Abstract
Hereditary hemochromatosis is caused by mutations in the hereditary hemochromatosis protein (HFE), transferrin-receptor 2 (TfR2), hemojuvelin, hepcidin, or ferroportin genes. Hepcidin is a key iron regulator, which is secreted by the liver, and decreases serum iron levels by causing the down-regulation of the iron transporter, ferroportin. Mutations in either HFE or TfR2 lower hepcidin levels, implying that both HFE and TfR2 are necessary for regulation of hepcidin expression. In this study, we used a recombinant adeno-associated virus, AAV2/8, for hepatocyte-specific expression of either Hfe or Tfr2 in mice. Expression of Hfe in Hfe-null mice both increased Hfe and hepcidin mRNA and lowered hepatic iron and Tf saturation. Expression of Tfr2 in Tfr2-deficient mice had a similar effect, whereas expression of Hfe in Tfr2-deficient mice or of Tfr2 in Hfe-null mice had no effect on liver or serum iron levels. Expression of Hfe in wild-type mice increased hepcidin mRNA and lowered iron levels. In contrast, expression of Tfr2 had no effect on wild-type mice. These findings suggest that Hfe is limiting in formation of the Hfe/Tfr2 complex that regulates hepcidin expression. In addition, these studies show that the use of recombinant AAV vector to deliver genes is a promising approach for studying physiologic consequences of protein complexes.
Introduction
Hereditary hemochromatosis (HH) is an autosomal recessive disease of iron metabolism characterized by increased intestinal iron absorption and hepatic iron overload.1 HH is caused by mutations in genes encoding proteins involved in iron homeostasis, including the HH protein, HFE2 ; hemojuvelin3 ; hepcidin4 ; transferrin-receptor 2 (TfR2)5 ; and ferroportin.6 Accumulation of excessive iron results in hepatic cirrhosis, hepatocellular carcinoma, cardiomyopathy, arrhythmias, diabetes, arthritis, and hypogonadotropic hypogonadism.7
HH type 1, the most common form of HH, is caused by a missense mutation in HFE resulting in a C282Y (numbering system includes the first 23 amino acid signal peptide) substitution and accounts for 85% of HH.2 HFE encodes an atypical major histocompatibility complex class I protein. Like the major histocompatibility complex class I proteins, HFE is a membrane protein that consists of a signal sequence, α1-α3 domains followed by a transmembrane domain, and a short cytoplasmic domain. HFE also forms a heterodimeric complex with β2-microglobulin.8 The C282Y mutation in HFE disrupts a disulfide bond in the α3 domain, leading to misfolding, lack of association with β2-microglobulin, and failure to traffic to the cell surface.9 The Hfe knockout mouse (Hfe−/−) also develops iron overload, confirming that the HFE-C282Y mutation confers loss of rather than gain of function.10 Although the importance of HFE in iron regulation is apparent in patients with HH and murine models,10,11 the underlying mechanism by which HFE regulates iron metabolism is only beginning to be understood.
Type 3 HH is caused by a variety of missense and nonsense mutations in TfR2.5 The most common mutation is the nonsense mutation (Y250X), which results in a truncation of TfR2 at amino acid 250. The equivalent mutation in the mouse is Y245x.12 TfR2, a homolog of TfR1, is expressed predominantly in the liver, in contrast to the ubiquitously expressed TfR1.13,14 Tf binds to TfR2 with a lower affinity than it binds to TfR1.15-19 Unlike TfR1, which is inversely regulated at the level of mRNA stability by intracellular iron,20,21 TfR2 is regulated at the level of protein stability by a novel mechanism, involving the stabilization of TfR2 on Tf binding.22,23 Although TfR2 has been investigated with respect to Tf-mediated iron uptake,24 binding affinity to Tf, tissue-specific expression, and posttranslation regulation by Tf, its role in regulation of iron homeostasis still remains to be clarified.
