotes cycling of cerebellar neuronal precursors,23 we examined the cell-cycle status of sorted LKS and LK subsets by staining with Hoechst and Pyronin dyes (Figure 3D). This analysis revealed a 2-fold increase in the number of cycling MxPtch1Δ/Δ LKS cells compared with MxPtch1+/Δ LKS, and a corresponding reduction in quiescent (G0) cells. In contrast, deletion of Ptch1 had no effect on the cell-cycle status of the more mature LK cell fraction. The increased cycling of MxPtch1Δ/Δ LKS cells was associated with a 4-fold increase in expression of the major D-type cyclin expressed in LKS, cyclin D1 (Figure 3E). Overall, these results show that homozygous deletion of Ptch1 by the MxCre transgene led to increased numbers of proliferating LKS cells and mobilization of myeloid progenitors.
Deletion of Ptch1 does not affect HSC activity
To address the functional consequences of deleting Ptch1 on HSC activity, we performed competitive repopulation transplantation experiments. Donor mice (MxPtch1fl/fl, MxPtch1fl/+, Ptch1fl/fl) were treated with poly(I:C) 6 weeks before transplantation. Competitor and recipient BM cells were distinguished from donor cells by the CD45.1 congenic marker. Despite increased numbers of LKS, homozygous deletion of Ptch1 in the adult had no effect on short-term (4 weeks) repopulating activity of peripheral blood leukocytes (Figure 4A). Furthermore, analysis of BM repopulation 16 weeks after transplantation did not reveal any change in long-term HSC activity (Figure 4B). Secondary transplantation of donor MxPtch1Δ/Δ HSCs was also normal, indicating that Ptch1 does not regulate self-renewal of HSCs (Figure 4C). Thus, deletion of Ptch1 in HSCs had no significant effect on HSC activity despite the altered cycling status of LKS cells.
Competitive repopulation assays of Ptch1-null hematopoietic stem cells. (A) Donor leukocyte contribution in peripheral blood of recipient mice 4 weeks after transplantation with a 1:1 ratio of donor (Ptchfl/fl, MxPtch1+/Δ, or MxPtch1Δ/Δ) and wild-type CD45.1 competitor BM cells. Mean plus SD calculated from 4 recipients of each donor genotype. (B) Donor contribution 16 weeks after transplantation. Mice were killed to determine donor contribution to the myeloid lineage (Mac-1+ BM cells), B-cell lineage (B220+ BM cells), and T-cell lineage (CD4+CD8+ DP thymocytes). Mean plus SD calculated from 4 recipients of each donor genotype. (C) Donor contribution to hematopoiesis of secondary recipients that received transplants of BM cells from primary recipients shown in panel B. Mice were analyzed 12 weeks after transplantation. Mean plus SD calculated from 4 recipients of each donor genotype.
Competitive repopulation assays of Ptch1-null hematopoietic stem cells. (A) Donor leukocyte contribution in peripheral blood of recipient mice 4 weeks after transplantation with a 1:1 ratio of donor (Ptchfl/fl, MxPtch1+/Δ, or MxPtch1Δ/Δ) and wild-type CD45.1 competitor BM cells. Mean plus SD calculated from 4 recipients of each donor genotype. (B) Donor contribution 16 weeks after transplantation. Mice were killed to determine donor contribution to the myeloid lineage (Mac-1+ BM cells), B-cell lineage (B220+ BM cells), and T-cell lineage (CD4+CD8+ DP thymocytes). Mean plus SD calculated from 4 recipients of each donor genotype. (C) Donor contribution to hematopoiesis of secondary recipients that received transplants of BM cells from primary recipients shown in panel B. Mice were analyzed 12 weeks after transplantation. Mean plus SD calculated from 4 recipients of each donor genotype.
Hematopoietic-specific deletion of Ptch1 demonstrates redundancy
Although the MxCre transgene efficiently deletes “floxed” genes in HSCs, it can also be induced in a wide range of nonhematopoietic cell types, including the liver, brain, and endothelial cells.16,24 To examine the effects of deleting Ptch1 specifically in hematopoietic cells, we used transgenic mice carrying the tamoxifen-inducible Cre-ER recombinase under the control of the SCL stem cell enhancer [HSC-SCL-Cre-ER(T)].17 Genomic Southern blot analysis demonstrated more than 90% deletion of the Ptch1fl allele in BM cells (Figure 5A). Despite this, SclPtch1Δ/Δ mice displayed normal numbers of LKS cells (Figure 5B), suggesting that the increased LKS cells seen in MxPtch1 mice was not cell intrinsic. To achieve B cell–specific deletion of Ptch1, we used CD19Cre transgenic mice, which constitutively express Cre from the pre-B cell stage.18 PCR genotyping confirmed more than 80% deletion of Ptch1 in BM pre-BII cells (Figure 5C). Consistent with a cell-extrinsic B-cell defect in MxPtch1Δ/Δ mice, CD19Ptch1Δ/Δ mice had normal numbers of pre-B cells at 6 months of age (Figure 5D). To examine the effects of deleting Ptch1 specifically in T cells, we used the LckCre transgenic mice.19 Despite highly efficient deletion of Ptch1 in the thymus of LckPtch1Δ/Δ mice, these animals exhibited normal thymic cellularity (Figure 5E) and no change in the proportion of T-cell subsets at 6 months of age (Figure 5F).
