Abstract
DLK1 is an imprinted gene on chromosome 14. Using informative coding single nucleotide polymorphisms, we found DLK1 expression to be monoallelic in normal bone marrow, whereas it was biallelic in 76% of acute myeloid leukemia (AML) overexpressing DLK1 (61% of all AML). Quantitative methylation analysis of 7 cytosine-phosphate-guanosine-rich areas (3 upstream of or within DLK1, the putative intergenic-differentially methylated region and 3 upstream of or within MEG3) revealed a strong association between biallelic DLK1 expression and hypermethylation of a cytosine-phosphate-guanosine-rich region 18 kb upstream of DLK1. Allele-specific methylation analysis of this region revealed the alleles to be differentially methylated in normal bone marrow and monoallelic DLK1 AML, whereas there was increased methylation of both alleles in AML with biallelic expression. Moreover, chromatin immunoprecipitation analysis revealed that CCTC-binding factor binds to this region in monoallelic but not biallelic expression samples. Taken together, our data indicate that an insulator located 18 kb upstream of DLK1 plays an important role in regulating DLK1 imprinting.
Introduction
DLK1 is a transmembrane protein of the epidermal growth factor–like family. Generally, DLK1 functions as a growth factor that maintains proliferating cells in an undifferentiated state.1 In hematopoiesis, DLK1 is important in supporting the proliferation of early progenitors.2 DLK1 overexpression has been found in many cancers, including myelodysplastic syndrome and acute myeloid leukemia (AML).3-5
The DLK1 gene is located within an imprinted region of chromosome 14q32 and expressed only from the paternal allele in normal cells.6 In addition to DLK1, this region contains other paternally expressed genes, including Dio3 and RTL1, and a set of maternally expressed ones, including Meg3, C/D snoRNA, and Mirg.7 We previously showed IGF-2 overexpression in some cases of AML was the result of loss of imprinting.8 The IGF-2 locus at 11p15 has many similarities with the imprinted DLK1 14q32 locus.9,10 In this study, we found that DLK1 loss of imprinting is associated with increased DLK1 expression and hypermethylation of a region located approximately 18 kb upstream of DLK1. Furthermore, we found that acquired methylation of this region inhibits CTCF binding to this region, suggesting that this region is an insulator element and is important in regulating DLK1 imprinting.
Methods
DLK1 transcript levels were quantified by real-time reverse-transcribed polymerase chain reaction. To determine whether DLK1 expression is monoallelic or biallelic, we studied 3 coding single nucleotide polymorphisms (SNPs; rs#1802710, rs#2295660, and rs#1058009) of 11 normal bone marrows (NBMs), 3 cell lines overexpressing DLK1 (OCI/AML5, K562, and NB4), and 40 AML patient samples with DLK1 overexpression. The Mann-Whitney test was used to compare DLK1 and MEG3 transcript levels. Quantitative methylation was performed using the MassARRAY EpiTYPER.11 Seven cytosine-phosphate-guanosine (CpG)–rich regions were analyzed (Figure 2A): 3 upstream of or within DLK1 (D1, D2, and D3), one corresponding to the putative intergenic differentially methylated region (IG-DMR; named IG), and 3 upstream or within MEG3 (M1, M2, and M3). The methylation values at each CpG dinucleotide were compared using analysis of variance. Further, the methylation data were subjected to unsupervised hierarchical cluster analysis, using MeV 4.1.01 software. Informative SNPs were identified in the D1 region by sequencing and the allelic methylation pattern of the D1 region analyzed, using bisulfite conversion and sequencing of at least 18 individual clones. Chromatin immunoprecipitation (ChIP) assay was used to analyze whether CTCF interacts in vivo with the D1 region in a methylation-dependent manner.
