Abstract
Somatic mutations in TET2 occur in patients with myeloproliferative neoplasms and other hematologic malignancies. It has been suggested that TET2 is a tumor suppressor gene and mutations in TET2 precede the acquisition of JAK2-V617F. To examine the order of events, we performed colony assays and genotyped TET2 and JAK2 in individual colonies. In 4 of 8 myeloproliferative neoplasm patients, we found that some colonies with mutated TET2 carried wild-type JAK2, whereas others were JAK2-V617F positive, indicating that TET2 occurred before JAK2-V617F. One of these patients carried a germline TET2 mutation. However, in 2 other patients, we obtained data compatible with the opposite order of events, with JAK2 exon 12 mutation preceding TET2 mutation in one case. Finally, in 2 of 8 patients, the TET2 and JAK2-V617F mutations defined 2 separate clones. The lack of a strict temporal order of occurrence makes it unlikely that mutations in TET2 represent a predisposing event for acquiring mutations in JAK2.
Introduction
TET2, a member of the ten-eleven-translocation (TET) family of genes,1,2 can be mutated in various hematopoietic disorders, including myeloprolieferative neoplasms (MPNs), myelodysplastic syndromes, acute myeloid leukemia, and chronic myelomonocytic leukemia.3-11 TET2 is mutated in 13% of the MPN cases, with the highest frequency occurring in patients with primary myelofibrosis (PMF) and polycythemia vera (PV; 17% and 16%, respectively) and lowest in patients with essential thrombocythemia (ET; 5%).3 The mutations in TET2 do not cluster in a particular region and show a very diverse pattern of frame shift, nonsense, and missense mutations. The TET2 mutations are generally present in a heterozygous state, and only a minority of patients displays mutations in both TET2 alleles. A recent study showed that loss of heterozygosity in TET2 might occur through mitotic recombination or gene copy number changes.9 The loss of both gene copies through mutations and chromosomal aberrations is compatible with a potential tumor suppressor activity of TET2. However, the mechanism of how TET2 is involved in disease initiation and/or progression is unknown.
Methods
Patients
The collection of blood samples was performed at the study center in Basel, Switzerland, and was approved by the Ethik Kommission Beider Basel. Written consent was obtained from all patients in accordance with the Declaration of Helsinki. The diagnosis of MPN was established according to the criteria of the World Health Organization.12-14
Cells and DNA analysis
Purification of granulocytes, peripheral mononuclear cells, extraction of DNA, and colony assays were performed as described.15-17 Methylcellulose-based media (no. 4431 and no. 4434) containing erythropoietin and methylcellulose based media without erythropoietin (no. 4531; StemCell Technologies) were used. The cells were plated at a density of 100 000 cells/mL, which provided an optimal density of colonies for picking, without the danger of contamination by neighboring colonies. Primers used for sequencing, copy number analysis, and X chromosomal inactivation analyses are provided in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) and were performed as described.17-20
Results and discussion
To study the order of events in the clonal evolution of TET2 and JAK2 mutations, we examined individual colonies derived from peripheral blood of 8 MPN patients with TET2 mutations (6 PV, 1 ET, and 1 PMF) that were identified by sequencing of DNA from 57 MPN patients. All patients with TET2 mutation were also positive for JAK2-V617F (Table 1).