A characteristic finding in HH caused by mutations either in HFE or in TFR2 is a lower level of hepcidin than in either humans or mice with the same degree of iron loading from other causes.25-29 Hepcidin, a key iron regulator, is a peptide hormone synthesized by the liver and secreted into the circulation. Hepcidin modulates serum iron levels, by binding to and causing the down-regulation of the iron transporter, ferroportin.30 In murine studies, hepcidin expression (Hamp1) increases in response to iron loading, thus preventing further iron uptake. Conversely, during iron deficiency, hepcidin expression decreases.31,32
The observation that mutations in either HFE or TfR2 lower hepcidin levels implies that both HFE and TfR2 play a role in the regulation of hepcidin expression. HFE, TfR2, and hepcidin are expressed predominantly in hepatocytes, suggesting that HFE and TfR2 regulate iron metabolism upstream of hepcidin.33 The findings that macrophage/granulocyte-specific depletion of HFE in mice (Hfe) has no detectable effect on Hamp1 expression, whereas hepatic-specific depletion of Hfe is sufficient to decrease Hamp1 expression reinforces the importance of Hfe in regulating hepcidin.34 TfR2 and HFE are capable of forming a complex,35,36 suggesting that the complex is involved in hepcidin regulation.37
In this study, we used a recombinant serotype 2 adeno-associated virus vector pseudotyped with serotype 8 capsid (AAV2/8) and carrying a hepatic-specific promoter38 to express either Hfe or Tfr2 in mice to test the role of the HFE/TfR2 complex in the regulation of hepcidin. Our results indicate that the virally encoded Hfe and Tfr2 are expressed in mouse livers. Expression of Hfe in Hfe-null mice both lowered hepatic iron levels and serum Tf saturation and increased hepcidin levels and HFE levels. Tfr2-deficient mice responded similarly when Tfr2 was expressed but not when Hfe was expressed. Expression of Hfe but not Tfr2 in wild-type mice also increased hepcidin levels and lowered iron levels in the liver. However, expression of Hfe in Tfr2-deficient mice or of Tfr2 in Hfe-null mice had no effect on liver or serum iron levels. These findings imply that the HFE/TfR2 complex regulates the expression of hepcidin and that HFE may be limiting in the formation of the HFE/TfR2 complex to regulate hepcidin expression. They also validate AAV-mediated gene delivery to the liver as a strategy for the reduction of iron overload.
Methods
Preparation of AAV2/8 Hfe and Tfr2 vectors
Full-length mouse Hfe open reading frame was amplified from a mouse liver cDNA library by polymerase chain reaction (PCR) with the use of the Expand High Fidelity PCR System (Roche Applied Science) with the following primers: forward, 5′-TCTAGAATGAGCCTATCAGCTGGGCTCCC-3′; reverse, 5′-TCTAGATCACTTGTCATCGTCGTCCTTGTAGTCTGACTCACAGTCTGTTAAGACATAGCC-3′. The reverse primer contains the FLAG sequence before the stop codon. The gel-purified PCR product was inserted into the pGEM-T vector (Promega) and isolated by cleaving with XbaI followed by insertion into pcDNA3. The Hfe-FLAG sequence was isolated from pcDNA3-Hfe-FLAG with KpnI and NheI and inserted adjacent to the liver-specific promoter (LSP) in a plasmid containing AAV serotype 2 inverted terminal repeats. The LSP contains promoter sequences from the thyroid hormone-binding globulin gene (−382 to 3) and 2 copies of the α1-microglobulin/bikunin enhancer sequence (−2804 to −2704).38 Tfr2 was amplified from mouse liver cDNA and inserted into the same AAV2 vector plasmid with the use of the KpnI/NheI sites. In the final constructs, the LSP-driven expression cassettes were flanked by AAV2 inverted terminal repeats. All constructs were verified by sequencing. Recombinant AAV2 vectors pseudotyped with AAV serotype 8 capsid were prepared by cotransfection of HEK293 cells with either the AAV2-Hfe plasmid or the AAV2-Tfr2 plasmid, along with an AAV packaging plasmid expressing serotype 8 capsid and pAdHelper as described previously.39 AAV2/8 viruses were purified from culture supernatants by cesium chloride density centrifugation. Virus titers were determined by quantitative PCR designed to measure the number of LSP-containing genomes per milliliter. All viral vector stocks were handled according to Biohazard Safety Level 2 guidelines published by the National Institutes of Health.