Results and discussion
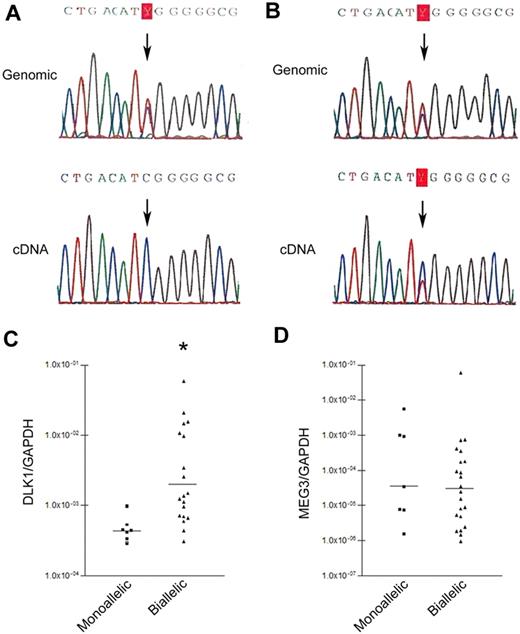
DLK1 overexpression was present in 3 cell lines (OCI/AML5, NB4, and K562) and 80% of patient cases (supplemental Figures 1-2, available on the Blood website; see the Supplemental Materials link at the top of the online article). Heterozygous SNPs (Figure 1A) were found in 6 of 11 NBMs, one cell line (K562), and 29 of 40 patient samples. The NBMs with informative SNPs had monoallelic expression, whereas biallelic expression was found in the K562 cell line and 22 of 29 (76%) patient samples (Figure 1A-B); the level of DLK1/glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was higher in cases with biallelic versus monoallelic expression (median, 0.002046 vs 0.000443, P < .002; Figure 1C).
DLK1 allelic expression patterns in AML. (A) Monoallelic DLK1 expression. Genomic DNA sequencing (top) showing informative SNP at rs#1802710; cDNA sequencing (bottom) shows monoallelic expression (part #8107). (B) Biallelic DLK1 expression. Genomic DNA sequencing (top) showing informative SNP at rs#1802710; cDNA sequencing (bottom) shows biallelic expression (part #8101). (C) Comparison of DLK1 expression in patients with monoallelic (7 patients) and biallelic (22 patients) DLK1 expression. *P < .002. DLK1 levels were normalized using GAPDH. (D) Comparison of MEG3 expression in patients with monoallelic and biallelic DLK1 expression (P = .52). MEG3 levels were normalized using GAPDH.
DLK1 allelic expression patterns in AML. (A) Monoallelic DLK1 expression. Genomic DNA sequencing (top) showing informative SNP at rs#1802710; cDNA sequencing (bottom) shows monoallelic expression (part #8107). (B) Biallelic DLK1 expression. Genomic DNA sequencing (top) showing informative SNP at rs#1802710; cDNA sequencing (bottom) shows biallelic expression (part #8101). (C) Comparison of DLK1 expression in patients with monoallelic (7 patients) and biallelic (22 patients) DLK1 expression. *P < .002. DLK1 levels were normalized using GAPDH. (D) Comparison of MEG3 expression in patients with monoallelic and biallelic DLK1 expression (P = .52). MEG3 levels were normalized using GAPDH.
Previous reports suggested that the expression of Meg3 and Dlk1 is inversely regulated in mice, with the noncoding Meg3 controlling Dlk1 transcription and imprinting.7 To determine whether this occurs in AML, we quantified MEG3 and DLK1 RNA levels in 6 NBMs, 29 patient samples (22 with biallelic and 7 with monoallelic DLK1 expression), and K562 cells. NBMs expressed low MEG3/GAPDH (median, 4.7 × 10−6; range, 4.6 × 10−5-1.9 × 10−6). MEG3/GAPDH levels were quite variable in AML patients with biallelic or monoallelic DLK1 expression. There was no significant difference between the 2 groups (median, 0.000035 vs 0.0000305, P = .52; Figure 1C). Further, an inverse correlation was not found between DLK1 and MEG3 levels in AML patients regardless of monoallelic or biallelic DLK1 expression (r = −0.1212; supplemental Figure 3). Finally, K562, characterized by high DLK1 expression, showed high MEG3 expression (MEG3/GAPDH of 0.064). Our data indicate that DLK1 transcription is not controlled by MEG3 in human AML.