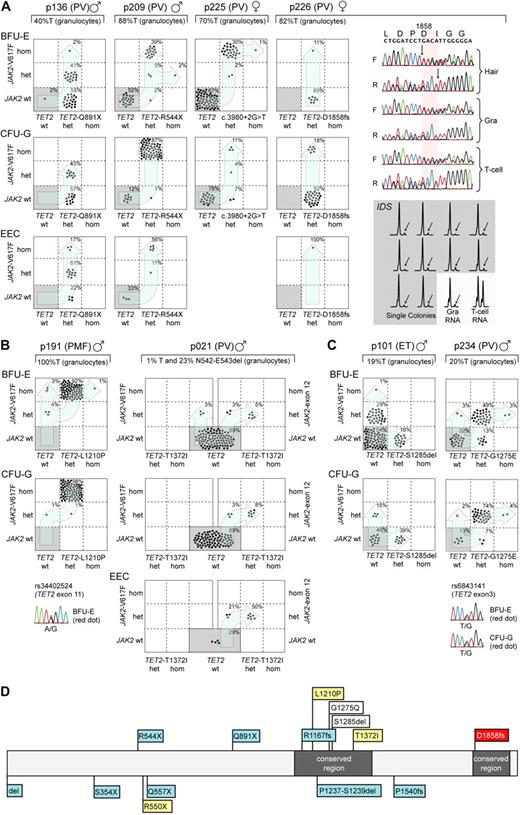
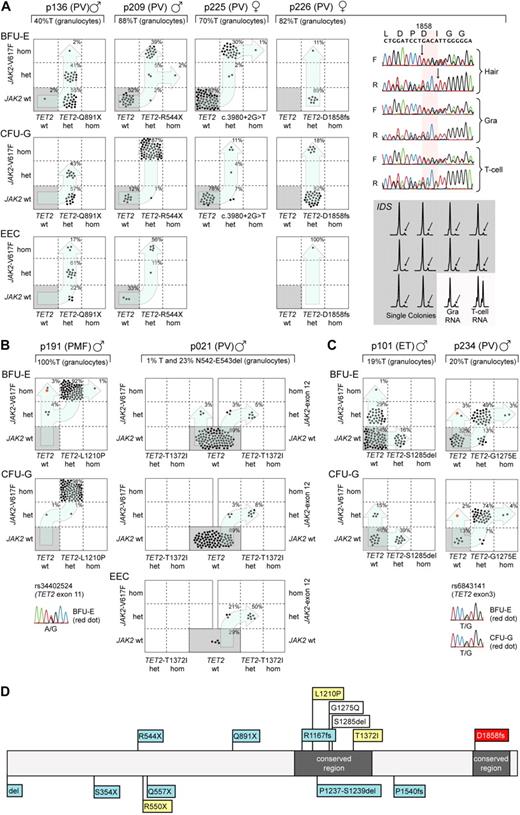
Mutational analysis of single colonies allowed us to distinguish 3 different patterns of mutation accumulation (Figure 1): In 4 of 8 MPN patients, we found that some colonies with mutated TET2 carried wild-type JAK2, whereas others were JAK2-V617F+, indicating that TET2 occurred before the acquisition of JAK2-V617F (Figure 1A). These 4 patients confirm previous data by Delhommeau et al,3 who showed that TET2 occurs before JAK2-V617F. A second group showed a pattern compatible with the inverse order of events, ie, JAK2 mutations occurred before the acquisition of TET2 mutations (Figure 1B): patient p191 displayed colonies positive for JAK2-V617F without the TET2 mutation, and all colonies with mutated TET2 were also positive for JAK2-V617F. Unfortunately, 9 single nucleotide polymorphisms (SNPs) in the vicinity of JAK2-V617F were noninformative to provide evidence for a possible biclonal acquisition of JAK2-V617F. The transition from heterozygous to homozygous JAK2-V617F appears to have independently occurred twice. Indeed, the analysis of individual colonies homozygous for JAK2-V617F revealed the presence of 2 subclones with different sizes of the 9p uniparental disomy (UPD) region (supplemental Figure 1A). By analyzing an informative SNP in exon 11 of TET2 (Figure 1B), we can exclude the possibility that the JAK2-V617F positive and TET2-L1210P-negative colonies arose through mitotic recombination, in which the TET2-L1210P was lost. In patient p021, 2 independent clones were present: a smaller clone positive for JAK2-V617F and a larger clone positive for a JAK2 exon 12 mutation (JAK2-N542-E543del).21,22 Interestingly, the TET2 mutation in this patient occurs only in combination with the JAK2 exon 12 mutation, and the TET2 mutation in this patient was acquired after the JAK2 exon 12 mutation. A third pattern consisted of biclonal disease, as illustrated in patient p101 with colonies positive either for JAK2-V617F or TET2, but absence of double-positive colonies (Figure 1C). A similar biclonal pattern was previously described in one patient with del20q and JAK2-V617F, consistent with the presence of a predisposition to independently acquire 2 otherwise rare somatic events.23 Patient p234 also shows a pattern compatible with biclonal disease. However, to progress to the double-positive stage, either JAK2 or TET2 must have mutated twice independently. Again, SNP analysis excluded the possibility that the JAK2-V617F+ and TET2-G1275E− colonies arose through mitotic recombination (Figure 1C). Because it is unlikely that TET2 independently mutated twice at the exact same position (TET2-G1275E), we conclude that the majority of colonies first acquired TET2-G1275E followed by JAK2-V617F. A second independent event produced the subclone that is positive for JAK2-V617F only. The SNPs in close proximity (rs10974944 and rs12343867) were noninformative and were not part of the recently described GGCC or 46/1 JAK2 haplotype (data not shown).24,25
Analysis of single colonies for mutations in TET2 and JAK2. Mononuclear cells from peripheral blood were grown in methylcellulose in the presence or absence of erythropoietin. Single burst-forming units erythroid (BFU-E), endogenous erythroid colonies (EEC), and colony-forming units granulocyte (CFU-G) were picked and analyzed individually for the presence of TET2 and JAK2-V617F mutations by DNA sequencing and allele-specific polymerase chain reaction (PCR), respectively. Each colony is represented by a dot that is placed into one of 6 quadrangles representing the 6 possible genotypes: wild-type (wt), heterozygous (het), and homozygous (hom) for JAK2-V617F on the vertical axis, and for TET2 mutations on the horizontal axis. The unique patient numbers, the diagnoses (PMF, ET, and PV) and the allelic ratio of JAK2-V617F in purified granulocytes (%T) are shown above the corresponding boxes. Light blue arrows indicate the suggested order of mutation events. (A) Patterns compatible with TET2 mutations occurring before JAK2-V617F. The sequencing chromatograms for patient p226 show the presence of TET2 mutation in DNA from hair follicles, T cells, and granulocytes, demonstrating