Abstract
Hepcidin is the master regulatory hormone of systemic iron metabolism. Hepcidin deficiency causes common iron overload syndromes whereas its overexpression is responsible for microcytic anemias. Hepcidin transcription is activated by the bone morphogenetic protein (BMP) and the inflammatory JAK-STAT pathways, whereas comparatively little is known about how hepcidin expression is inhibited. By using high-throughput siRNA screening we identified SMAD7 as a potent hepcidin suppressor. SMAD7 is an inhibitory SMAD protein that mediates a negative feedback loop to both transforming growth factor-β and BMP signaling and that recently was shown to be coregulated with hepcidin via SMAD4 in response to altered iron availability in vivo. We show that SMAD7 is coregulated with hepcidin by BMPs in primary murine hepatocytes and that SMAD7 overexpression completely abolishes hepcidin activation by BMPs and transforming growth factor-β. We identify a distinct SMAD regulatory motif (GTCAAGAC) within the hepcidin promoter involved in SMAD7-dependent hepcidin suppression, demonstrating that SMAD7 does not simply antagonize the previously reported hemojuvelin/BMP-responsive elements. This work identifies a potent inhibitory factor for hepcidin expression and uncovers a negative feedback pathway for hepcidin regulation, providing insight into a mechanism how hepcidin expression may be limited to avoid iron deficiency.
Introduction
Hepcidin is an iron-regulated hepatic peptide hormone that controls systemic iron homeostasis. Iron excess or inflammatory cytokines stimulate hepcidin expression, leading to reduced plasma iron levels as the result of iron retention in macrophages and reduced intestinal iron absorption. Hypoxia, high erythropoietic activity, and iron deficiency inhibit hepcidin expression by largely unknown mechanisms to mobilize iron stores and increase iron absorption.1 Hepcidin exerts its function by binding to the iron efflux channel ferroportin, which is predominantly expressed on macrophages, intestinal enterocytes, and hepatocytes, causing ferroportin internalization and degradation.2 Hepcidin levels are inappropriately low in hereditary hemochromatosis, a disease caused by mutations in HFE,3 transferrin receptor 2,4 hemojuvelin (HJV, HFE2),5 or hepcidin itself.6 By contrast, constant induction of hepcidin by inflammatory cytokines is implicated in the pathogenesis of the anemia of inflammation, a disease commonly observed in hospitalized patients.7
Two major signaling pathways communicate systemic stimuli to activate hepcidin mRNA expression in hepatocytes. One is induced by bone morphogenetic proteins (BMPs), a group of cytokines of the transforming growth factor-β (TGF-β) family.8 BMP-mediated hepcidin activation involves BMP receptors at the cell surface, as well as the BMP coreceptor HJV.9,10 BMP-receptor interaction induces phosphorylation of receptor activated (R)-SMAD proteins and subsequent formation of active transcriptional complexes involving the co-SMAD factor, SMAD4. Two sequence motifs (the proximal BMP-RE1 and the distal BMP-RE2) within the human and murine hepcidin promoters are critical for the stimulation of hepcidin via HJV, BMP, and SMAD4.11,12 The BMP signaling pathway communicates systemic iron levels,13-15 maintains steady-state hepcidin expression, and contributes to the activation of hepcidin by inflammatory stimuli at the level of SMAD4.12,16,17 In addition, proinflammatory cytokines stimulate hepcidin transcription via the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway and a STAT binding motif proximal to the transcription start site.18,19
Although the hepcidin activating pathways are beginning to be understood, comparatively little is known about how hepcidin expression is suppressed by hypoxia and active erythropoiesis to allow adequate iron uptake. Growth differentiation factor-15, twisted gastrulation 1, and erythropoietin have been implicated in mediating hepcidin suppression in response to augmented hematopoietic activity,20-23 but their mode of involvement remains to be defined. Sensing of iron deficiency recently was linked to TMPRSS6, a protease shown to cleave HJV,24 and to the von Hippel-Lindau–hypoxia-inducible factor pathway.25 A further study by Braliou et al26 suggested that hypoxia-mediated hepcidin suppression requires 2-oxoglutarate–dependent oxygenases but is independent of hypoxia-inducible factor-1α. For all of the implicated negative regulators, it is unclear how repressive signals reach the hepcidin promoter.
In this work, we identify SMAD7 as a potent repressor of hepcidin transcription and define its mechanism of action. In addition, our data assign functional importance to the previously reported observation that SMAD7 and hepcidin are coregulated in the liver of iron-loaded mice,27 identifying a negative feedback loop that is initiated by activating signals.
Methods
Cell culture
The human hepatocarcinoma cell line Huh7 was cultured in Dulbecco modified Eagle medium (high glucose; Invitrogen) supplemented with 10% heat-inactivated low-endotoxin fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin (PAA). Primary murine hepatocytes were cultured in Williams medium E (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Invitrogen), 2mM l-glutamine (Cambrex), 100mM dexamethasone (Sigma-Aldrich), and 1% penicillin/streptomycin (Cambrex). Cell cultures were maintained at 37°C under 5% CO2.
