Abstract
The cell of origin of chronic lymphocytic leukemia (CLL) has long been sought, and immunoglobulin gene analysis provides new clues. In the unmutated subset (U-CLL), there is increased usage of the 51p1-related alleles of the immunoglobulin heavy chain variable 1-69 gene, often combined with selected genes and with immunoglobulin heavy chain diversity IGHJ6. Stereotypic characteristics of the HCDR3 result and suggest antigen selection of the leukemic clones. We have now analyzed 51p1/IGHJ6 combinations in normal blood B cells from 3 healthy persons for parallel sequence patterns. A high proportion (33.3% of sequences) revealed stereotypic patterns, with several (15.0%) being similar to those described in U-CLL. Previously unreported CLL-associated stereotypes were detected in 4.8%. Stereotypes (13.6%) not detected in CLL also were found. The HCDR2-IGHJ6 sequences were essentially unmutated. Junctional amino acids in normal B cells were heterogeneous, as in cases of stereotyped CLL. Phenotypically, normal B cells expressing 51p1-derived immunoglobulin M were naive. This snapshot of the naive B-cell repertoire reveals subsets of B cells closely related to those characteristic of CLL. Conserved patterns in the 51p1-encoded immunoglobulin M of normal B cells suggest a restricted sequence repertoire shaped by evolution to recognize common pathogens. Proliferative pressure on these cells is the likely route to U-CLL.
Introduction
Immunogenetic analysis provides an important key to understanding pathogenesis and behavior of B-cell tumors.1 Clinical relevance of the immunoglobulin heavy chain variable gene (IGHV) status has been dramatically illustrated for chronic lymphocytic leukemia (CLL), where 2 subsets, delineated by the absence or presence of somatic mutation, have strikingly different prognoses.2-4 The unmutated subset (U-CLL), of inferior prognosis, appears to derive from a pregerminal center B cell. In contrast, mutated CLL (M-CLL), of superior prognosis, is likely to be derived from a postgerminal center B cell.5 Although tumor cells of both U-CLL and M-CLL share phenotypic features of an activated B cell, U-CLL is distinguished by frequent expression of significant levels of ZAP-70, a tyrosine kinase involved in several signaling pathways.6 The B-cell receptor of U-CLL also tends to be more responsive to engagement by anti-Ig in vitro than that of M-CLL, leading to phosphorylation of p72Syk and other mediators and an increase in intracellular [Ca(2+)](i).7-9 These findings point to the B-cell receptor as a potentially critical molecule in determining disease progression.
Analysis of IG variable (IGV) gene use in B-cell tumors also can reveal bias indicative of antigenic (or superantigenic) pressure on the cell of origin. Expansion of B cells expressing particular IGV genes may occur after binding of antigen to framework regions.10 Because antigens binding in this way are able to bind to all B cells expressing that IGV gene, they are considered to be superantigens. Recognition of expansions has required knowledge of the repertoire of normal B cells, where, although not all IGV genes available in the germline are expressed equally, the relative levels in healthy subjects have been reported, providing a baseline for comparison.11,12 In CLL, the major asymmetry concerns the 51p1-related alleles of the IGHV1-69 gene.13,14 These are expressed by 4.7% plus or minus 0.45% of normal circulating B cells in young adults, with levels dependent on the gene copy number.14 Similar levels are found in the healthy elderly (4.3% ± 0.44%)13 but increased to 10% to 20% in CLL, almost all within U-CLL.15,16
There is also a tendency in these cases for 51p1 to be combined with certain diversity (immunoglobulin heavy chain diversity; IGHD) genes in specific reading frames (RFs), and with selected joining (immunoglobulin heavy chain joining; IGHJ) genes, often generating long and rather similar HCDR3 regions.17 Cases of CLL derived from the IGHD3-16 gene (RF 2) combined with IGHJ3 can show particularly strong similarity in HCDR3 sequences, with nontemplated codon insertions tending to generate similar amino acids.16 Shared sequences, clearly evident in 51p1-encoded cases, are found in other IGV gene combinations, for example, in rearrangements involving the IGV3-21 gene, and have given rise to the concept of stereotypic sequences.18 Not only is there conservation of IGHVDJ rearrangements, but these can also be combined with particular IG light chain-variable and joining gene rearrangements, strongly suggesting that there may be common (super)antigens binding to subsets of B cells found in CLL. Although this finding could reflect the selective stimulation of the B cell of origin, there also exists the possibility that antigenic drive continues after transformation.5
However, the question of whether such conserved sequences could be detected in significant numbers among normal B cells remained. Analysis of “non-CLL” sequences from the databases found very few,18,19 but deposited sequences are not generally derived from naive unmutated B cells, the real comparator for U-CLL. In view of this, we have now analyzed the 51p1-encoded sequence repertoire from 3 healthy persons in the older age group. To reflect U-CLL, we have biased the analysis toward the commonly used IGHJ6 gene, knowing that most of these sequences are likely to be unmutated.20 Phenotypic analysis confirmed that the majority of the 51p1-encoded population is naive. Among these normal B cells, which are found in the naive B-cell population, we can detect a repertoire containing many CLL-like subsets and some not found in CLL. Clearly this part of the B-cell population has the preferential combinations and conserved sequences that later emerge in CLL.