the germline origin of the mutation. Allele-specific PCR assay for the X-chromosomal gene IDS is shown for p226. The genomic DNA from patient p226 was heterozygous for a C/T single nucleotide polymorphism (not shown). The relative expression of the 2 IDS alleles was determined by comparing the C and T peak intensities obtained by the allele-specific reverse-transcribed PCR assay in T cells and granulocytes. The skewing of expression toward the C-allele is shown for 10 individual colonies (gray area). The inactivated IDS allele is marked with an arrow. (B) Patterns compatible with JAK2 mutations occurring before TET2 was mutated. Patient p021 carries 2 JAK2 mutations, JAK2-V617F and JAK2 N542-E543del, but the TET2 mutation can be only found together with the deletion in exon 12 of JAK2. In patient p191, the sequencing chromatogram for the SNP rs34402524 located in TET2 is shown for one BFU-E colony marked in red. The presence of a heterozygous SNP excludes the possibility that this colony is the product of a mitotic recombination event. (C) Patterns compatible with a biclonal state of the disease. In patient p234, the sequencing chromatogram for the SNP rs6843141 located in TET2 is shown for one BFU-E and one CFU-G colony marked in red. Again, the presence of a heterozygous SNP excludes the possibility that these colonies are the product of a mitotic recombination event. (D) Location of mutations in the Tet2 protein in patients from whom data on single colonies are available. Mutations from this study are shown above the protein strand, and mutations analyzed in previous publications3,26 are shown below. The gray boxes represent regions conserved between the different TET family members; blue boxes, TET2 mutations that occur before JAK2 mutations; yellow boxes, TET2 mutations that occurred after JAK2; white boxes, TET2 and JAK2 mutations compatible with biclonal disease; and red box, germline TET2 mutation.
Analysis of single colonies for mutations in TET2 and JAK2. Mononuclear cells from peripheral blood were grown in methylcellulose in the presence or absence of erythropoietin. Single burst-forming units erythroid (BFU-E), endogenous erythroid colonies (EEC), and colony-forming units granulocyte (CFU-G) were picked and analyzed individually for the presence of TET2 and JAK2-V617F mutations by DNA sequencing and allele-specific polymerase chain reaction (PCR), respectively. Each colony is represented by a dot that is placed into one of 6 quadrangles representing the 6 possible genotypes: wild-type (wt), heterozygous (het), and homozygous (hom) for JAK2-V617F on the vertical axis, and for TET2 mutations on the horizontal axis. The unique patient numbers, the diagnoses (PMF, ET, and PV) and the allelic ratio of JAK2-V617F in purified granulocytes (%T) are shown above the corresponding boxes. Light blue arrows indicate the suggested order of mutation events. (A) Patterns compatible with TET2 mutations occurring before JAK2-V617F. The sequencing chromatograms for patient p226 show the presence of TET2 mutation in DNA from hair follicles, T cells, and granulocytes, demonstrating the germline origin of the mutation. Allele-specific PCR assay for the X-chromosomal gene IDS is shown for p226. The genomic DNA from patient p226 was heterozygous for a C/T single nucleotide polymorphism (not shown). The relative expression of the 2 IDS alleles was determined by comparing the C and T peak intensities obtained by the allele-specific reverse-transcribed PCR assay in T cells and granulocytes. The skewing of expression toward the C-allele is shown for 10 individual colonies (gray area). The inactivated IDS allele is marked with an arrow. (B) Patterns compatible with JAK2 mutations occurring before TET2 was mutated. Patient p021 carries 2 JAK2 mutations, JAK2-V617F and JAK2 N542-E543del, but the TET2 mutation can be only found together with the deletion in exon 12 of JAK2. In patient p191, the sequencing chromatogram for the SNP rs34402524 located in TET2 is shown for one BFU-E colony marked in red. The presence of a heterozygous SNP excludes the possibility that this colony is the product of a mitotic recombination event. (C) Patterns compatible with a biclonal state of the disease. In