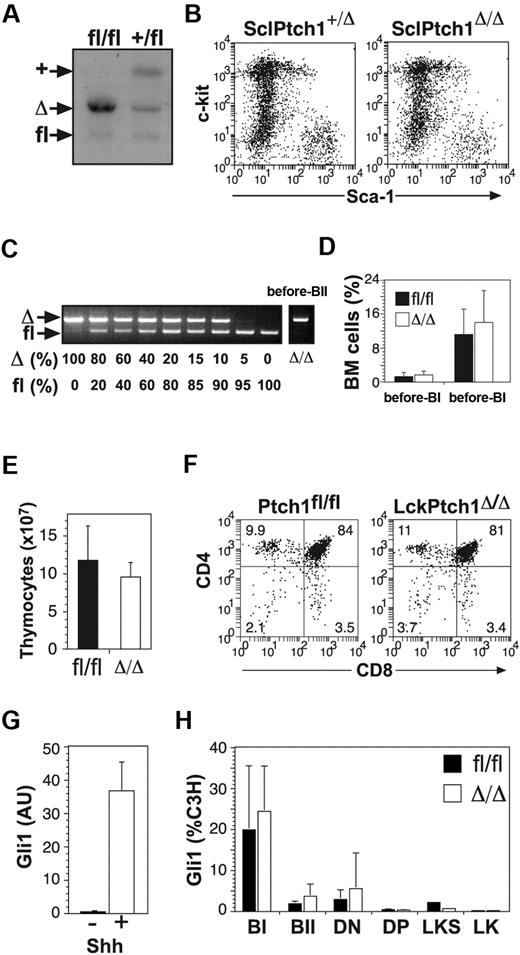
Hematopoietic-specific deletion of Ptch1. (A) Ptch1 Southern blot of BM cells from SclPtch1fl/fl and SclPtch1+/fl mice 8 weeks after tamoxifen. Unlike all other blots, the probe used for this blot distinguishes Ptch1 wild-type (+) and Ptch1fl alleles. (B) Representative dot-plots of lineage-negative BM cells from SclPtch1-heterozygous (+/Δ) and SclPtch1-null (Δ/Δ) mice 8 weeks after tamoxifen. (C) PCR for efficiency of Ptch1fl deletion in pre-BII cells FACS-isolated from a CD19Ptch1Δ/Δ mouse. Deletion efficiency is at least 80%, as determined by the dose titration curve shown on the left. (D) Pre-B cell subsets in BM from Ptch1fl/fl (fl/fl) and CD19Ptch1-null (Δ/Δ) mice. Mean plus SD calculated from 4 mice of each genotype. (E) Thymocyte cellularity of Ptch1fl/fl (fl/fl) and LckPtch1-null (Δ/Δ) mice. Mean plus SD calculated from 4 mice of each genotype. (F) Representative dot-plots of T-cell subsets. The mean percentage shown for each subset was calculated from 4 mice of each genotype. (G) Quantitative RT-PCR for Gli1 mRNA expression in the mesenchymal cell line C3H treated with or without conditioned media containing Shh. Mean plus SD for 3 independent experiments is shown. (H) Quantitative RT-PCR for Gli1 mRNA expression in FACS-isolated BM fractions (pre-BI, pre-BII, LKS, and LK) and thymocyte fractions (DN and DP) from control (Ptchfl/fl) and MxPtch1-null (Δ/Δ) mice. Results for B- and T-cell subsets are mean plus SD of 2 independent sorts, and LKS and LK results are from a single sort from a pool of 5 mice of each genotype.