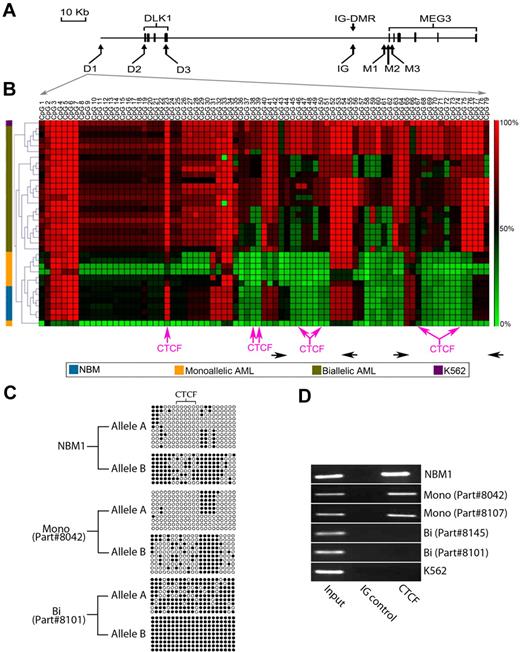
In mice, the DMRs located between Dlk1 and Meg3, particularly the IG-DMR, are important for Dlk1 imprinting; when unmethylated, they bind to the insulator protein CTCF, blocking access of the Dlk1 promoter to downstream enhancers. In contrast, their hypermethylation leads to loss of binding, permitting the interaction between the Dlk1 promoter and downstream enhancers.7,12,13 To determine whether changes in methylation of the IG-DMR and other CpG islands upstream and downstream of DLK1 were associated with the loss of DLK1 imprinting, we assessed the methylation of 7 CpG-rich regions (Figure 2A). Similar methylation patterns of all areas were observed in all 6 NBMs. In contrast, AML patient samples showed variability of methylation in most areas tested. The greatest difference in methylation between NBM, monoallelic, and biallelic patient samples was evident in region D1, with analysis of variance showing significant difference at all CpG dinucleotides, except dinucleotides 3 to 7, 33, 34, and 57 (P < .001 in most of them). Moreover, in cluster analysis, the D1 region only showed patterns that distinguish between biallelic from monoallelic patients and NBM samples (Figure 2B; supplemental Figures 4-9).
Differential methylation of the upstream of DLK1 insulator. (A) Map of the CpG-rich areas analyzed. The D1, D2, and D3 regions are located at −18 210 to −17 139 bp, −306 to +203 bp, and +7287 to +7704 bp relative to the DLK1 start site, respectively. The IG area represents the putative IG-DMR, located approximately 15 kb upstream of the MEG3 start site. The M1, M2 and M3 regions are located −2014 to −1516 bp, −299 to +289 bp, and +1219 to +1919 bp relative to the MEG3 start site, respectively. The bold boxes represent the exons in DLK1 and MEG3 genes. (B) Cluster analysis of the methylation of the region D1 in 6 NBMs (blue), 7 AML patient samples with DLK1 monoallelic expression (yellow), 22 AML patient samples with biallelic (olive green), and the K562 cell lines (purple). Pink arrows indicate the site of predicted CTCF-binding sites (detailed in supplemental Figure 14). Horizontal black arrows indicate the location of primers used for DNA amplification after ChIP. (C) This figure shows allele-specific methylation of an assessable region within the D1 region (located between CpG40 and CpG60) in one NBM (NBM1), one AML case with monoallelic DLK1 expression (part #8042), and one AML case with biallelic DLK1 expression (part #8101). The relatively hypomethylated allele was designated as allele A, whereas the hypermethylated allele was designated as allele B. The different alleles are differentially methylated with the CTCF-binding site completely unmethylated in allele A and partially methylated in allele B in the NBM sample (NBM1) and in the monoallelic DLK1 patient (part #8042). In contrast, there is increased methylation of both alleles in the patient with biallelic DLK1 expression (part #8101) with acquisition of methylation in the CTCF site of the hypomethylated allele. (D) ChIP with CTCF antibody showing that the CTCF antibody was able to precipitate the D1 region in one NBM (NBM1) and 2 AML samples with monoallelic DLK1 expression (indicated as Mono 1 and 2 with the parts #8042 and #8107, respectively), whereas there was no precipitation in 2 patient samples with biallelic DLK1 expression (indicated as Bi 1 and 2 with the parts #8145 and #8101, respectively) and the K562 cell line. Two DNA areas were amplified after ChIP as shown in panel B, and similar results were obtained from both areas.