Animal analysis
Hfe−/− mice on 129/SvEvTac (129/S) background were obtained from Dr Nancy Andrews (Duke University). TfR2245x/245x mice were on an FVB/NJ (FVB) background. Both mutant and wild-type mice were bred and maintained in the Laboratory Animal Facility of Oregon Health & Science University. The studies were approved by the institutional committee. For AAV treatment, 8-week-old male mice were injected with 5 × 1011 genome equivalents/mouse of AAV-Hfe or with 2 × 1011 genome equivalents per mouse of AAV-Tfr2 by the tail vein because no differences in Tfr2 protein levels could be detected when either 5 × 1011 or 2 × 1011 genome equivalents/mouse were used. Two weeks later, mice were anesthetized by intraperitoneal injection of mouse cocktail (ketamine 7.5 mg, xylazine 1.5 mg, and acepromazine 0.25 mg/mL). Blood was collected by cardiac puncture. Serum was collected from clotted samples after centrifugation for 10 minutes at 10 000g at 4°C. Liver tissue was harvested and stored in liquid nitrogen for assays.
Quantitative reverse transcription PCR
Total RNA was isolated from mouse liver with the use of the RNAeasy RNA isolation kit (QIAGEN) and treated with DNase (Roche Applied Science) to remove any contaminating genomic DNA as previously described.33 Oligo (dT) primers and Superscript II reverse transcriptase were used to synthesize cDNA according to the manufacturer's instructions. Hamp1, Hfe, Tfr2, and Gapdh mRNA were measured with the use of the mouse primers listed in Table 1 as previously described.33,40 Results were expressed as the level relative to the corresponding Gapdh. All primers were verified for linearity of amplification.
Immunoblots
Murine liver pieces were solubilized on ice in 1% NET-Triton buffer (150mM NaCl, 5mM EDTA, 10mM Tris, 1% Triton X-100, pH 7.4) with Complete Mini Protease Inhibitor Cocktail (Roche Diagnostic) and was cleared by centrifugation at 16 000g for 30 minutes, and the supernatant was collected. Protein concentrations of the cell extracts were measured with the use of the BCA Protein Assay (Pierce Chemical). The liver tissue extracts (50 or 100 μg) were reduced and denatured with 3.6× Laemmli buffer for 5 minutes at 95°C and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis on 10% or 12% gels. Proteins were transferred to nitrocellulose. Immunoblot analysis was carried out with the use of monoclonal anti–FLAG M2 and mouse antiactin antibodies from Sigma-Aldrich, and rabbit anti-mTfR2 from Alpha Diagnostic International. Bands were detected by horseradish peroxidase–coupled secondary antibody and enhanced chemiluminescence (SuperSignal WestPico; Pierce Chemical) or by fluorescently labeled secondary antibodies as described previously.23
Nonheme iron assay
Nonheme iron in liver was determined as previously described41 with the following modifications. Briefly, 50 to 150 mg of wet liver were incubated in 250 to 750 μL of acid buffer41 at 65°C for 72 hours. The supernatant was collected after centrifugation at 10 000g for 5 minutes. The chromogen (1.86mM bathophenanthroline sulfonate, 143mM thioglycolic acid in water) was added to the supernatants. Each sample was measured in triplicate.
Blood iron analysis
Enzyme-linked immunoabsorbent assay of interleukin-6
Serum samples were tested for interleukin-6 (IL-6) according to the manufacturer's instructions (R&D Systems).
Statistical analysis
The standard deviation and the paired 2-tailed Student t test were used to compare 2 sets of data. The 1-way analysis of variance and Tukey test was used to compare 3 sets of matched data. The Kruskal-Wallis test was used to compare 3 sets of unmatched data.