High-throughput siRNA screening
For the siRNA screen we adapted the previously established cell-based dual luciferase assay12,16,18 to the 384-well plate format and high-throughput conditions. Optimization was performed to obtain sufficiently high luciferase counts to monitor both decreased and increased hepcidin promoter activity. For the screen we used the Protein Kinase siRNA library ThermoFisher siGenome (Dharmacon) targeting protein kinases and other related genes (779 genes). The library was arrayed in 384-well white plates (Greiner Bio-One), each well containing 2.5 pmol of a pool of 4 synthetic siRNA duplexes (final concentration in wells, 50nM). Positive controls included an siRNA pool directed against HFE (Dharmacon) and one single siRNA duplex targeting SMAD4 (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). As a negative control, scrambled siRNA pool (Dharmacon) was used. We performed reverse transfection of Huh7 cells by dispensing 15 μL of RPMI medium (Invitrogen) together with 0.05 μL of Dharmafect1 reagent (Dharmacon) to the siRNA-containing 384-well plates.
After 50 minutes of incubation at ambient temperature, Huh7 cells (6 × 103 per well) were added to the siRNA transfection mix in a 30-μL volume of complete culture medium. Twenty-four hours after siRNA transfection, the reporter plasmids were transfected by use of the TransIT reagent (Mirus). Transfection reagent and reporter plasmids were first diluted in RPMI, then mixed together and incubated for 30 minutes at room temperature. Each well was transfected by the use of 0.2 μL of TransIT together with 90 ng of wild-type [WT]_2.7kb hepcidin promoter construct and 20 ng of control plasmid containing the Renilla gene under the control of the CMV promoter. After 48 hours, the cells were lysed for 30 minutes by the use of 0.2% Triton X100 (Fluka Analytical). Afterward substrate solutions for Firefly (D-luciferin; Biosynth) and Renilla (Coelenterazine; Biosynth) luciferases were added to the plates, and luciferase activity was measured by use of the Mithras LB 940 luminometer (Berthold Technologies). All dispensing steps were performed with the use of Multidrop Combi dispensing systems (Thermo Scientific).
Hepcidin promoter analysis
Luciferase reporter construct containing a 2762-bp (2.7 kb; WT_2.7kb) fragment of the human hepcidin promoter as well as mutant derivatives with mutations in BMP-responsive element (BMP-RE) 1 (position −84/−79; BMP_RE1_2.7 kb), BMP-RE2 (position −2255/−2250; BMP_RE2_2.7 kb), in BMP-RE1 and BMP-RE2 (BMP_RE1_BMP_RE2_2.7 kb), or the nonconserved SMAD_RE2 (previously called BMP_RE3; position −2301/−2296 bp; SMAD_RE2_2.7kb) were previously described.12,18 In the present study we generated a luciferase reporter construct that contained mutations in 2 conserved BMP-REs and the nonconserved SMAD_RE2 (BMP_RE1_BMP_RE2_SMAD_RE2_2.7kb). Furthermore, site-directed mutagenesis was applied to the reporter vectors WT_2.7kb and BMP_RE1_BMP_RE2_2.7kb to introduce mutations within the SMAD-binding motif at position −1414/−1406 (as detected by the motif search program MatInspector; Genomatix, http://www.genomatix.de/), to exchange the sequence TCAAGAC to TATAAGC. These new constructs were named SMAD_RE1_2.7kb and BMP_RE1_BMP_RE2_SMAD_RE1_2.7kb, respectively.
Transfection of siRNAs, reporter plasmids, and luciferase assay
Huh7 cells were seeded onto 12-well plates and transfected by Oligofectamine Reagent (Invitrogen), according to the manufacturer's instructions, with 100nM siRNA directed against SMAD7, SMAD4, or scrambled siRNA (Dharmacon) as a control. Forty-eight hours after siRNA transfection, 200 ng of hepcidin promoter reporter constructs were transfected into the same cells together with a control plasmid containing the Renilla gene under the control of the CMV promoter. Plasmid transfections were performed by the use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 24 hours, the cells were harvested, and the luciferase activity was measured by use of the Dual-Luciferase-Reporter assay system (Promega) and a Centro LB 960 luminometer (Berthold Technologies).
For the siRNA-mediated double knockdown of SMAD4 and SMAD7, Huh7 cells (6 × 104 per well) were seeded onto 12-well plates. After 24 hours, cells were transfected by twice the amount of Oligofectamine Reagent (Invitrogen) and 200nM total siRNA. SiRNA directed against SMAD4 (100nM; Dharmacon) and siRNA directed against SMAD7 (100nM; Dharmacon) were mixed together for the double knockdown. For the single knockdowns the specific siRNAs were supplemented with 100nM scrambled siRNA (Dharmacon), and as a control 200nM scrambled siRNA was used. Seventy-two hours after siRNA transfection cells were harvested for the extraction of total RNA.
To validate the screening data, Huh7 cells were reverse transfected with single siRNA duplexes directed against SMAD7 or a pool of siRNAs directed against SMAD7 or JAK1. Subsequently, the cells were transfected with the reporter plasmids, and luciferase activities were measured according to the same protocol as described for the high-throughput screen. For endogenous gene-expression analysis Huh7 cells were reverse transfected with the same single or pooled siRNAs targeting the SMAD7 or JAK1 genes. Dharmafect1 (3 μL) diluted in 97 μL of RPMI was added to each well of a 12-well plate containing 1μM siRNA in 100 μL of RPMI and incubated 50 minutes at room temperature. After incubation, 2 × 105 cells were seeded on top of the transfection mix, cultured for 72 hours, and then harvested for extraction of total RNA.