Methods
Donors and blood samples
Blood was taken from 3 healthy persons: D1 (a British woman, 69 years), D2 (an Italian man, 69 years), and D3 (a British woman, 51 years), and age-matched to that of patients with CLL. All donors were healthy at time of blood collection. Blood was layered over Lymphoprep (Axis-Shield), and lymphocytes were isolated following manufacturer's instructions. The cells were lysed in Tri-reagent (Sigma-Aldrich), and RNA was isolated according to the manufacturer's instructions. cDNA was prepared by the use of the Transcriptor High Fidelity cDNA synthesis kit (Roche). The study was approved by the institutional review boards of the University of Southampton and the University of Siena, and all donors provided informed consent before inclusion into the study in accordance with the Declaration of Helsinki.
Amplification of IGHVDJ rearrangements by the use of IGHV1-69 and IGHJ6 genes
IGHV gene rearrangements were amplified from cDNA, cloned, and sequenced independently from each donor in 2 different laboratories (Siena, Italy, and Southampton, United Kingdom). IGHV1-69 51p1-related gene rearrangements were amplified by polymerase chain reaction (PCR) with a forward primer specific for 51p1-related alleles with common HCDR2 sequences (5′-AGGGATCATCCCTATCTT-3′). These alleles correspond to IGHV1-69*01, *03, *05, *06, *12, and *13 in the IMGT database. Reverse primers included Cμ100 (5′-GGAGAAAGTGATGGAGTCGG-3′), JH6o (5′-TGAGGAGACGGTGACCGTGGTCCCTTG-3′), and JH6n (5′-CCCAGACGT CCATACCGTA-3′). The PCR products were cloned into the pGEMT vector (Promega) and transformed into Escherichia coli XL-1 Blue. Isolated colonies were picked, grown overnight, and the DNA was isolated by the use of the QIAGEN Mini-prep system (QIAGEN). Sequencing was performed with the T7 and SP6 primers and an ABI 3130X or ABI 310 DNA Sequencer (Applied Biosystems).
Analysis of 51p1-DJ rearrangements
Sequences were aligned to the ImMunoGeneTics (IMGT) sequence directory (http://imgt.cines.fr/) and analyzed for IGHV1-69 allele and IGHD and IGHJ gene usage. The IGHD germline gene was assigned according to the IMGT/Junction Analysis tool (http://www.imgt.org/IMGT_jcta/jcta), following established IMGT criteria.21 HCDR3 length was determined according to IMGT numbering.21 Rearrangements that used 51p1 and IGHJ6 were identified among all clones amplified from HCDR2 to either IGHCM or IGHJ6, and frequencies of 51p1-DJ rearrangements that used IGHJ6 were calculated from the clones of these PCRs.