Results
Expression of Hfe in Hfe−/− mice results in lower iron in the liver, lower Tf saturation in the blood, and increased hepcidin expression
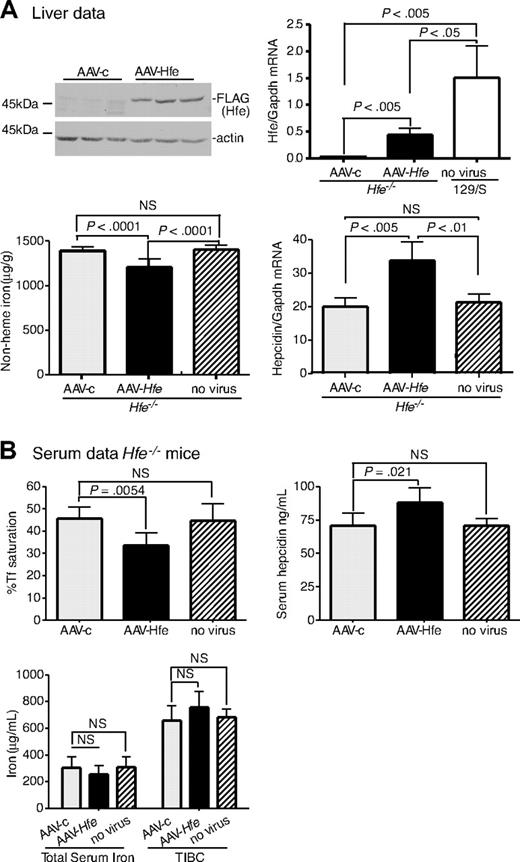
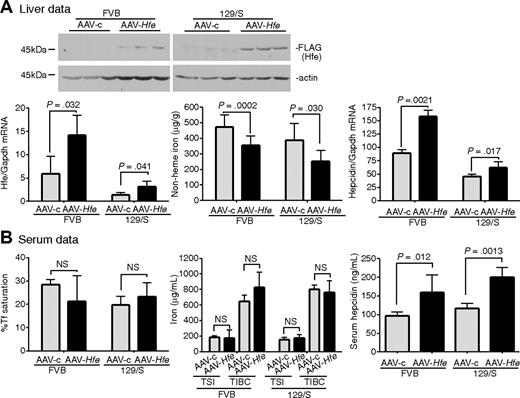
To determine the feasibility of using the AAV2/8 virus to introduce genes into mice, we tested whether AAV-Hfe could correct low levels of hepcidin and iron overload in Hfe−/− mice. AAV-Hfe was injected through the tail vein of 8-week-old male Hfe−/− mice on a 129/SvEvTac (129/S) background. The AAV2/8 vector used in our experiments is specifically targeted to liver and muscle,43 and the promoter/enhancing elements limit gene expression to hepatocytes.38 Mice injected with a virus (AAV-c) encoding glutaryl CoA-dehydrogenase, which is unrelated to iron metabolism, and animals not injected served as controls to test whether the vector injection increased hepcidin levels by causing inflammation. All mice were fed with standard mouse chow for 2 weeks and then killed. The timing was based on previous studies showing that expression of target proteins was achieved over this period.44 Antibodies to Hfe were insufficiently sensitive to detect endogenous Hfe in wild-type 129/S mice; therefore, the AAV-Hfe virus included a FLAG-epitope on the C-terminus of Hfe to allow for immunodetection.
Hfe−/− mice injected with AAV-Hfe showed hepatic expression of Hfe approximately 3-fold lower than in wild-type 129/S mice. Yet this subphysiologic level of Hfe expression was sufficient to decrease liver nonheme iron levels and to increase hepcidin mRNA compared with AAV-c–injected mice. Hepcidin mRNA did not change in response to the AAV-c virus, indicating that increased hepcidin mRNA in the mice injected with AAV-Hfe did not simply reflect an inflammatory response (Figure 1A, AAV-c vs no virus hepcidin mRNA). Measurements on the serum from Hfe−/− mice injected with AAV-Hfe showed that the Tf saturation decreased and hepcidin increased. Although no significant changes were seen in total serum iron levels and total iron-binding capacity, these results showed systemic effects on iron homeostasis in the injected mice compared with mice injected with a control virus and uninjected mice (Figure 1B). Thus, vector-mediated Hfe expression was sufficient to increase hepcidin levels and to decrease the iron overload in Hfe−/− mice.