Preparation of total RNA, reverse transcription, and quantitative real-time polymerase chain reaction analysis
Isolation of total RNA and the conditions for reverse transcription and real-time quantitative polymerase chain reaction (PCR) were described previously.12 Primers were designed to specifically amplify the human hepcidin, GAPDH, JAK1, SMAD4, and SMAD7 as well as murine Smad7, hepcidin1, or Gapdh cDNAs. Sequences of the primers are shown in supplemental Table 2. The mRNA/cDNA abundance of each gene was calculated relative to the expression of a housekeeping gene, glyceraldehyde-3-phosphate-dehydrogenase (GAPDH).
Treatment of Huh7 cells with BMP-2, BMP-6, BMP-9, and interleukin-6
Huh7 cells (1.5 × 105 per well) were seeded onto 6-well plates. Twenty-four hours later, the culture medium was exchanged to fetal calf serum-free medium. After 24 hours the cells were treated with BMP-2 (50 ng/mL, 15 hours; R&D Systems), BMP-6 (50 ng/mL, 24 hours; R&D Systems), BMP-9 (5 ng/mL, 24 hours; R&D Systems), or interleukin-6 (IL-6, 5 ng/mL and 20 ng/mL, 24 hours; R&D Systems) and then harvested for the extraction of total RNA.
SMAD7 overexpression in primary murine hepatocytes
Primary hepatocytes were isolated from livers of C57/BL-6 male mice as described previously.28 The same day cells were transduced with adenoviral vectors constructed as described before29 and generously provided by Prof Heldin and Dr Moustakas (Uppsala University) expressing the full-length murine Smad7 gene under the control of the CMV promoter (AdSmad7) and the β-galactosidase gene as a control (AdLacZ) in doses ranging between 5 × 106 and 6 × 107 infectious particles per milliliter. Forty-eight hours later the primary cultures were treated with IL-6 (20 ng/mL, 24 hours), BMP-6 (50 ng/mL, 24 hours), BMP-9 (5 ng/mL, 24 hours; R&D Systems), or TGF-β (5 ng/mL, 6 hours; PeproTech) and then harvested for isolation of total RNA.
Statistical analysis
For analysis of the screening data, the CellHTS2 package (Bioconductor)30 was used. Each Firefly/Renilla ratio was transformed logarithmically and then the median of the transformed values was calculated for each plate individually. To obtain z-scores, this median was subtracted from each logarithmic value and divided by the median absolute deviation of a whole plate. Mean z-scores for control siRNAs were first calculated within each replicate and then between replicates. For the screening data the mean z-score of 2 replicates was calculated. All the other results from this study are expressed as a mean of at least 3 independent experiments plus or minus standard deviation (SD). Student t test was used for estimation of statistical significance.
Results
High-throughput siRNA screen identifies regulators of hepcidin transcription
Cell-based RNAi screens represent powerful genetic means to systematically identify pathway-specific genes.31 To discover novel regulators of hepcidin expression we adapted a cell-based assay established in our previous work12,16,18 to high-throughput conditions. In this assay, Huh7 cells are reverse transfected with a pool of 4 synthetic siRNAs from an siRNA library (Dharmacon/ThermoFisher siGenome) that targets 779 genes encoding for kinases and other signaling molecules and with a luciferase reporter vector driven by the 2.7-kb human hepcidin promoter together with a vector encoding an internal standardization control. After 48 hours, the cells were lysed, and dual luciferase activities were measured (Figure 1A). SiRNAs directed against SMAD4 (strong activator) and HFE (weak activator) were included as positive controls and a scrambled siRNA pool (containing unrelated sequences) as a negative control.
High-throughput siRNA screening identifies SMAD7 as a suppressor of hepcidin expression. (A) Schematic representation of the high-throughput siRNA screening. Huh7 cells were seeded in 384-well plates containing siRNAs from the ThermoFisher siGenome siRNA library (Dharmacon) as well as siRNAs directed against HFE and SMAD4 as positive controls and scrambled siRNAs as a negative control. Twenty-four hours later, cells were transfected with a firefly luciferase reporter construct containing the 2.7-kb human hepcidin promoter (WT_2.7kb) and a renilla luciferase control vector. Dual luciferase activities were measured 48 hours later. (B) The results of the knockdown of SMAD4, HFE, SMAD7, JAK1, and BMPR1A (ALK3) genes are presented as a mean of the z-scores from 2 replicates. (C-D) Validation of the screening results. SiRNA-mediated knockdown of JAK1 (C) and SMAD7 (D). Results are presented as ratios between the luciferase activity (± SD of Firefly/Renilla) obtained from cells transfected with specific siRNAs and cells transfected with scrambled siRNA. The knockdown efficiencies for JAK1 or SMAD7 also are shown together with their consequences for endogenous hepcidin mRNA expression. Levels of mRNA expression were normalized to the house-keeping gene GAPDH. Results are presented as fold change (± SD) compared with samples transfected with control siRNA. The mean of 3 independent experiments is shown. Significant changes are marked by *(P < .05) or **(P < .005).
High-throughput siRNA screening identifies SMAD7 as a suppressor of hepcidin expression. (A) Schematic representation of the high-throughput siRNA screening. Huh7 cells were seeded in 384-well plates containing siRNAs from the ThermoFisher siGenome siRNA library (Dharmacon) as well as siRNAs directed against HFE and SMAD4 as positive controls and scrambled siRNAs as a negative control. Twenty-four hours later, cells were transfected with a firefly luciferase reporter construct containing the 2.7-kb human hepcidin promoter (WT_2.7kb) and a renilla luciferase control vector. Dual luciferase activities were measured 48 hours later. (B) The results of the knockdown of SMAD4, HFE, SMAD7, JAK1, and BMPR1A (ALK3) genes are presented as a mean of the z-scores from 2 replicates. (C-D) Validation of the screening results. SiRNA-mediated knockdown of JAK1 (C) and SMAD7 (D). Results are presented as ratios between the luciferase activity (± SD of Firefly/Renilla) obtained from cells transfected with specific siRNAs and cells transfected with scrambled siRNA. The knockdown efficiencies for JAK1 or SMAD7 also are shown together with their consequences for endogenous hepcidin mRNA expression. Levels of mRNA expression were normalized to the house-keeping gene GAPDH. Results are presented as fold change (± SD) compared with samples transfected with control siRNA. The mean of 3 independent experiments is shown. Significant changes are marked by *(P < .05) or **(P < .005).