Hematopoietic-specific deletion of Ptch1. (A) Ptch1 Southern blot of BM cells from SclPtch1fl/fl and SclPtch1+/fl mice 8 weeks after tamoxifen. Unlike all other blots, the probe used for this blot distinguishes Ptch1 wild-type (+) and Ptch1fl alleles. (B) Representative dot-plots of lineage-negative BM cells from SclPtch1-heterozygous (+/Δ) and SclPtch1-null (Δ/Δ) mice 8 weeks after tamoxifen. (C) PCR for efficiency of Ptch1fl deletion in pre-BII cells FACS-isolated from a CD19Ptch1Δ/Δ mouse. Deletion efficiency is at least 80%, as determined by the dose titration curve shown on the left. (D) Pre-B cell subsets in BM from Ptch1fl/fl (fl/fl) and CD19Ptch1-null (Δ/Δ) mice. Mean plus SD calculated from 4 mice of each genotype. (E) Thymocyte cellularity of Ptch1fl/fl (fl/fl) and LckPtch1-null (Δ/Δ) mice. Mean plus SD calculated from 4 mice of each genotype. (F) Representative dot-plots of T-cell subsets. The mean percentage shown for each subset was calculated from 4 mice of each genotype. (G) Quantitative RT-PCR for Gli1 mRNA expression in the mesenchymal cell line C3H treated with or without conditioned media containing Shh. Mean plus SD for 3 independent experiments is shown. (H) Quantitative RT-PCR for Gli1 mRNA expression in FACS-isolated BM fractions (pre-BI, pre-BII, LKS, and LK) and thymocyte fractions (DN and DP) from control (Ptchfl/fl) and MxPtch1-null (Δ/Δ) mice. Results for B- and T-cell subsets are mean plus SD of 2 independent sorts, and LKS and LK results are from a single sort from a pool of 5 mice of each genotype.
Despite efficient deletion of Ptch1 by hematopoietic-specific Cre transgenics, no hematopoietic effects were observed, suggesting that either activation of Hh signaling in hematopoiesis had no consequence or that, alternatively, deletion of Ptch1 did not lead to inappropriate activation of Hh signaling in hematopoietic cells. To address these 2 alternatives, we examined the expression of Gli1 in our target cell populations by quantitative RT-PCR. As a positive control for activation of the Hh pathway, we used the mesenchymal cell line C3H treated with Shh-conditioned media, which produced a 50-fold increase in Gli1 mRNA (Figure 5G). Surprisingly, no significant increase in Gli1 expression was detected in freshly isolated pre-B cells from CD19Ptch1Δ/Δ mice or DP thymocytes from LckPtch1Δ/Δ mice (supplemental Figure 3). More importantly, Gli1 expression was not increased in sorted B-cell progenitors, thymocyte subsets, LKS cells, or LK cells from phenotypic MxPtch1Δ/Δ mice (Figure 5H). Thus, deletion of Ptch1 did not lead to activation of the Hh signaling pathway in hematopoietic cells. Furthermore, the hematopoietic phenotype in MxPtch1Δ/Δ mice could not be attributed to cell-intrinsic activation of the Hh pathway.
Hematopoietic abnormalities are due to Ptch1 deletion in nonhematopoietic cells
The absence of a phenotype in hematopoietic-specific Cre transgenics suggested that the phenotype in MxPtch1Δ/Δ mice was a cell-extrinsic defect due to activation of the Hh signaling pathway in nonhematopoietic cells. To address this possibility, we generated BM chimeras: lethally irradiated MxPtch1fl/fl mice were reconstituted with wild-type BM cells, and lethally irradiated wild-type mice were reconstituted with MxPtch1fl/fl BM cells (Figure 6A). At 4 to 8 weeks after transplantation, reconstituted mice were treated with poly(I:C) to delete Ptch1, either in nonhematopoietic cells including the cell niche (MxPtch1fl/fl recipient mice), or hematopoietic cells (MxPtch1fl/fl donor BM). Consistent with a cell-extrinsic mechanism, MxPtch1Δ/Δ mice reconstituted with wild-type BM displayed 3-fold more LKS, reduced pre-B cell numbers and marked loss of thymic cellularity compared with wild-type mice reconstituted with MxPtch1Δ/Δ BM (Figure 6B). Thus, deletion of Ptch1 in nonhematopoietic cells explains both the lymphoid and myeloid abnormalities of MxPtch1Δ/Δ mice.