Liver-specific expression of Hfe lowers iron levels, increases hepcidin expression, and lowers Tf saturation in Hfe−/− mice. Hfe−/− mice (n = 3) were injected with AAV-Hfe (5 × 1011 genome equivalents per mouse; ■ in all panels) or with the same amount of AAV-c virus (▩). Two weeks later, the mice were killed. (A) Liver extracts (100 mg) were analyzed by immunoblots with the use of anti–FLAG-M2 antibody or antiactin. Bands were detected by horseradish peroxidase secondary antibodies and enhanced chemiluminescence. Hfe mRNA was determined by quantitative reverse transcription–PCR (qRT-PCR) and normalized to Gapdh. The levels of Hfe mRNA in AAV-c– and in AAV-Hfe–injected Hfe−/− mice were compared with strain-, sex-, and age-matched control wild-type 129/SvTac (129/S) mice (□) with the use of qRT-PCR. Nonheme iron in livers of uninjected Hfe−/− mice ( ) and of AAV-c– and AAV-Hfe–injected Hfe−/− mice is described in “Nonheme iron assay.” Each sample was measured in triplicate. Hepcidin mRNA in Hfe−/− murine liver was determined by qRT-PCR. (B) Serum data. Percentage of Tf saturation was calculated from total serum iron and total iron-binding capacity (TIBC). The latter 2 parameters were measured as described in “Blood iron analysis.” Three to 4 mice were used in each group. All mice were on a 129/S background. The experiments were repeated with 2 more sets of mice with similar results. Error bars represent SD.
) and of AAV-c– and AAV-Hfe–injected Hfe−/− mice is described in “Nonheme iron assay.” Each sample was measured in triplicate. Hepcidin mRNA in Hfe−/− murine liver was determined by qRT-PCR. (B) Serum data. Percentage of Tf saturation was calculated from total serum iron and total iron-binding capacity (TIBC). The latter 2 parameters were measured as described in “Blood iron analysis.” Three to 4 mice were used in each group. All mice were on a 129/S background. The experiments were repeated with 2 more sets of mice with similar results. Error bars represent SD.
Liver-specific expression of Hfe lowers iron levels, increases hepcidin expression, and lowers Tf saturation in Hfe−/− mice. Hfe−/− mice (n = 3) were injected with AAV-Hfe (5 × 1011 genome equivalents per mouse; ■ in all panels) or with the same amount of AAV-c virus (▩). Two weeks later, the mice were killed. (A) Liver extracts (100 mg) were analyzed by immunoblots with the use of anti–FLAG-M2 antibody or antiactin. Bands were detected by horseradish peroxidase secondary antibodies and enhanced chemiluminescence. Hfe mRNA was determined by quantitative reverse transcription–PCR (qRT-PCR) and normalized to Gapdh. The levels of Hfe mRNA in AAV-c– and in AAV-Hfe–injected Hfe−/− mice were compared with strain-, sex-, and age-matched control wild-type 129/SvTac (129/S) mice (□) with the use of qRT-PCR. Nonheme iron in livers of uninjected Hfe−/− mice ( ) and of AAV-c– and AAV-Hfe–injected Hfe−/− mice is described in “Nonheme iron assay.” Each sample was measured in triplicate. Hepcidin mRNA in Hfe−/− murine liver was determined by qRT-PCR. (B) Serum data. Percentage of Tf saturation was calculated from total serum iron and total iron-binding capacity (TIBC). The latter 2 parameters were measured as described in “Blood iron analysis.” Three to 4 mice were used in each group. All mice were on a 129/S background. The experiments were repeated with 2 more sets of mice with similar results. Error bars represent SD.