The knockdown of HFE and SMAD4 both diminish luciferase activity and accurately reflect the relative potency of SMAD4 and HFE as hepcidin activators (Figure 1B). Within the 779 genes that we screened, the high-throughput assay implicated approximately 70 hits as candidate effectors (data not shown) and identified 3 genes (JAK1, BMPR1A, and SMAD7) that significantly alter luciferase activity and belong to the BMP or JAK/STAT signaling pathways (Figure 1B). Of these, the BMP receptor 1A (BMPR1A, also known as ALK3) was already shown in earlier work10 to mediate the BMP response of hepcidin, further validating the screening experiments. Therefore, we focused the secondary assays on JAK1 and SMAD7.
In Huh7 cells the siRNA pool directed against JAK1 significantly diminishes JAK1 mRNA expression (to 13% of the control) and luciferase activity (to 29%). Importantly, endogenous hepcidin mRNA expression is also strongly reduced (to 40%; Figure 1C). This finding complements earlier data that implicate STAT3 as a positive regulator of basal, steady-state hepcidin mRNA expression18 by demonstrating that JAK1, the kinase that phosphorylates STAT proteins, is also involved in this process.
The most interesting hit of our screen was SMAD7 because its knockdown leads to a profound increase in luciferase activity, suggesting that SMAD7 may represent an elusive hepcidin suppressor. To validate the SMAD7 data, we transfected either the siRNA pool directed against SMAD7 or each single siRNA contained within the siRNA pool, separately, into Huh7 cells. Although only 2 of 4 siRNAs increased luciferase activity, all of them significantly enhanced endogenous hepcidin expression, the latter consistently being more pronounced than the former (Figure 1D); the efficiency of the knockdown for each siRNA (to 75%, 90%, 59%, 49%, and 54% of the control for a, b, c, d, and the pool, respectively) correlates well with the change in luciferase activity and hepcidin mRNA expression (Figure 1D). We conclude that SMAD7 negatively regulates hepcidin expression in Huh7 cells via its 2.7-kb promoter.
SMAD7 expression is increased by BMPs and decreased by IL-6
SMAD7 is an inhibitory SMAD protein that antagonizes TGF-β signaling32,33 and that is itself induced by the TGF-β pathway, acting as a suppressor by a negative feedback loop.34-37 Interestingly, SMAD7 is induced by BMP-7, epidermal growth factor,36 interferon-γ,38 and tumor necrosis factor-α39 and, more recently, SMAD7 expression was shown to be regulated by dietary iron levels in mice.27 To compare the induction of SMAD7 by cytokines known to activate the hepcidin promoter to that of hepcidin, Huh7 cells (Figure 2A-B) or primary mouse hepatocytes (Figure 2C) were treated with BMPs 2, 6, or 9; TGF-β; or IL-6. The hepcidin response to TGF-β was only analyzed in primary hepatocytes because TGF-β has been reported not to significantly activate hepcidin mRNA expression in a hepatoma cell line.8 These data reveal that BMPs (strong hepcidin inducers) and TGF-β (weaker hepcidin inducer) both activate SMAD7 mRNA expression, whereas IL-6 stimulates hepcidin mRNA expression as expected but slightly suppresses SMAD7. Whether or not this effect is of biologic relevance remains to be established. Small reductions of SMAD7 mRNA expression after siRNA-mediated knockdown do, however, induce a significant increase in hepcidin mRNA levels (Figure 1D). It is thus possible that the decrease of SMAD7 mRNA expression in IL-6–treated Huh7 cells may contribute to elevated hepcidin mRNA levels and may in part explain interactions between STAT- and SMAD-mediated signaling events in this experimental setting.
SMAD7 expression is induced by BMPs and inhibited by IL-6. Huh7 cells were treated with (A) BMP-2, BMP-6, and BMP-9 or (B) IL-6. Cells were harvested 24 hours later for isolation of total RNA. (C) Primary murine hepatocytes were treated with BMP-6 (50 ng/mL) or BMP-9 (5 ng/mL) for 24 hours or with TGF-β (5 ng/mL) for 6 hours. Levels of mRNA expression were determined by quantitative real-time PCR for SMAD7 and hepcidin and then normalized to mRNA expression of GAPDH. Results are presented as fold change (± SD) compared with untreated cells. The mean of 3 (A), 5 (B), and 4 (C) independent experiments is presented. Significant changes are marked by *(P < .05) or **(P < .005).