HCDR3 clustering
All putative HCDR3 amino acid sequences derived from the functional 51p1-DJ rearrangements were aligned to the HCDR3 from a preestablished CLL reference database (n = 312) and to each other. The CLL reference database included the published IGHV1–69DJ rearrangements assigned (n = 123) or not assigned (n = 135) to subsets15 or the unpublished IGHV1-69-DJ rearrangements from the University of Southampton (n = 27) and the University of Siena (n = 27). The normal HCDR3 amino acid sequences were analyzed for identity with the HCDR3 subsets from the public CLL subsets and for the presence of new subsets. For HCDR3-driven clustering, all in-frame IGHV-D-J rearrangements were converted into amino acid sequences and aligned according to the putative HCDR3 amino acid sequences by use of the multiple sequence alignment software ClustalW2 (http://www.ebi.ac.uk/tools/clustalw2/index.html). HCDR3 amino acid sequences with the same IGHD-J genes, an alignment ClustalW2 score of 60 or greater, and an amino acid identity of 60% or greater were assigned to identical subsets and defined as stereotyped, in concordance with the previously established procedures.15,19 A further restriction for normal B cell–derived sequences was that they should match the length of the nearest CLL-derived sequence to within 3 amino acids.22 Subset nomenclature was according to Murray et al.15 For the purpose of this study, subsets not previously included in the Murray nomenclature system were assigned a supplemental “Sx” number (x = 1, 2, 3, etc).
Phenotypic analysis of the 51p1-expressing normal B cells
Peripheral-blood mononuclear cells were purified from the blood of normal donor 1 (D1) by the use of Lymphoprep (Axis-Shield), and the surface expression of immunoglobulin M (IgM), immunoglobulin D (IgD), CD5, CD23, CD27, and CD38 in G6+ and G6− B cells was determined by 4-color flow cytometry. Aliquots of 106 cells were labeled with 5 μg/mL G6 antibody (mouse monoclonal immunoglobulin G1; anti-IGHV1-69 IMGT allele set 01, 03, 05, 06, 12, and 13, previously known as 51p1-related alleles; from Roy Jeffries, University of Birmingham) then washed and stained with anti–mouse immunoglobulin G fluorescein isothiocyanate (rabbit F(ab′)2 Dako). The cells were washed and blocked with 10 μL of normal mouse serum for 10 minutes, followed by the addition of mouse anti-CD19 PerCP-Cy5.5, anti-CD27 APC (Biolegend), and anti-CD5, -CD23, or -CD38 phycoerythrin (BD Biosciences). Staining with anti-IgM or -IgD phycoerythrin (rabbit F(ab′)2; Dako) was performed before labeling with G6 to prevent steric hindrance from the G6 antibody. All incubations were for 30 minutes on ice. Data were acquired on a BD FACS CantoII flow cytometer and analyzed with FlowJo software (TreeStar Inc).
Results
51p1-derived sequences in normal B cells
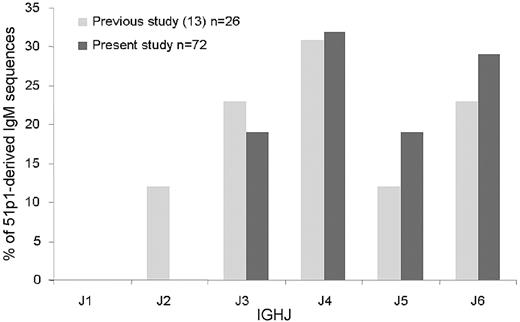
To assess the incidence of IGHJ6 gene use among 51p1-encoded IgM-positive normal B cells in the study, a 51p1-Cμ100 primer pair was used for amplification. Sequencing of the clones obtained revealed that IGHJ6 was used by 21 (29%) of 72 of the 51p1-derived IgM-positive B-cell population (Figure 1), similar to our previous findings.13 Because this combination is common in U-CLL, we then focused our analysis on the normal 51p1-IGHJ6 sequences. A total of 141 potentially functional sequences with the 51p1-IGHJ6 combination was obtained from the 3 healthy persons, with 62 from donor D1, 9 from donor D2, and 70 from donor D3. Nonfunctional sequences were not detected, possibly because of the use of RNA as a source. Neither were repeated, clonally related sequences indicative of monoclonal lymphocytosis observed in any of the donors. Six sequences from our previous published study13 were included in the overall analysis (donor AY), giving a total of 147.
IGHJ use in normal B cells with IGHV1-69-DJ-Cμ rearrangements involving 51p1-related alleles. Light gray bars indicate the percentage of IGHJ genes in the previously published 51p1-derived sequences13 ; dark gray bars, percentage of IGHJ genes in the present new 51p1-derived sequences.
IGHJ use in normal B cells with IGHV1-69-DJ-Cμ rearrangements involving 51p1-related alleles. Light gray bars indicate the percentage of IGHJ genes in the previously published 51p1-derived sequences13 ; dark gray bars, percentage of IGHJ genes in the present new 51p1-derived sequences.