) and of AAV-c– and AAV-Hfe–injected Hfe−/− mice is described in “Nonheme iron assay.” Each sample was measured in triplicate. Hepcidin mRNA in Hfe−/− murine liver was determined by qRT-PCR. (B) Serum data. Percentage of Tf saturation was calculated from total serum iron and total iron-binding capacity (TIBC). The latter 2 parameters were measured as described in “Blood iron analysis.” Three to 4 mice were used in each group. All mice were on a 129/S background. The experiments were repeated with 2 more sets of mice with similar results. Error bars represent SD.
Animals were examined for cell-specific expression of AAV2/8-derived Hfe by comparing Hfe in the livers, spleens, and intestine of Hfe−/− mice treated with AAV-Hfe (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All of these tissues contain macrophages. If the liver promoter was not specific for hepatocyte expression, then macrophages might express the AAV-derived Hfe. No Hfe could be detected in nonliver tissues.
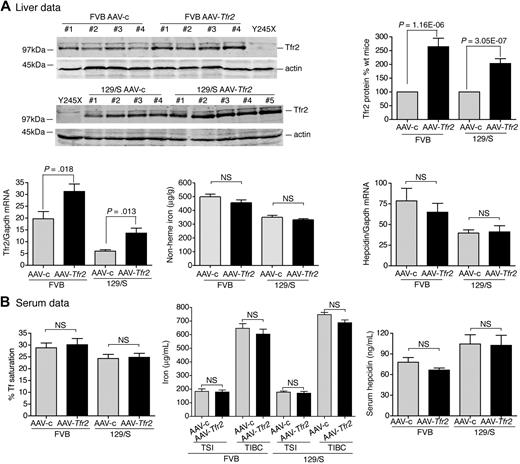
Expression of TfR2 in TfR2245x/245x mice lowers liver iron levels and serum Tf saturation and increases hepcidin expression
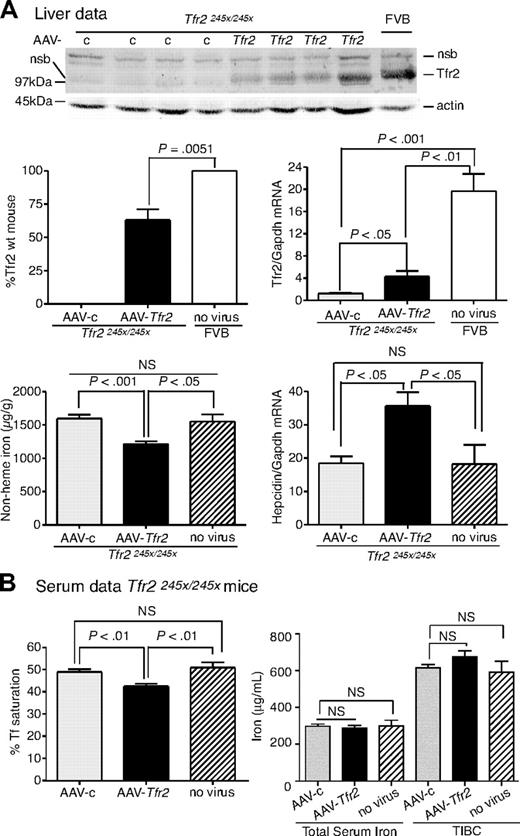
To test the ability of viral vector to rescue iron overload further, we also generated an AAV2/8 virus that encodes mouse Tfr2 (AAV-Tfr2). Tfr2-deficient mice (Tfr2245x/245x) on a FVB/NJ (FVB) background have a stop codon at amino acid 245 of Tfr2 and have iron overload.12 The equivalent mutation in humans produces HH.5 The mice have lower amounts of Tfr2 mRNA compared with wild-type mice and no detectable TfR2 protein.12
The effects of TfR2 expression on the iron overload in Tfr2245x/245x mice were examined. Tfr2245x/245x mice were injected with AAV-Tfr2 and killed 2 weeks later, similar to the procedures described for the injection of AAV-Hfe into the Hfe−/− mice. The average amount of liver Tfr2, measured by quantitative fluorescent immunoblots, was 53% of the levels in sex-, age-, and strain-matched wild-type controls (Figure 2A). This amount of Tfr2 was sufficient to lower nonheme iron in the liver by 23%, increase hepcidin mRNA by 1.9-fold, and lower serum Tf saturation by 13% (Figure 2B), indicating that the expression of Tfr2 in Tfr2-deficient mice partially corrected the iron overload. The mRNA of Tfr2 in AAV-c–injected mice was 15.5-fold lower than that of the wild-type FVB mice, suggesting that the Tfr2245x/245x transcript is less stable than the wild-type transcript. The lower amount of mRNA in the Tfr2245x/245x mice was noted previously and attributed to nonsense-mediated mRNA decay.12 The Tfr2 mRNA in the AAV-Tfr2–injected mice increased 3.5-fold compared with mice injected with AAV-c, but this was still less than wild-type strain-matched mice. Animals