Hematopoietic abnormalities in MxPtch1Δ/Δ mice are cell-extrinsic. (A) Schematic of transplantation experiment. Donor BM cells from MxPtch1fl/fl or wild-type mice (+/+) were transplanted into lethally irradiated recipients. At 4 to 8 weeks after reconstitution, recipients were injected with poly(I:C) to generate either Ptch1-null hematopoietic cells or a Ptch1-null nonhematopoietic cells including the cell niche. (B) Numbers of LKS and B220loCD25+ pre-BII BM cells and thymocytes were measured 4 to 6 weeks after poly(I:C). Mean plus SD calculated from 4 recipients of each genotype. *P < .05 by 2-tailed Mann-Whitney test. (C) Mean cortical thickness plus SD through the mid-shaft of femurs from 3 mice of each genotype. *P < .05 by unpaired 2-tailed t test. (D) Numbers of osteoblasts along femoral shaft of MxPtch1 mice. Mean plus SD from 3 mice of each genotype. *P < .05 by unpaired 2-tailed t test. (E) Histologic features of periosteum from trabecular bone of femurs from MxPtch1 mice. OB indicates osteoblast. (F) Quantitative RT-PCR for RANKL mRNA expression in BM from MxPtch1 mice. Mean plus SD from 2 mice of each genotype. (G) PCR for Ptch1fl deletion in sorted osteoblasts from MxPtch1+/Δ mice (lane 1). As a control for nondeleted Ptch1fl allele, BM cells from a Ptch1+/fl mouse is shown (lane 2). (H) Quantitative RT-PCR for Gli1 mRNA expression in whole bone samples from control (Ptchfl/fl) and MxPtch1-null (Δ/Δ) mice. Results are mean plus SD of 3 mice of each genotype.
Hematopoietic abnormalities in MxPtch1Δ/Δ mice are cell-extrinsic. (A) Schematic of transplantation experiment. Donor BM cells from MxPtch1fl/fl or wild-type mice (+/+) were transplanted into lethally irradiated recipients. At 4 to 8 weeks after reconstitution, recipients were injected with poly(I:C) to generate either Ptch1-null hematopoietic cells or a Ptch1-null nonhematopoietic cells including the cell niche. (B) Numbers of LKS and B220loCD25+ pre-BII BM cells and thymocytes were measured 4 to 6 weeks after poly(I:C). Mean plus SD calculated from 4 recipients of each genotype. *P < .05 by 2-tailed Mann-Whitney test. (C) Mean cortical thickness plus SD through the mid-shaft of femurs from 3 mice of each genotype. *P < .05 by unpaired 2-tailed t test. (D) Numbers of osteoblasts along femoral shaft of MxPtch1 mice. Mean plus SD from 3 mice of each genotype. *P < .05 by unpaired 2-tailed t test. (E) Histologic features of periosteum from trabecular bone of femurs from MxPtch1 mice. OB indicates osteoblast. (F) Quantitative RT-PCR for RANKL mRNA expression in BM from MxPtch1 mice. Mean plus SD from 2 mice of each genotype. (G) PCR for Ptch1fl deletion in sorted osteoblasts from MxPtch1+/Δ mice (lane 1). As a control for nondeleted Ptch1fl allele, BM cells from a Ptch1+/fl mouse is shown (lane 2). (H) Quantitative RT-PCR for Gli1 mRNA expression in whole bone samples from control (Ptchfl/fl) and MxPtch1-null (Δ/Δ) mice. Results are mean plus SD of 3 mice of each genotype.
The BM cell niche contains cell types important for regulation of both HSCs and B-cell progenitors, including osteoblasts lining the endosteum.25,26 We examined the bones of MxPtch1Δ/Δ mice for abnormalities that might explain the hematopoietic phenotype. Histologic examination of long bones from MxPtch1Δ/Δ mice revealed extensive thinning of cortical bone (Figure 6C), which was confirmed by peripheral quantitative computer-aided tomography (supplemental Figure 4). Loss of cortical bone was associated with marked changes in osteoblasts: MxPtch1Δ/Δ bones had 4-fold fewer osteoblasts (Figure 6D), which were flat and elongated rather than the normal cuboidal shape (Figure 6E). In addition to impaired bone formation, MxPtch1Δ/Δ BM cells had increased expression of RANKL (Figure 6F), a cytokine that promotes bone resorption and mobilization of BM progenitors.27
MxCre is expressed in cells of the BM niche, although the specific cell types are poorly defined.28,29 To determine whether Ptch1 is deleted in osteoblasts, we isolated CD45−Lin−CD31−CD51+ osteoblasts by fluorescence-activated cell sorter (FACS) from MxPtch1-heterozygous (MxPtch1+/Δ) mice, which had normal numbers of osteoblasts. PCR genotyping demonstrated that the MxCre transgene deleted the Ptch1fl allele in osteoblasts (Figure 6G).
Initial attempts to isolate osteoblasts from MxPtch1Δ/Δ mice yielded insufficient RNA for Gli1 expression. However, RNA prepared from whole bones revealed 3-fold more Gli1 in MxPtch1Δ/Δ bones (Figure 6H). Thus, deletion of Ptch1 by MxCre leads to activation of the Hh signaling pathway in the BM cell niche.