Analysis of somatic mutational levels in the limited sequences obtained by the use of the HCDR2 primer can only provide an indicator. Within this restriction, following the 98% germline homology cutoff value, the majority (143 of 147, 97%) of the 51p1-IGHJ6 rearrangements from the 4 normal donors were unmutated. Specifically, they were either completely unmutated (100% homology to germline sequence, 124 of 143, 87%) or had very low levels (≤ 2%) of mutation (19 of 143, 13%). A total of 4 of 147 (3%) were mutated (range, 92.2%-97.6% homology to germline; median, 96.5%).
CLL-like stereotypic 51p1-IGHJ6 sequences in normal B cells
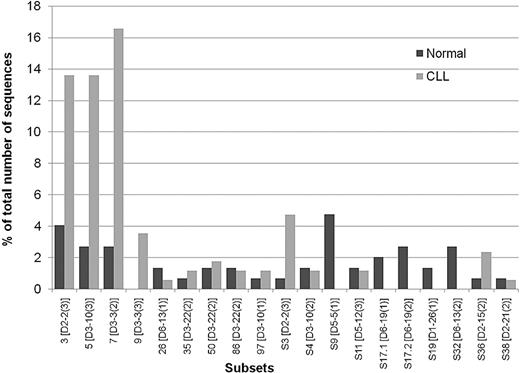
To provide a basis of comparison between normal B cells and CLL, the CLL IGHVDJ rearrangement database15 was mined for 51p1-IGHJ6 sequences, and 139 cases were identified. An additional 30 sequences from Southampton and Siena were added, giving a total of 169 (Table 1 and supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). By using the ClustalW2 alignment program, we found that 90 (53.2%) of the CLL sequences could be assigned to previously described subsets. The majority of these (80 of 90; 88.9%) were in subsets 3, 5, 7, and 9. The remaining sequences (10 of 90, 11.1%) were distributed among other subsets (Figure 2). HCDR3 length ranged from 18 to 27 amino acids (median, 22; supplemental Table 1a). Applying the same criteria to the 147 sequences from normal B cells revealed that 22 (15%) were assignable to these CLL subsets. Supplemental Table 1a lists the subsets of CLL to which normal B-cell sequences could be assigned. The data are summarized in Table 1. The comparison of the subsets within the normal B cells with those in CLL is shown in Figure 2 and includes the IGHD gene use and the reading frame for each subset.
Stereotypic 51p1-IGHJ6 sequences detected in normal B cells and in CLL. Sequences were assigned to known subsets or to new subsets (prefix S). Dark gray bars indicate the percentage of normal B-cell sequences assigned to subsets; light gray bars, percentage of CLL sequences assigned to subsets. For each subset, code, IGHD gene, and reading frame are indicated.
Stereotypic 51p1-IGHJ6 sequences detected in normal B cells and in CLL. Sequences were assigned to known subsets or to new subsets (prefix S). Dark gray bars indicate the percentage of normal B-cell sequences assigned to subsets; light gray bars, percentage of CLL sequences assigned to subsets. For each subset, code, IGHD gene, and reading frame are indicated.
Conservation of 51p1-IGHJ6 HCDR3 sequences arises from selective use of IGHD genes in specific RFs. In CLL, 51p1-derived sequences are preferentially combined (89%) with the IGHD2 and IGHD3 subgroups.17,23 Our study of 51p1-IGHJ6 combinations within subsets indicates an even greater association, with 107 (97.2%) of 110 involving D2 or D3. Although the normal B cells also show this preference, it is less strong, accounting for only 25 (51%) of 49. Major CLL IGHD gene–associated stereotypes have been described: IGHD2-2 (RF 3) for subset 3, IGHD3-10 (RF 3) for subset 5, and IGHD3-3 (RF 2) for subset 7. Normal B cells showed a similar pattern of subset-associated IGHD gene use (Figure 2). However, not all CLL stereotypes were found, with, for example, no normal sequences assignable to subset 9 (Figure 2).