SMAD7 expression is induced by BMPs and inhibited by IL-6. Huh7 cells were treated with (A) BMP-2, BMP-6, and BMP-9 or (B) IL-6. Cells were harvested 24 hours later for isolation of total RNA. (C) Primary murine hepatocytes were treated with BMP-6 (50 ng/mL) or BMP-9 (5 ng/mL) for 24 hours or with TGF-β (5 ng/mL) for 6 hours. Levels of mRNA expression were determined by quantitative real-time PCR for SMAD7 and hepcidin and then normalized to mRNA expression of GAPDH. Results are presented as fold change (± SD) compared with untreated cells. The mean of 3 (A), 5 (B), and 4 (C) independent experiments is presented. Significant changes are marked by *(P < .05) or **(P < .005).
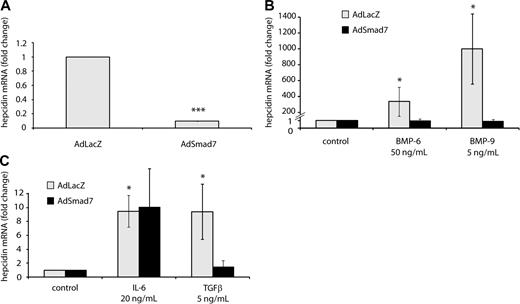
Smad7 overexpression strongly reduces hepcidin mRNA levels and completely blocks the hepcidin response to BMPs and TGF-β
To investigate whether SMAD7 can suppress hepcidin transcription in a dominant way, we overexpressed Smad7 in primary hepatocytes by adenovirus-mediated gene delivery.40 Compared with hepatocytes infected by a control virus, hepcidin mRNA expression is 10-fold suppressed upon Smad7 overexpression (Figure 3A). Even more strikingly, Smad7 gene delivery completely eliminates hepcidin activation by BMP-6, BMP-9 (Figure 3B), and TGF-β (Figure 3C). Demonstrating the specificity of these responses, hepcidin induction by IL-6 is essentially unaffected by Smad7 overexpression (Figure 3C). These data further corroborate the role of SMAD7 as a potent suppressor of hepcidin transcription via the BMP signaling.
Smad7 overexpression inhibits steady-state hepcidin expression and abrogates the hepcidin response to BMP and TGF-β. (A) Smad7 overexpression under control and (B-C) stimulatory conditions. Primary murine hepatocytes were transduced with adenoviral vectors expressing Smad7 (AdSmad7) or β-galactosidase (AdLacZ) as a control. Forty-eight hours later, the cells were treated with IL-6 (20 ng/mL, 24 hours), BMP-6 (50 ng/mL, 24 hours), BMP-9 (5 ng/mL, 24 hours), or TGF-β (5 ng/mL, 6 hours) and harvested for the isolation of total RNA. Hepcidin mRNA levels were determined by quantitative real-time PCR and normalized to Gapdh mRNA expression. (A) Under control conditions results are presented as fold change (± SD) compared with samples transduced with control AdLacZ vectors. (B-C) Under stimulatory condition results are presented as fold change (± SD) compared with untreated cells. Results represent a mean of 4 independent experiments. Significant changes are marked by *(P < .05) or ***(P < .001).
Smad7 overexpression inhibits steady-state hepcidin expression and abrogates the hepcidin response to BMP and TGF-β. (A) Smad7 overexpression under control and (B-C) stimulatory conditions. Primary murine hepatocytes were transduced with adenoviral vectors expressing Smad7 (AdSmad7) or β-galactosidase (AdLacZ) as a control. Forty-eight hours later, the cells were treated with IL-6 (20 ng/mL, 24 hours), BMP-6 (50 ng/mL, 24 hours), BMP-9 (5 ng/mL, 24 hours), or TGF-β (5 ng/mL, 6 hours) and harvested for the isolation of total RNA. Hepcidin mRNA levels were determined by quantitative real-time PCR and normalized to Gapdh mRNA expression. (A) Under control conditions results are presented as fold change (± SD) compared with samples transduced with control AdLacZ vectors. (B-C) Under stimulatory condition results are presented as fold change (± SD) compared with untreated cells. Results represent a mean of 4 independent experiments. Significant changes are marked by *(P < .05) or ***(P < .001).
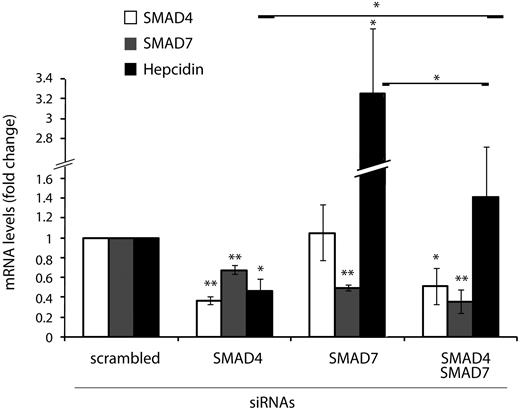
SMAD4 and SMAD7 antagonistically control hepcidin mRNA expression
SMAD4 is an essential component of activating SMAD complexes that induce the transcription of target genes of TGF-β and BMPs.32 Stimulation of SMAD7 expression by TGF-β also involves SMAD4.35,37 Hepatocyte-specific Smad4 ablation in mice as well as siRNA-mediated knockdown of SMAD4 in cultured hepatocyte cell lines decreases hepcidin mRNA levels (Figure 1B).12,17 To investigate the interrelationship of SMAD7 and SMAD4 in regulating hepcidin mRNA expression, we performed siRNA-mediated knockdown of these 2 opposing factors in Huh7 cells. As expected, hepcidin and SMAD7 expression requires appropriate levels of SMAD4 (Figure 4). By contrast, SMAD4 expression is unaffected by the knockdown of SMAD7, whereas hepcidin mRNA expression is strongly elevated. The knockdown of the hepcidin activator SMAD4 and the hepcidin repressor SMAD7 compensate each other, suggesting that the 2 proteins act in a mutually antagonistic way to control hepcidin expression.