Lymphoid defects are caused by activation of the Hh signaling pathway in epithelial cells
Ptch1 was deleted not only in osteoblasts but also in the basal cells of the skin. 6 to 8 weeks after poly(I:C), MxPtch1Δ/Δ mice developed ruffled coats that progressed to typical basal cell carcinoma of the skin (Figure 7A). These skin changes were due to marked activation of the Hh pathway: Gli1 mRNA levels were 10-fold more than that seen in Shh-treated C3H cells (Figure 7B). As the skin disease progressed, MxPtch1Δ/Δ mice became unwell and were killed by 8 to 10 weeks after poly(I:C). To explore the relationship between the hematopoietic abnormalities and skin disease, we examined hematopoiesis in mice where Ptch1 was deleted using the keratin-specific Cre transgenic mouse strain K14Cre. This mouse strain expresses Cre recombinase in the basal cells of the epidermis and other stratified epithelia including the thymic epithelium.20,30 As expected, K14Ptch1Δ/Δ mice developed widespread basal cell carcinomas by 3 to 4 weeks of age, which was more extensive than that seen in MxPtch1Δ/Δ mice (Figure 7C). Remarkably, K14Ptch1Δ/Δ mice developed lymphoid defects similar to but more severe than those seen in MxPtch1Δ/Δ mice: K14Ptch1Δ/Δ mice had a 16-fold loss of BM pre-BII cells (Figure 7D) and a 7-fold loss of thymocytes (Figure 7E). In contrast, K14Ptch1Δ/Δ mice did not develop increased numbers of LKS (Figure 7F), splenomegaly, or neutrophilia (data not shown). Thus, activation of the Hh signaling pathway in epithelial cells is the likely mechanism of the lymphoid but not myeloid cell defects observed in MxPtch1Δ/Δ mice.
Deletion of Ptch1 in epithelial cells causes the B- and T-cell defects. (A) Characteristic histologic appearance in skin of MxPtch1Δ/Δ mice. (B) Quantitative RT-PCR for Gli1 mRNA expression in skin from MxPtch1Δ/Δ mice. Mean plus SD from 3 mice of each genotype. (C) Characteristic histologic changes in skin of K14Ptch1Δ/Δ mice. (D) Numbers of pre-BII cells and (E) thymocytes in K14Ptch1 mice. Analyses were performed at 4 to 6 weeks of age. Mean plus SD from 5 mice of each genotype. (F) Numbers of LKS cells in BM from K14Ptch1 mice. Mean plus SD from 7 mice of each genotype. (G) Numbers of osteoblasts along femoral shaft of K14Ptch1 mice. Mean plus SD from 3 mice of each genotype. (H) Quantitative RT-PCR for TSLP mRNA expression in thymus and skin of control (Ptchfl/fl) and MxPtch1-null (Δ/Δ) mice. Results were normalized for HPRT and expressed as the mean plus SD (arbitrary units [AU]) from 3 mice of each genotype. (I) Serum TSLP levels in MxPtch1Δ/Δ mice (n = 4) and K14Ptch1Δ/Δ mice (n = 4). Serum TSLP levels in control Ptch1fl/fl mice (n = 8) were below the detectable limit of the immunoassay (4 pg/mL).
Deletion of Ptch1 in epithelial cells causes the B- and T-cell defects. (A) Characteristic histologic appearance in skin of MxPtch1Δ/Δ mice. (B) Quantitative RT-PCR for Gli1 mRNA expression in skin from MxPtch1Δ/Δ mice. Mean plus SD from 3 mice of each genotype. (C) Characteristic histologic changes in skin of K14Ptch1Δ/Δ mice. (D) Numbers of pre-BII cells and (E) thymocytes in K14Ptch1 mice. Analyses were performed at 4 to 6 weeks of age. Mean plus SD from 5 mice of each genotype. (F) Numbers of LKS cells in BM from K14Ptch1 mice. Mean plus SD from 7 mice of each genotype. (G) Numbers of osteoblasts along femoral shaft of K14Ptch1 mice. Mean plus SD from 3 mice of each genotype. (H) Quantitative RT-PCR for TSLP mRNA expression in thymus and skin of control (Ptchfl/fl) and MxPtch1-null (Δ/Δ) mice. Results were normalized for HPRT and expressed as the mean plus SD (arbitrary units [AU]) from 3 mice of each genotype. (I) Serum TSLP levels in MxPtch1Δ/Δ mice (n = 4) and K14Ptch1Δ/Δ mice (n = 4). Serum TSLP levels in control Ptch1fl/fl mice (n = 8) were below the detectable limit of the immunoassay (4 pg/mL).