Mining the growing database of CLL sequences revealed additional subsets that fulfilled the established criteria (Table 1, Figure 2, and data not shown). Some CLL-derived sequences not previously assigned15 could then be placed in minor subsets. As indicated in supplemental Table 1 and summarized in Table 1, normal B-cell 51p1-IGHJ6 sequences from all 3 donors also could be assigned to these newly identified subsets, accounting for a further 4.8% of stereotypic sequences. Normal B cell–derived sequences assigned to new CLL-like subsets are shown in supplemental Table 1b and Figure 2. The total numbers of sequences assignable to each new subset ranged from 2 to 9 (supplemental Table 1b). Again, there is overlap but not sequence identity within the subsets.
Non-CLL–like stereotypic 51p1-IGHJ6 sequences in normal B cells
Conserved sequences not reported in CLL so far were also evident in the normal 51p1-IGHJ6 sequences (supplemental Table 1c; Figure 2). These non-CLL subsets accounted for a further 13.6%, bringing the total of stereotypic sequences in the normal donors to 33.3% (Table 1).
Although the frequency of normal 51p1-IGHJ6–derived B cells expressing stereotypic sequences (33.3%) is less than in CLL (65.0%), the phenomenon appears relatively common. Importantly, subsets were identifiable in all 3 normal donors, even though the distribution varied. Several of the stereotypes reflect those characteristic of CLL, whereas others appear to be found only in normal B cells. Subsets of normal B cells not found so far in CLL were derived from IGHD5-5 (RF 1), IGHD6-19 (RF 1 and RF 2), IGHD1-26 (RF1), and IGHD6-13 (RF 2; Figure 2).
Comparison of the HCDR3 regions in 51p1-IGHJ6 rearrangements derived from CLL and normal B cells
Comparative analysis of the HCDR3 sequences was focused on the 3 major subsets described for CLL and confirmed to be frequently expressed in our series.
Subset 3.
Cases of CLL assigned to subset 3 consist of 51p1 combined with the IGHD2-2 gene segment (RF 3) and IGHJ6. The length of the HCDR3 in CLL is in the range of 20 to 25 codons (median, 22 codons), and normal B-cell sequences were similar at 20 to 24 (median, 22) codons (supplemental Table 1a). The restricted IGHVDJ gene combinations inevitably lead to common sequence motifs in HCDR3, which in subset 3 is DIVVVPAA(I/M), the I/M being a recognized polymorphism of the IGHD2-2 gene.24 This full sequence was found in 13 of 23 CLL-derived sequences and in a truncated form in the remaining 10 of 23 (supplemental Table 2). Among the 6 normal B cell–derived sequences, all were truncated. Nontemplated codon insertions at the IGHV-D junction varied in length and composition in both normal and CLL cells. However, the acquisition of valine or aspartic acid amino acids following the CAR C-terminus of the 51p1 gene, often found in CLL sequences, also was evident in normal B cells (supplemental Table 2). Similarities also were evident in the nontemplated codon insertions at the IGHD-J junctions.
Subset 5.
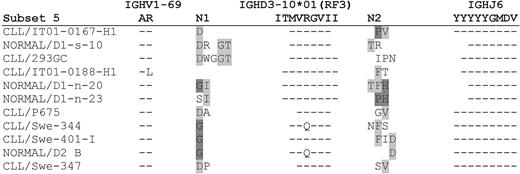
Cases of CLL assigned to subset 5 consist of 51p1 combined with the IGHD3-10 gene segment (RF 3) and IGHJ6. The lengths of the HCDR3 sequences in CLL or normal B cells were 20 to 25 (median, 20) and 18 to 23 (median, 21), respectively (supplemental Tables 1a,2). Common sequence motifs arising from the use of the IGHD3-10 gene were evident in both CLL and normal B cells. Overall, nontemplated codons generated heterogeneous amino acids at the IGHV-D and IGHD-J junctions in both CLL and normal B cells. The HCDR3 sequences from normal B cells are compared with the most similar CLL-derived sequences in Figure 3. Where some similarities occur in cases of CLL, for example, the N1 sequence of CLL/Swe-344 and CLL/Swe-401-I, this was also evident in the normal D2 B sequence (Figure 3).
Comparison of the HCDR3 sequences of CLL and normal B cells in the 51p1-IGHJ6–derived subset 5. Amino acid sequences of the HCDR3 of each normal sequence aligned to the closest CLL HCDR3 are represented. Dashes indicate homology to the germline IGHV1-69, IGHD3-10 in reading frame (RF) 3, and IGHJ6 genes at the top of the figure. Identical N1 and N2 amino acids between different sequences are highlighted in shades of gray.