SMAD7 and SMAD4 antagonistically control hepcidin mRNA expression. Huh7 cells were transfected with siRNAs targeting SMAD4, SMAD7, or SMAD4 and SMAD7 or a scrambled siRNA as a control. Seventy-two hours after siRNA transfection, total RNA was isolated. SMAD4, SMAD7, and hepcidin mRNA levels were determined by quantitative real-time PCR analysis and normalized to the housekeeping gene GAPDH. Results are presented as fold change (± SD) compared with samples transfected with control siRNA. The mean of 3 independent experiments is shown. Significant changes are marked by *(P < .05) or **(P < .005).
SMAD7 and SMAD4 antagonistically control hepcidin mRNA expression. Huh7 cells were transfected with siRNAs targeting SMAD4, SMAD7, or SMAD4 and SMAD7 or a scrambled siRNA as a control. Seventy-two hours after siRNA transfection, total RNA was isolated. SMAD4, SMAD7, and hepcidin mRNA levels were determined by quantitative real-time PCR analysis and normalized to the housekeeping gene GAPDH. Results are presented as fold change (± SD) compared with samples transfected with control siRNA. The mean of 3 independent experiments is shown. Significant changes are marked by *(P < .05) or **(P < .005).
SMAD7 antagonizes SMAD4-mediated responses by use of a novel SMAD binding motif in the hepcidin promoter
Suppression of the TGF-β pathway by SMAD7 is exerted at multiple levels33 : (1) SMAD7 triggers the dephosphorylation and/or degradation of TGF-β and BMP receptors41-43 ; (2) it blocks the phosphorylation of R-SMADs involved in TGF-β signaling34 ; (3) it mediates SMAD4 degradation44 ; and (4) it directly binds to the promoter of target genes to compete with activating SMAD complexes for binding to the same motifs.45 In principle, either of these regulatory mechanisms could be operational in hepatic cells to mediate hepcidin suppression.
The highly conserved BMP-RE1 and BMP-RE2, located in the proximal and distal regions of the hepcidin promoter, respectively, are critical for the BMP/HJV/SMAD4-mediated hepcidin responses.11,12,16 If SMAD7 antagonizes the activity of one or more proteins involved in BMP/HJV/SMAD4-mediated hepcidin activation, the hepcidin response to the siRNA-mediated SMAD7 knockdown should be abolished when BMP-RE1 and BMP-RE2 are mutated.
Unexpectedly, mutation of BMP-RE1 (BMP_RE1_2.7kb), BMP-RE2 (BMP_RE2_2.7kb), or the nonconserved SMAD-responsive element 2 (SMAD_RE2_2.7kb; formerly called BMP-RE311,12 because of its sequence identity to BMP-RE1 and BMP-RE2) does not prevent hepcidin promoter activation upon siRNA-mediated SMAD7 knockdown compared with the WT_2.7kb reporter construct (Figure 5A,C). Luciferase activity is partially reduced when BMP_RE1 and BMP_RE2 (BMP_RE1_BMP_RE2_2.7kb) are jointly mutated. To our surprise, complete suppression of hepcidin activation upon SMAD7 knockdown requires triple mutations. SMAD-RE2 is only present in the human hepcidin promoter, and mutations in this element do not impair BMP/HJV/SMAD4-mediated hepcidin activation per se, but the BMP_RE1_BMP_RE2_SMAD_RE2_2.7kb construct completely loses activation by SMAD7 knockdown. A motif search with MatInspector yielded one additional SMAD4-binding motif (SMAD-responsive element 1 [SMAD-R1]) at position −1414/−1406 of the human hepcidin 2.7-kb promoter. Although similar putative SMAD-binding motifs also are detected in the murine hepcidin promoter, their localization differs from the SMAD-RE1 (data not shown). Mutation of the human SMAD-RE1 neither interferes with hepcidin promoter activity under basal conditions nor with the activation of the hepcidin promoter in SMAD7-deficient cells (Figure 5B-C). Interestingly, combined mutations within the BMP-RE1, BMP-RE2, and the SMAD-RE1 (BMP_RE1_BMP_RE2_SMAD_RE1_2.7kb) also completely abolish hepcidin activation upon SMAD7 knockdown (Figure 5C).
SMAD7 antagonizes SMAD4-mediated responses. (A) The name of each construct refers to the elements that have been mutated compared with the WT_2.7kb construct. B1 and B2 indicate BMP-RE1 and BMP-RE2, respectively, and S1 and S2 indicate SMAD-RE1 and SMAD-RE2, respectively. (B) Steady-state hepcidin promoter activity was measured from the SMAD_RE1_2.7kb luciferase reporter construct, compared with WT_2.7kb promoter, and is presented as a fold change ± SD of Firefly/Renilla ratios. (C-D) Huh7 cells were transfected with siRNAs directed against SMAD7 (C), SMAD4 (D), or scrambled siRNA as a control. Forty-eight hours later, cells were transfected with luciferase reporter vectors. Luciferase activity was measured 24 hours later. Results are presented as ratios between the luciferase activity (± SD of Firefly/Renilla) obtained from samples transfected with specific siRNA and samples transfected with control siRNA. Results represent a mean of 4 independent experiments. Statistically significant changes between the WT promoter and the mutated promoter (C) or between the SMAD4 siRNA transfection and scrambled controls (C) are marked by an asterisk (*P < .05, **P < .005, and ***P < .001). (E) SMAD7, a potent hepcidin suppressor, counteracts the BMP/SMAD-mediated induction of hepcidin expression. SMAD7 is coregulated with hepcidin via SMAD4-dependent signaling pathways and thus antagonizes SMAD4-mediated responses of the hepcidin promoter. SMAD7 expression is further modulated by cytokines independent of SMAD4 signaling.