Although loss of osteoblasts may explain the B-cell defect seen in MxPtch1Δ/Δ mice, this was not the case in K14Ptch1Δ/Δ mice, where osteoblasts numbers were normal (Figure 7G). The T-cell defect observed in K14Ptch1Δ/Δ mice was also unlikely to be explained by an abnormality of the thymic cell niche because, unlike epithelial cells of the skin, thymic epithelial cells of K14Ptch1Δ/Δ mice were not increased (supplemental Figure 5). One possible mechanism for the lymphoid defects is elevated levels of epithelial-derived TSLP, a cytokine implicated in regulating B- and T-cell numbers. TSLP transgenic mice have very similar lymphoid defects to that seen in MxPtch1Δ/Δ and K14Ptch1Δ/Δ mice.31 Consistent with this hypothesis, TSLP mRNA expression was increased 6-fold in the skin, but not thymus, of MxPtch1Δ/Δ mice (Figure 7H). Increased epithelial expression of TSLP led a 10- to 30-fold increase in circulating TSLP. Thus, activation of the Hh pathway in epithelial cells leads to markedly increased levels of circulating TSLP.
Discussion
We have used inducible and tissue-specific deletion of Ptch1 to determine the cell-intrinsic and cell-extrinsic hematopoietic effects of activating the Hh pathway. Surprisingly, we found that deletion of Ptch1 in hematopoietic cells, including HSCs, did not lead to activation of the Hh signaling pathway and consequently had no phenotypic effect. In contrast, deletion of Ptch1 in nonhematopoietic cells produced 2 dramatic hematopoietic phenotypes: increased numbers of cycling LKS with increased circulating myeloid progenitors and apoptosis of B- and T-cell precursors. Deletion of Ptch1 with an epithelial-specific Cre demonstrated that these 2 hematopoietic phenotypes are mediated by distinct cell-extrinsic mechanisms. Activation of the Hh signaling pathway in the BM cell niche led to loss of osteoblasts and increased LKS numbers, whereas activation of Hh signaling in epithelial cells led to increased circulating TSLP and death of lymphoid precursors.
Using a tamoxifen-inducible deletion of Ptch1 in adult mice, Uhmann et al reported rapid loss of common lymphoid progenitors (CLPs), but they suggested that this defect was cell-extrinsic because Ptch1-null CLPs could grow in immunodeficient Rag2−/− γ−/− mice.14 Our results provide significant insight into the mechanism of the lymphoid defects reported by Uhmann et al. First, we show that the loss of B and T cells is due to apoptosis of early lymphoid progenitors rather than defects in proliferation or differentiation. The normal numbers of pro-B cells (B220loCD19−) in the BM of MxPtch1Δ/Δ mice suggest that Ptch1 is required for survival of pre-B cells rather than CLPs. Second, demonstration that wild-type HSCs have severe lymphoid abnormalities whentransplanted into MxPtch1Δ/Δ mice definitively proves the cell-extrinsic nature of these defects. Finally, identical lymphoid defects in K14Ptch1Δ/Δ mice strongly suggest that the cell-extrinsic mechanism is due to deletion of Ptch1 in epithelial cells.