Comparison of the HCDR3 sequences of CLL and normal B cells in the 51p1-IGHJ6–derived subset 5. Amino acid sequences of the HCDR3 of each normal sequence aligned to the closest CLL HCDR3 are represented. Dashes indicate homology to the germline IGHV1-69, IGHD3-10 in reading frame (RF) 3, and IGHJ6 genes at the top of the figure. Identical N1 and N2 amino acids between different sequences are highlighted in shades of gray.
Subset 7.
Cases of CLL assigned to subset 7 consist of 51p1 combined with the IGHD3-3 gene (RF 2) and IGHJ6. The lengths of the HCDR3 sequences in CLL or normal B cells were 22 to 27 (median, 24) and 20 to 25 (median, 23), respectively (supplemental Tables 1a,2). A common sequence motif was evident in both CLL and normal B cells arising from full or truncated sequence of the IGHD gene. A particular sequence reported previously to be common in 51p1-expressing CLL samples is GGYDFWSGYY, where the GG amino acids are considered to derive from insertion of guanosine and cytidine by the terminal deoxynucleotidyl transferase.25 This feature was rather less common inour CLL database, being in only 3 of 29 sequences. However, it was found in 1 of 4 normal B-cell sequences in this subset (supplemental Table 2).
Phenotypic profile of 51p1-expressing normal B cells
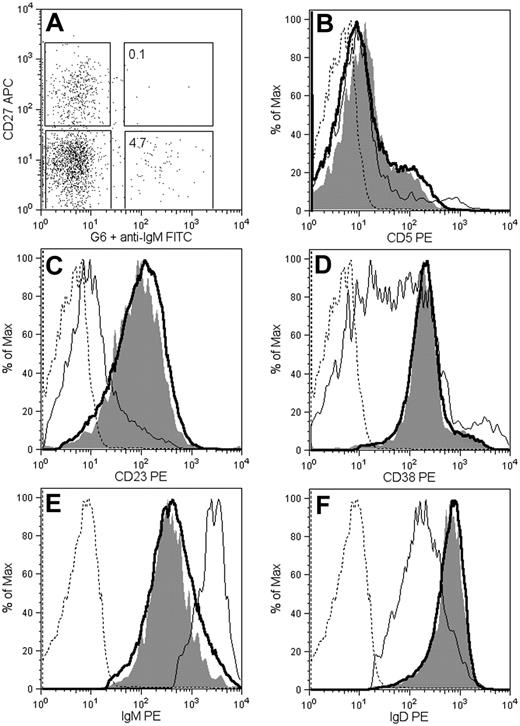
The monoclonal antibody G6 was used to determine the cellular origin of the 51p1-encoded sequences obtained from normal blood. This monoclonal antibody reacts with an idiotypic determinant derived from the HCDR2 of the 51p1-related alleles, regardless of the IGHJ gene involved.14,26,27 Immunofluorescent staining of the CD19+ blood B cells of donor D1 revealed that 4.8% were positive for G6 (Figure 4A). The majority of G6+ cells (98%) were CD27 negative, indicative of naive B cells.
Phenotypic profile of G6+ B cells from healthy donor D1. Peripheral blood mononuclear cells were analyzed by 4-color fluorescence-activated cell sorting. (A) Distribution of G6 reactivity between CD27− and CD27+ B lymphocytes (CD19 selected). The histogram plots compare the surface expression of CD5 (B), CD23 (C), CD38 (D), IgM (E), and IgD (F) in G6+ and G6− cells, gated as in panel A (thin line, G6− CD27+; thick line, G6− CD27−; shaded, G6+; dashed line, isotype controls). Histograms are normalized to the maximum for each peak.
Phenotypic profile of G6+ B cells from healthy donor D1. Peripheral blood mononuclear cells were analyzed by 4-color fluorescence-activated cell sorting. (A) Distribution of G6 reactivity between CD27− and CD27+ B lymphocytes (CD19 selected). The histogram plots compare the surface expression of CD5 (B), CD23 (C), CD38 (D), IgM (E), and IgD (F) in G6+ and G6− cells, gated as in panel A (thin line, G6− CD27+; thick line, G6− CD27−; shaded, G6+; dashed line, isotype controls). Histograms are normalized to the maximum for each peak.