SMAD7 antagonizes SMAD4-mediated responses. (A) The name of each construct refers to the elements that have been mutated compared with the WT_2.7kb construct. B1 and B2 indicate BMP-RE1 and BMP-RE2, respectively, and S1 and S2 indicate SMAD-RE1 and SMAD-RE2, respectively. (B) Steady-state hepcidin promoter activity was measured from the SMAD_RE1_2.7kb luciferase reporter construct, compared with WT_2.7kb promoter, and is presented as a fold change ± SD of Firefly/Renilla ratios. (C-D) Huh7 cells were transfected with siRNAs directed against SMAD7 (C), SMAD4 (D), or scrambled siRNA as a control. Forty-eight hours later, cells were transfected with luciferase reporter vectors. Luciferase activity was measured 24 hours later. Results are presented as ratios between the luciferase activity (± SD of Firefly/Renilla) obtained from samples transfected with specific siRNA and samples transfected with control siRNA. Results represent a mean of 4 independent experiments. Statistically significant changes between the WT promoter and the mutated promoter (C) or between the SMAD4 siRNA transfection and scrambled controls (C) are marked by an asterisk (*P < .05, **P < .005, and ***P < .001). (E) SMAD7, a potent hepcidin suppressor, counteracts the BMP/SMAD-mediated induction of hepcidin expression. SMAD7 is coregulated with hepcidin via SMAD4-dependent signaling pathways and thus antagonizes SMAD4-mediated responses of the hepcidin promoter. SMAD7 expression is further modulated by cytokines independent of SMAD4 signaling.
Finally, we tested the response of the triple mutant constructs BMP_RE1_BMP_RE2_SMAD_RE2_2.7kb and BMP_RE1_BMP_RE2_SMAD_RE1_2.7kb to the knockdown of SMAD4. Although the BMP_RE1_BMP_RE2_2.7kb reporter construct could not completely relieve repression induced by the knockdown of SMAD4 compared with the WT_2.7kb construct, BMP_RE1_BMP_RE2_SMAD_RE2_2.7kb and BMP_RE1_BMP_RE2_SMAD_RE1_2.7kb completely abrogated the effect of the SMAD4 knockdown (Figure 5D). This result suggests that SMAD-RE2 and the newly identified SMAD-RE1 positively interact with SMAD4 and that SMAD7 antagonizes this SMAD4-mediated effect.
Discussion
Hepcidin mRNA expression is precisely controlled to adjust systemic iron levels to physiologic conditions. The BMP signaling pathway plays an important role in this process by responding to systemic iron availability,13-15 regulating steady-state hepcidin expression, and partially mediating the hepcidin response to inflammation.16,17 The BMP coreceptor hemojuvelin, the protease TMPRSS6,24 and the cytokine twisted gastrulation 123 modulate the BMP/SMAD-mediated hepcidin response. In addition, hepcidin mRNA expression also responds to TGF-β treatment.17 Here, we report a direct inhibitory mechanism for hepcidin expression to prevent excessive hepcidin synthesis and subsequent iron deficiency.
SMAD7 emerged as a hit for a candidate hepcidin suppressor from a high-throughput siRNA screen (Figure 1). Although the primary screening data have not yet been validated as a complete dataset, we specifically selected hits related to the inflammatory JAK/STAT or the SMAD signaling pathways for validation in secondary assays (Figure 1). Among these, JAK1, the kinase activating STAT proteins, shows the phenotype of a hepcidin activator, whereas SMAD7 stands out as a hepcidin repressor. Moreover, SMAD7 is coregulated with hepcidin via SMAD4 in response to altered iron availability in vivo27 and mediates negative feedback to both TGF-β and BMP signaling.34,41 Therefore, we analyzed the role of SMAD7 in hepcidin suppression in more detail.
We show that SMAD7 mRNA expression is activated by BMP-2, BMP-6, and BMP-9 (Figure 2), suggesting that BMPs are potent activators of SMAD7 in Huh7 cells and/or primary hepatocytes. In other cell types SMAD7 activation is predominantly induced by TGF-β.35,37,46 To the best of our knowledge, this is the first report demonstrating that SMAD7 is induced by BMPs in hepatic cells. Interestingly, the SMAD7 promoter contains 4 putative BMP-REs, with identical sequences to the ones previously identified in the hepcidin promoter,11,12 suggesting that activation of SMAD7 by BMPs occurs via these elements and that the transcriptional control mechanisms regulating hepcidin and SMAD7 mRNA expression are similar, at least in part. In the hepcidin promoter, BMP-RE1 and BMP-RE2 are critical for the response to BMP-6,12 a crucial regulator of iron levels in mice.14,15 The response of SMAD7 to BMP-6 treatment together with the observation that this gene is regulated by iron levels in vivo27 strongly suggest a role for SMAD7 in the iron-controlled regulation of hepcidin.