were examined for cell-specific expression of Tfr2 by comparing Tfr2 in the livers and spleens of Tfr2245x/245x mice treated with AAV-Tfr2 (supplemental Figure 1B). No Tfr2 was detected in the spleens, indicating that other splenic cell types, including macrophages, do not express Tfr2. The difference in levels of Tfr2 mRNA and protein is consistent with the posttranslational control of Tfr2 observed previously.22,23 These results indicate that expression of Tfr2 in hepatocytes was able to increase hepcidin levels and to decrease Tf saturation and nonheme iron levels within 2 weeks.
Liver-specific expression of Tfr2 lowers iron levels, increases hepcidin expression, and lowers Tf saturation in Tfr2245x/245x mice. (A-B) A similar protocol was used as outlined in Figure 1, except mice were injected with 2 × 1011 genome equivalents per mouse of AAV-Tfr2. In this case AAV-Tfr2 virus was injected into mice lacking functional Tfr2. The highest and lowest bands on the TfR2 immunoblot are nonspecific bands (nsb). Similar measurements were made as in Figure 1; ■ represent AAV-Tfr2 virus injected into Tfr2245x/245x mice; ▩, AAV-c virus injected into Tfr2245x/245x mice;  , uninjected Tfr2245x/245x mice; □, strain-, age-, and sex-matched wild-type mice. All mice were on a FVB/NJ (FVB) background. Four mice were used in each set of measurements. The experiment was repeated once with similar results. Error bars represent SD.
, uninjected Tfr2245x/245x mice; □, strain-, age-, and sex-matched wild-type mice. All mice were on a FVB/NJ (FVB) background. Four mice were used in each set of measurements. The experiment was repeated once with similar results. Error bars represent SD.
Liver-specific expression of Tfr2 lowers iron levels, increases hepcidin expression, and lowers Tf saturation in Tfr2245x/245x mice. (A-B) A similar protocol was used as outlined in Figure 1, except mice were injected with 2 × 1011 genome equivalents per mouse of AAV-Tfr2. In this case AAV-Tfr2 virus was injected into mice lacking functional Tfr2. The highest and lowest bands on the TfR2 immunoblot are nonspecific bands (nsb). Similar measurements were made as in Figure 1; ■ represent AAV-Tfr2 virus injected into Tfr2245x/245x mice; ▩, AAV-c virus injected into Tfr2245x/245x mice;  , uninjected Tfr2245x/245x mice; □, strain-, age-, and sex-matched wild-type mice. All mice were on a FVB/NJ (FVB) background. Four mice were used in each set of measurements. The experiment was repeated once with similar results. Error bars represent SD.
, uninjected Tfr2245x/245x mice; □, strain-, age-, and sex-matched wild-type mice. All mice were on a FVB/NJ (FVB) background. Four mice were used in each set of measurements. The experiment was repeated once with similar results. Error bars represent SD.