The phenotype of the G6+ population was homogeneous and similar to that of G6− naive (CD27−) B cells, that is, IgM+ IgD+ CD23+ CD5−, and CD38+ (Figure 4). Although the majority of the naive and G6+ populations did not express CD5, each had a small percentage of CD5+ B cells not found in the memory B-cell subset (Figure 4B). Expression of CD23 (Figure 3C) distinguishes the naive/G6+ B cells from tonsillar or splenic marginal zone B cells.28 CD38 expression was similarly high in naive and G6+ populations and more heterogeneous in the CD27+ population (Figure 4D). The level of IgM expression in G6+ B cells was similar to G6− naive B cells and lower than CD27+ B cells (Figure 4E). In contrast, IgD expression was greater than CD27+ B cells and again similar to the G6− naive B cells (Figure 4F). Light chain expression was 69% Igκ and 35% Igλ, comparable with normal B cells and 51p1+ CLL (data not shown). Activation markers (CD25 and CD69) were absent (data not shown). It appears therefore that the 51p1-expressing B cells in blood are part of the conventional resting naive B-cell population.
Discussion
There have been many attempts to find the B-cell of origin of CLL by the use of phenotypic and immunogenetic analyses, a search that became even more challenging when 2 subsets were described on the basis of the mutational status of the IGHV genes.2,4 The more recent discoveries of common conserved sequences in the IGV regions, expressed in a significant proportion of cases of CLL, apparently distanced CLL further from normal B cells because similar sequences in the latter were reported to be vanishingly rare.29,30 It appeared that CLL was derived from antigen-driven B cells with no clear counterpart in the normal B-cell repertoire. However, one of the reasons for this is that IGHV sequences in the public databases are rarely from normal B cells and almost never from naive B cells, limiting comparative analysis.
The majority of conserved (stereotypic) sequences is found in U-CLL, where biased usage of IGHV genes, together with certain IGHD genes and selected IGHJ gene segments, leads to similar sequences in a proportion of cases.15 The most obvious example of bias in CLL is the 51p1 allele of the IGHV1-69 gene, which is rarely mutated and constitutes 13% of all CLL and 25% to 30% of U-CLL.15 Combination with certain IGHD genes, in particular RFs, and with IGHJ6 generates highly conserved stereotypic sequences of similar HCDR3 length.15,16,19,31,32 These features have not been noted in the database of normal B-cell sequences.18,19 Specific attempts to find counterparts in normal B cells have been made, commonly by amplifying 51p1 sequence together with mixed IGHJ or Cμ primers. The HCDR3 sequences produced were shorter than those characteristic of CLL, with a different repertoire of IGHD gene use, leading to the conclusion that CLL cells were different from normal B cells.30 Our own study used the same strategy to investigate 51p1 gene use in healthy elderly persons, and although HCDR3 lengths appeared to define 2 populations, the overall conclusion was again that normal B cells did not reflect CLL.13 An exception to this was a small study of 6 51p1-derived sequences from normal B cells where all happened to be derived from IGHJ6 and where HCDR3 lengths were similar to those in CLL.20 Taken together, the CLL-like sequences were found mainly where IGHJ6 was used, and because these account for only 28% of 51p1-derived sequences, combinations with other IGHJ genes, especially IGHJ4, were dominating the analysis.13
We have now focused only on the 51p1-derived sequences combined to IGHJ6 in age-matched healthy subjects, and it is immediately clear that this larger database reveals the sequence counterparts of CLL. Stereotypic sequences of several of the major subsets described in CLL are identifiable, as well as counterparts of new stereotypes. In addition, stereotypic sequences not yet described in CLL are detectable. In total, 33.3% of the sequences could be assigned to subsets. This finding demonstrates that conserved sequences are characteristic of this fraction of the normal blood B cells and are a likely source of transformation to U-CLL.