Our data further show that the hepcidin activator SMAD4 and the hepcidin suppressor SMAD7 control hepcidin mRNA expression antagonistically (Figure 4) via the same promoter elements (Figure 5). Consistent with previous observations,17,27 hepcidin and SMAD7 mRNA expression both require appropriate levels of SMAD4, whereas the knockdown of SMAD7 does not affect SMAD4 expression but strongly elevates hepcidin mRNA levels. Importantly, decreased hepcidin mRNA expression as a consequence of low SMAD4 levels can be rescued by the simultaneous knockdown of SMAD7, establishing that the opposing functions of these 2 proteins indeed balance hepcidin mRNA expression. Smad7 overexpression in primary murine hepatocytes shifts this equilibrium toward hepcidin repression and completely abolishes hepcidin activation by BMPs and TGF-β (Figure 3). These results motivate future investigation of SMAD7 expression in patients and animal models with decreased hepcidin levels.
We further aimed to define the mechanism of SMAD7-mediated hepcidin suppression by identifying the regulatory motifs for this response within the hepcidin promoter (Figure 5). Recent work suggests that Smad7 overexpression in primary hepatocytes reduces the phosphorylation of Smad1, Smad2, and Smad3 both under steady-state conditions and upon stimulation with TGF-β.47 Consistently, we observe a partial requirement for BMP-RE1 and BMP-RE2 that control the SMAD4-dependent response of the hepcidin promoter (Figure 5C-D) in basal steady-state conditions as well as in response to BMP-2, BMP-6, and HJV.12 However, to our surprise, SMAD7-dependent repression of hepcidin promoter activity is not limited to those promoter elements (Figure 5C). Hence, SMAD7 regulation of hepcidin expression appears to reach beyond inhibition of the HJV/BMP pathway.
In addition to BMP-RE1 and BMP-RE2, we identify SMAD-REs that are equally important for repression of the hepcidin promoter by SMAD7. These promoter motifs include the nonconserved SMAD-RE2 (at position −2301/−2296 of the hepcidin promoter11,12 ) and the newly identified SMAD-RE1 (GTCAAGAC; Figure 5) at position −1414/−1406 of the hepcidin promoter. These SMAD-REs are functionally different from BMP-RE1 and BMP-RE2 because they appear not to contribute significantly to basal hepcidin promoter activity (Figure 5B).12 The data presented in Figure 5 show that these elements are important in 2 experimental settings: (1) to fully abrogate hepcidin activation by the siRNA-mediated knockdown of SMAD7 and (2) to fully prevent hepcidin repression upon siRNA-mediated SMAD4 knockdown. These findings suggest that these SMAD-REs positively engage SMAD4 and that SMAD7 antagonizes these effects. Interestingly, SMAD7 functions as a direct repressor of gene transcription in Hep3B cells by competing with activating SMAD complexes for binding to AGAC sequence motifs.45 SMAD7 may thus (at least in part) directly act on the hepcidin promoter to prevent the binding of SMAD4-containing activator complexes to the SMAD responsive motifs, a hypothesis that is supported by the data presented in Figure 5.
We and others12,17 have previously shown that the inflammatory response of hepcidin involves SMAD4 and requires an intact BMP-RE1 within the hepcidin promoter. Intriguingly, Smad7 cannot antagonize hepcidin induction in response to IL-6. In addition, mutation of BMP-RE1 alone is insufficient to affect SMAD7-mediated hepcidin repression. These data suggest that it is unlikely that SMAD7 affects SMAD4 protein stability, which is in accordance with the observation that Smad4 levels are not changed upon Smad7 overexpression (data not shown).
Our data further imply that SMAD4-dependent hepcidin expression may not be restricted to the BMP signaling pathway. SMAD-RE1 (GTCAAGAC) differs by a single nucleotide from a functional SMAD binding motif located within the SMAD7 promoter (GTCTAGAC), which binds SMAD2, SMAD3, and SMAD4 upon TGF-β stimulation.35,37 As the BMP and the TGF-β signaling pathways converge at the level of SMAD4 (co-SMAD), this finding highlights the possibility that the classical TGF-β signaling pathway also controls hepcidin transcription.
Taken together, the identification of SMAD7 as a potent hepcidin suppressor uncovers a module that counteracts the BMP/SMAD-mediated induction of hepcidin expression (Figure 5E). We hypothesize that this mechanism prevents overshooting hepcidin responses and iron deficiency.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Research in the laboratory of M.M., M.H., and S.D. was supported by the BMBF (HepatoSys-Iron_liver). Work in the laboratory of M.M. and M.H. also was supported by the EEC Framework 6 (LSHM-CT-2006037296 EuroIron1). Research in the laboratory of M.B. was supported by a grant of the DFG and the Helmholtz Alliance for Systems Biology.
Authorship
Contribution: M.W.H. and M.U.M. designed the project and supervised the research; K.M.-S. and G.C. designed research; K.M.-S., G.C., A.R., K.B., and A.M. performed experiments; M.B and S.D. contributed to the experiments; K.M.-S., G.C., M.U.M., A.R., K.B., and A.M. analyzed data; K.M.-S., M.W.H., and M.U.M. wrote the manuscript; and G.C., A.R., K.B., M.B., and S.D. edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Martina U. Muckenthaler, University of Heidelberg, Department of Pediatric Oncology, Hematology and Immunology, Im Neuenheimer Feld 153, 69120 Heidelberg, Germany; e-mail: martina.muckenthaler@med.uni-heidelberg.de; or Matthias W. Hentze, European Molecular Biology Laboratory (EMBL), Meyerhofstr 1, 69117 Heidelberg, Germany; e-mail: hentze@embl.de.