The phenotype of the K14Ptch1Δ/Δ mice suggests that the B- and T-cell defects are not due to defects of the cell niches regulating B- and T-cell progenitors. With respect to the B-cell defect, K14Ptch1Δ/Δ mice had normal numbers of osteoblasts. Furthermore, there is no evidence that the K14Cre transgene is expressed in the BM cell niche. With respect to the T-cell defects, we observed no expansion of thymic epithelial cells (supplemental Figure 5) despite the ability of the K14Cre to delete “floxed” genes in thymic epithelium.20 One possible explanation for the lymphoid defects is an elevated level of epithelial-derived TSLP, which was observed in both MxPtch1Δ/Δ and K14Ptch1Δ/Δ mice (Figure 7H,I). TSLP transgenic mice have very similar lymphoid defects to that seen in MxPtch1Δ/Δ and K14Ptch1Δ/Δ mice.31 However, it remains to be determined whether Hh signaling is a direct regulator of TSLP in epithelial cells and, if so, whether it is the direct cause of the lymphoid defects. Despite markedly elevated levels of circulating TSLP, we cannot exclude the possibility that apoptosis of pre-B cells and thymocytes are due to high levels of lymphotoxins observed in acute illness such as cortisol, TNF-α, and IFN-γ.32
Similar to the lymphoid defects, we show that expansion of LKS and mobilization of myeloid progenitors is caused by deletion of Ptch1 in nonhematopoietic cells. Despite these changes, highly quantitative, competitive transplantation assays in congenic mice did not reveal any expansion or loss of HSC activity. Deletion of a single Ptch1 allele in the adult had no demonstrable effects. This result contrasts with germline Ptch1-heterozygous mice, where elevated Gli1 mRNA in Ptch1-heterozygous LKS associated with HSC exhaustion were reported.13 Possible explanations for these differences include the timing of Ptch1 deletion (embryonic vs adult), the cell types affected (all cells compared with MxCre targeted cells) and the targeting strategy (start codon compared with first transmembrane domain). A similar discrepancy between germline and inducible deletion of Ptch1 was reported for bone homeostasis.33,34
Our results do not definitively identify the target cell mediating the increased LKS and circulating myeloid progenitors in MxPtch1Δ/Δ mice. However, the loss of osteoblasts in MxPtch1Δ/Δ mice, but not K14Ptch1Δ/Δ mice, that have no LKS abnormality, makes osteoblasts a good candidate. It is hypothesized that more quiescent HSCs reside in the endosteal niche while more active cycling HSCs “primed” for mobilization reside in the vascular niche.35 The increased cell cycling and mobilization of LKS in MxPtch1Δ/Δ mice would be consistent with this hypothesis. We also found increased expression of RANKL, which may contribute to the loss of bone and increased progenitor cell mobilization in these mice.27 The relative importance of the bone and vascular cell niches remains controversial although the normal HSC activity in MxPtch1Δ/Δ BM, despite the marked loss of osteoblasts, supports the recent studies of biglycan-deficient mice, which have reduced osteoblasts and trabecular bone yet normal HSC function.36
Loss of osteoblasts in MxPtch1Δ/Δ mice may be a direct effect of activation of the Hh signaling pathway because MxCre can excise the Ptch1fl allele in osteoblasts (Figure 6G), and activation of Hh signaling in mesenchymal cells inhibits osteoblast differentiation.37 Mak et al have also reported loss of postnatal bone after deletion of Ptch1 in mature osteoblasts.33 Our data suggest that targeting Hh signaling in bone might be a method of regulating HSC cell cycle without impairing long-term stem cell activity. This may be useful for enhancing hematopoietic recovery after chemotherapy or for sensitizing quiescent leukemic stem cells that reside in the endosteal cell niche.38
The observation that deleting Ptch1 in hematopoietic cells did not lead to activation of the Hh signaling pathway has important implications for therapies that target Ptch1. Given that Hh proteins act by inhibiting Ptch1,39 our results raise the possibility that the reported hematopoietic effects of Hh proteins are indirect through their effects on the microenvironment. For example, the enhanced ability of Ihh-expressing stroma to support HSCs may be indirect through changes to the stromal cells.11 A similar effect of Hh proteins on the stromal microenvironment was shown to be important for maintaining the growth of epithelial tumors.40 Alternatively, Ptch1 redundancy in hematopoiesis may be due to expression of Ptch2, which was expressed at normal levels in MxPtch1Δ/Δ myeloid and lymphoid progenitors (supplemental Figure 6), and can bind Hh proteins with similar affinity to Ptch1.41 Hh agonists have been proposed for expansion of HSCs and improving BM regeneration after transplantation or chemotherapy. Although the HSC effects seen in MxPtch1Δ/Δ support this notion in part, our results highlight the importance of distinguishing direct from indirect effects of activating the Hh signaling pathway in hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Lan Ta, Rebecca Bowyer, and Jenny Davis for animal husbandry; Dean Hewish for cell sorting; and Natalie Sims for advice with analyses of bone, including histomorphometry.
This study was supported in part by research funding from National Health & Medical Research Council (NHMRC) to D.J.C. (Project Grant No. 435107; RD Wright Fellows 382904) and S.M.J. (Principal Research Fellow).
Authorship
Contribution: S.L.S. designed research, performed research, analyzed data, and wrote the manuscript; M.P.M. designed research, performed research, and analyzed data; S.M.J. performed research and wrote the manuscript); N-Y.N.N. performed research and analyzed data; S.V. performed research; D.J.C. performed research and wrote the manuscript; and R.V. and B.J.W. contributed new reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David J. Curtis, Rotary Bone Marrow Research Laboratories, Royal Melbourne Hospital PO, Grattan St, Melbourne VIC 3050, Australia; e-mail: dcurtis@wehi.edu.au.