However, many other non-IGHJ6 subsets may exist. Because we focused on IGHJ6, the striking stereotypic sequence in CLL derived from the IGHD3-16 gene (RF 2) combined with IGHJ316 was not detected and is being sought separately. In IGHJ6-derived CLL stereotypes, there is less similarity in these junctional amino acids between cases of CLL, and this variability also was observed in normal B cells.15,16,19,32 It should be noted also that B cells using the IGHV1-69 gene, usually mutated, are expanded in other B-cell proliferative diseases, particularly those associated with hepatitis C infection.33-35 However, these appear rarely to involve IGHJ6 or to display the conserved sequence characteristic of CLL.33-35
Conserved sequences in U-CLL could imply a common antibody activity. Autoreactivity has been noted for 51p1-derived IgM obtained from CLL cases, with 1 CLL case in subset 5 (IGHV1-69/IGHD3-10/IGHJ6) reacting with stomach chief cells and proline-rich acidic protein 1 expressed by apoptotic cells.36 IgM from another 3 CLL cases derived from subset 7 (IGHV1-69/IGHD3-3/IGHJ6) also bound to apoptotic cells.37 Although autoreactivity is complex and polyreactivity is common among IgM from U-CLL,38 there is a suggestion that IgM from CLL cases within subsets may be recognizing similar cytoplasmic structures.38 Perhaps more importantly, at least one IGHV1-69–encoded CLL-derived IgM recognized polysaccharide derived from Streptococcus pneumoniae.36 If the closely similar IgMs from normal B cells mimic these reactivities, it would support the concept that the repertoire of conserved sequences is generated to deal with common pathogens or possibly apoptotic cell clearance.
It has been proposed previously that natural antibodies constitute an innate repertoire or “natural memory” able to recognize common bacteria.39 A panel of 7 IgM antibodies against capsular pneumococcal polysaccharides from healthy subjects was reported to express a range of unmutated IGHV genes with 6 of 7 combined to IGHJ6.40 IgM antibodies against pneumococcal polysaccharide are known to be produced by transitional B cells in the spleen via nonspecific stimulation with CpG.39 Human transitional B cells are considered to be precursors of naive B cells and possibly of IgM memory B cells. Taken together, there is a possibility that the precursors of at least the 51p1-derived subset of U-CLL could reside in this population. If that is the case, the cells must circulate as our analysis was confined to blood.
The phenotypic analysis of the G6+ population, which would include all 51p1-derived B cells, located these cells in the conventional resting naive, or possibly transitional, B-cell subset. Features reflect those of CD23+/CD27− normal B cells, which comprise approximately 35% of total B cells throughout life and include 20% to 25% of CD5+ B cells.41 The phenotype does not closely mirror that of CLL cells, especially with respect to CD5 expression, evident only on a small proportion of these normal B cells. Expression of CD5 is known to be modulated by B-cell receptor engagement and, in the mouse, is increased in anergic B cells, where it controls the signaling threshold.42 Regulation of expression of CD5 in human B cells is complex but is again influenced by B-cell receptor engagement and is down regulated by interleukin-6.43 Perhaps it is therefore not surprising that CD5 expression in naive G6+ normal B cells does not match that of CLL. Not only are these B cells transformed, but they are also apparently interacting with antigen.9 Whatever the origin of CLL, the normal B cells with similar features in their immunoglobulin genes are likely to have an evolutionary past that equips the human population to fight infection. We speculate that stimulation of these B cells can give rise to U-CLL, and we will seek confirmation in patients with specific infections. The question is whether further infection drives tumor cells, and suspicion that this is likely is already developing.44 The observation that CLL cells are engaging antigen in vivo with consequent down-regulation of sIgM suggests that there is a persistent “antigen” involved.9 It will be important to identify this antigen and to block the apparently stimulatory interaction which may be maintaining CLL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Prof Roy Jefferis for kindly supplying the G6 monoclonal antibody.
This study was supported by Tenovus United Kingdom, Tenovus Solentside, Cancer Research UK; Siena-AIL Onlus; Associazione Italiana per la Ricerca sul Cancro (AIRC); and the General Secretariat for Research and Technology of Greece (Program INA-GENOME).
Authorship
Contribution: F.K.S. supervised; F.F., K.P., and F.K.S. designed the study; F.F. and K.P. performed research; F.F., K.P., N.D., K.S., and F.K.S. performed analysis; E.S. and I.W. performed IGHV analysis; C.I.M. performed phenotypic analysis; C.I.M., F.F., K.P., N.D., K.S., and F.K.S. interpreted data; and F.F., K.P., G.P., and F.K.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Freda Stevenson, Genetic Vaccine Group, Cancer Sciences Division, Southampton University Hospitals Trust, Southampton General Hospital, Tremona Rd, Southampton, SO16 6YD, United Kingdom; e-mail: fs@soton.ac.uk.
References
Author notes
*F.F. and K.N.P. contributed equally to this work.