patient p234, the sequencing chromatogram for the SNP rs6843141 located in TET2 is shown for one BFU-E and one CFU-G colony marked in red. Again, the presence of a heterozygous SNP excludes the possibility that these colonies are the product of a mitotic recombination event. (D) Location of mutations in the Tet2 protein in patients from whom data on single colonies are available. Mutations from this study are shown above the protein strand, and mutations analyzed in previous publications3,26 are shown below. The gray boxes represent regions conserved between the different TET family members; blue boxes, TET2 mutations that occur before JAK2 mutations; yellow boxes, TET2 mutations that occurred after JAK2; white boxes, TET2 and JAK2 mutations compatible with biclonal disease; and red box, germline TET2 mutation.
In 4 of 8 patients, we observed a small number of colonies that were homozygous for the TET2 mutations. Gene copy number analysis revealed that p209 and p191 retained 2 copies of the TET2 gene, whereas in p225 loss of one copy of the TET2 gene was found, indicating that the normal TET2 allele was lost through a deletion (supplemental Figure 1B). In patient p234, homozygosity was achieved through UPD in some colonies and deletion in other colonies. Colonies homozygous for the TET2 mutations from all 4 patients displayed loss of heterozygosity of SNPs or microsatellite markers in the TET2 locus, as expected for a deletion or UPD at chromosome 4q (supplemental Figure 1B).
The heterozygous TET2-D1858fs mutation in patient p226 was also present in DNA from hair roots, indicating that the mutation was germline. This 4-bp deletion, located in the C-terminal conserved domain of TET2, results in a frame shift and premature stop and is probably functionally relevant. The same mutation was also found in buccal DNA from an asymptomatic sister of p226 (data not shown). This appears to be the first report of a germline mutation in TET2. Accordingly, all colonies in this patient were positive for the TET2 mutation. The X-chromosome inactivation pattern in individual colonies from p226 with wild-type JAK2, as determined by scoring a C/T polymorphism in the 3′-untranslated region of the IDS mRNA,18-20 revealed a strong skewing (10 of 10 expressed the C allele of IDS), indicating that these progenitors were of clonal origin (Figure 1A). The finding of clonality suggests that this patient has a second significant disease clone, which does not carry a mutation in JAK2.
We showed that a JAK2 exon 12 mutation preceded the TET2 mutation in p021. In addition, the data in p191 are compatible with JAK2-V617F preceding TET2. A similar conclusion was reached in one patient with familial MPN positive for TET2 and JAK2-V617F mutations.26 In 4 of 8 patients, some colonies carried a homozygous TET2 mutation that was the result of the loss of the wild-type allele through deletion or UPD. The percentage of such homozygous colonies in all 4 patients was very low (< 5%), opening the possibility that the loss of the wild-type TET2 may not provide an additional competitive advantage. The location of the TET2 mutation, which has been analyzed using single colony assays, is summarized in Figure 1D. Most of the TET2 frame shift and nonsense mutations occurred in patients in whom TET2 preceded the acquisition of JAK2. The lack of a strict temporal order of occurrence resembles the findings obtained for del20q and makes it unlikely that mutations in TET2 represent a predisposing event for acquiring JAK2.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Michael Medinger for contributing clinical data and Primo Schär and Jürg Schwaller for helpful comments on the manuscript.
This work was supported by the Swiss National Science Foundation (grant 310000-108006/1) and the Swiss Cancer League (grant OCS-01742-08-2005; R.C.S.).
Authorship
Contribution: F.X.S. performed research, analyzed data, and wrote the paper; R.L., S.L., and H.H.-S. performed research; T.L. and A.T. provided clinical data; and R.C.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Radek C. Skoda, Department of Biomedicine, Experimental Hematology, University Hospital Basel, Hebelstr 20, 4031 Basel, Switzerland; e-mail: radek.skoda@unibas.ch.