![Figure 7. Deletion of Ptch1 in epithelial cells causes the B- and T-cell defects. (A) Characteristic histologic appearance in skin of MxPtch1Δ/Δ mice. (B) Quantitative RT-PCR for Gli1 mRNA expression in skin from MxPtch1Δ/Δ mice. Mean plus SD from 3 mice of each genotype. (C) Characteristic histologic changes in skin of K14Ptch1Δ/Δ mice. (D) Numbers of pre-BII cells and (E) thymocytes in K14Ptch1 mice. Analyses were performed at 4 to 6 weeks of age. Mean plus SD from 5 mice of each genotype. (F) Numbers of LKS cells in BM from K14Ptch1 mice. Mean plus SD from 7 mice of each genotype. (G) Numbers of osteoblasts along femoral shaft of K14Ptch1 mice. Mean plus SD from 3 mice of each genotype. (H) Quantitative RT-PCR for TSLP mRNA expression in thymus and skin of control (Ptchfl/fl) and MxPtch1-null (Δ/Δ) mice. Results were normalized for HPRT and expressed as the mean plus SD (arbitrary units [AU]) from 3 mice of each genotype. (I) Serum TSLP levels in MxPtch1Δ/Δ mice (n = 4) and K14Ptch1Δ/Δ mice (n = 4). Serum TSLP levels in control Ptch1fl/fl mice (n = 8) were below the detectable limit of the immunoassay (4 pg/mL).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/5/10.1182_blood-2009-03-208330/5/m_zh89990939610007.jpeg?Expires=1767735760&Signature=CZj35dOhtWRabHppVvzGcKH0u3Q6CMcz3DXlReGkfSkoC6ojCPvWEQfq7Jlxzl2oiKngBW64Z6lAIJlC6B-pU~ZDFak2knsutVwPaePUKZRXkOZ7fD8AXqtncF7~bSyAlV7S3wwLgFaURFxe5pX3uNmaDPTmnGZ7DQnFXebqd2~LPPqPSIt4LJxLQ87fN3PHAYZj~QFkOcwV6NuggI1d7b2dup4-mGLX7eKOaDuMecbj~EypTvOXo-youywRvH9C4~M~lxpIM8r6adEfDdRN011GEaCQ9N9AkuSPo20s9EXNz1hsXAbWKTAXgL0qi0ELCUBL7dJrZ6k1ThAeoYSbew__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





![Figure 7. Deletion of Ptch1 in epithelial cells causes the B- and T-cell defects. (A) Characteristic histologic appearance in skin of MxPtch1Δ/Δ mice. (B) Quantitative RT-PCR for Gli1 mRNA expression in skin from MxPtch1Δ/Δ mice. Mean plus SD from 3 mice of each genotype. (C) Characteristic histologic changes in skin of K14Ptch1Δ/Δ mice. (D) Numbers of pre-BII cells and (E) thymocytes in K14Ptch1 mice. Analyses were performed at 4 to 6 weeks of age. Mean plus SD from 5 mice of each genotype. (F) Numbers of LKS cells in BM from K14Ptch1 mice. Mean plus SD from 7 mice of each genotype. (G) Numbers of osteoblasts along femoral shaft of K14Ptch1 mice. Mean plus SD from 3 mice of each genotype. (H) Quantitative RT-PCR for TSLP mRNA expression in thymus and skin of control (Ptchfl/fl) and MxPtch1-null (Δ/Δ) mice. Results were normalized for HPRT and expressed as the mean plus SD (arbitrary units [AU]) from 3 mice of each genotype. (I) Serum TSLP levels in MxPtch1Δ/Δ mice (n = 4) and K14Ptch1Δ/Δ mice (n = 4). Serum TSLP levels in control Ptch1fl/fl mice (n = 8) were below the detectable limit of the immunoassay (4 pg/mL).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/5/10.1182_blood-2009-03-208330/5/m_zh89990939610007.jpeg?Expires=1767772389&Signature=fQ0PoUQEQKaRU~0uMeSSsn5W7cJOlNlkLWV~NCKHI15CFhAL7r7gr03s0v1R2sb1w~0UzuDBB0GgGAk0X-ovLzQg6vbRIPuAX14haizB940F7p31MNsQZRvioTLtPQDSnHHB9qDSN7OFa~4O219F-IBflXR1lp1MFp6e~yUx4zVcThvgXxulo0WRgenhweLbZRx3QJ9oZwF45BQ1ZmHj9C~UxPMpIwL4rQ2FS2Wx7vvFaZZyWWNKOqc9GhiIbzrag66yB4YADA4fGjRmx2Y17kfTmcaaV4rY7n857Q9bwh8QJVHiU7MnV9f~JoVj9e9hqDNMBJIgiY3tRnlB04IIRA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)