Differential methylation of the upstream of DLK1 insulator. (A) Map of the CpG-rich areas analyzed. The D1, D2, and D3 regions are located at −18 210 to −17 139 bp, −306 to +203 bp, and +7287 to +7704 bp relative to the DLK1 start site, respectively. The IG area represents the putative IG-DMR, located approximately 15 kb upstream of the MEG3 start site. The M1, M2 and M3 regions are located −2014 to −1516 bp, −299 to +289 bp, and +1219 to +1919 bp relative to the MEG3 start site, respectively. The bold boxes represent the exons in DLK1 and MEG3 genes. (B) Cluster analysis of the methylation of the region D1 in 6 NBMs (blue), 7 AML patient samples with DLK1 monoallelic expression (yellow), 22 AML patient samples with biallelic (olive green), and the K562 cell lines (purple). Pink arrows indicate the site of predicted CTCF-binding sites (detailed in supplemental Figure 14). Horizontal black arrows indicate the location of primers used for DNA amplification after ChIP. (C) This figure shows allele-specific methylation of an assessable region within the D1 region (located between CpG40 and CpG60) in one NBM (NBM1), one AML case with monoallelic DLK1 expression (part #8042), and one AML case with biallelic DLK1 expression (part #8101). The relatively hypomethylated allele was designated as allele A, whereas the hypermethylated allele was designated as allele B. The different alleles are differentially methylated with the CTCF-binding site completely unmethylated in allele A and partially methylated in allele B in the NBM sample (NBM1) and in the monoallelic DLK1 patient (part #8042). In contrast, there is increased methylation of both alleles in the patient with biallelic DLK1 expression (part #8101) with acquisition of methylation in the CTCF site of the hypomethylated allele. (D) ChIP with CTCF antibody showing that the CTCF antibody was able to precipitate the D1 region in one NBM (NBM1) and 2 AML samples with monoallelic DLK1 expression (indicated as Mono 1 and 2 with the parts #8042 and #8107, respectively), whereas there was no precipitation in 2 patient samples with biallelic DLK1 expression (indicated as Bi 1 and 2 with the parts #8145 and #8101, respectively) and the K562 cell line. Two DNA areas were amplified after ChIP as shown in panel B, and similar results were obtained from both areas.
Using informative SNPs within the D1 region (supplemental Figure 10) and bisulfite sequencing, it was possible to determine the methylation pattern of each allele in 3 NBMs, 3 monoallelic AMLs, and 6 biallelic AMLs. The NBMs showed notable difference of the methylation between the 2 alleles (Figure 2C; supplemental Figure 11). A similar pattern was found in AML with monoallelic DLK1 expression (Figure 2C; supplemental Figure 12). Interestingly, the biallelic expression cases showed increased methylation of both alleles (Figure 2C; supplemental Figure 13). These methylation patterns suggest that the D1 area may function as a methylation-sensitive, cis-acting insulator that regulates DLK1 imprinting.
Because many insulator elements use the CTCF protein to achieve their function, we performed a bioinformatic search that revealed numerous potential CTCF-binding sites within the D1 region (Figure 2B; supplemental Figure 14). Of note, the CpG dinucleotides located within most of these binding sites were hypermethylated in patient samples with biallelic DLK1 expression and K562 cells compared with NBMs and monoallelic DLK1 expression AML (Figure 2B). Furthermore, the allele-specific methylation analysis showed that there was complete unmethylation of the CTCF-binding site of the hypomethylated allele in NBM and monoallelic AML cases, whereas at least one CpG was methylated within the CTCF-binding site of the other allele (Figure 2C; supplemental Figures 11-12). In contrast, there was increased methylation of the CTCF-binding site in both alleles of patient samples with biallelic DLK1 expression (Figure 2C; supplemental Figure 13).
To confirm that CTCF interacts in vivo with the D1 region in a methylation-dependent manner, we performed ChIP assays with CTCF antibody for 6 cases (3 with hypometlyation and 3 with hypermethylation of the D1 region). Indeed, CTCF binds to this region in cases with D1 hypomethylation (one NBM and 2 AML patients with monoallelic DLK1 expression; Figure 2D). In contrast, CTCF antibody did not coprecipitate the D1 region from 3 cases with D1 hypermethylation (2 AML patients with biallelic DLK1 expression and K562), suggesting that the insulator protein CTCF plays an important role in the regulation of DLK1 imprinting through methylation-dependent interaction with this region.
In conclusion, based on our observations reported herein, we propose that the CpG-rich area located approximately 18 kb upstream of DLK1 acts as an insulator of DLK1 imprinting. When it is hypomethylated, it binds to CTCF, preventing the interaction between DLK1 promoter and enhancers. In contrast, the hypermethylation of this region results in the loss of this binding, creating a condition more permissive for transcription.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Leukemia & Lymphoma Society of Canada and the Campbell Family Cancer Research Institute.
Authorship
Contribution: H.K. designed the research, performed the experiments, analyzed the data, and drafted the manuscript; F.S.-S. contributed to the data analysis; S.W. helped perform the experiments; and M.D.M. participated in the initiation and design of the research, provided supervision, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mark Minden, Department of Stem Cell Biology, Ontario Cancer Institute, 610 University Ave, Room, 9-111, Toronto, ON, Canada M5G 2M9; e-mail: mark.minden@uhn.on.ca.