We tried to estimate the percentage of the hepatocytes transduced by the AAV2/8 viruses. Using the AAV2/8 virus encoding Hfe or Tfr2, we were able to achieve 23% of the Hfe mRNA levels and 57% of Tfr2 protein levels compared with control mice. We were unable to determine the percentage of hepatocytes transduced by each virus with the antibodies currently available. Injection of an AAV-GFP virus encoding green fluorescent protein at a lower titer than we used for our other experiments resulted in GFP protein in 75% to 90% of the hepatocytes (supplemental Figure 1C). These results are consistent with the partial recovery of iron loading in the liver.
Lack of inflammatory response in AAV-infected mice
The expression of hepcidin is sensitive to inflammation as well as to iron.45 Using the AAV-c virus, we showed that the AAV2/8 virus itself does not affect iron homeostasis (Figure 1). Despite these results, expression of genes in animals lacking the endogenous protein could still produce an inflammatory response. In particular, IL-6 can stimulate hepcidin expression through activation of signal transducer and activator of transcription 3, which is an acute-phase response protein. We used 2 approaches to look for evidence that expression of Hfe in Hfe−/− or Tfr2 in Tfr2245x/245x mice could elicit an inflammatory response. First, the histology of livers of 4 Tfr2245x/245x mice was compared with that of 4 Tfr2245x/245x mice injected with AAV-Tfr2. No evidence of lymphocytic infiltrates or other histologic evidence of inflammation was detected in the AAV-Tfr2–injected mice (supplemental Figure 2). Comparison of Hfe−/− mice and AAV-Hfe–injected mice yielded similar results (data not shown). Second, we used an enzyme-linked immunoabsorbent assay to test for changes in serum IL-6. IL-6 values in 4 Hfe−/− mice and in the 4 AAV-Hfe–injected mice were all below levels of detection. The average value of IL-6 in noninflammatory states in mouse serum is less than 7.8 pg/mL. The enzyme-linked immunoabsorbent assay is sensitive to approximately 2 pg/mL IL-6. The Tfr2245x/245x mice had a maximum of 2.8 pg/mL IL-6 and an average value below the sensitivity of the assay. No increase in IL-6 could be detected in the mice injected with AAV-Tfr2. According to the manufacturer, a mouse injected with 15 μg of lipopolysaccharide has a level of 31 800 pg/mL 2 hours after injection. Thus, mice injected with AAV-Tfr2 or AAV-Hfe virus and killed 2 weeks later had no evidence of inflammatory processes that would cause the changes in hepcidin. These results are consistent with the lack of immune response detected when α-glucosidase was expressed in α-glucosidase knock-out mice using the same type of virus.38
Tfr2 and Hfe, individually, are insufficient to change the expression of hepcidin
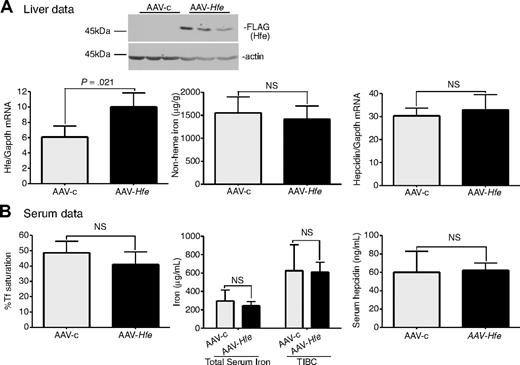
The partial correction of iron overload in Hfe-null mice by injection with AAV-Hfe virus, and in Tfr2-deficient mice with AAV-Tfr2 virus, led us to test whether overexpression of Tfr2 in Hfe-null mice and overexpression of Hfe in Tfr2-deficient mice could compensate for the lack of Hfe and Tfr2, respectively. A normal level of hepcidin expression requires the presence of both Hfe and Tfr2.25,26,37,41 Overexpression of one in the absence of the other should be unable to correct for the lower hepcidin expression. To test this model, TfR2245x/245x mice were injected with AAV-Hfe or AAV-c virus. Although increased expression of both Hfe protein and mRNA were observed (Figure 3A), no changes in iron parameters could be detected (Figure 3A-B).
Expression of Hfe in Tfr2245x/245x mice fails to increase hepcidin expression and to reduce iron levels. (A-B) An identical protocol was used as outlined in Figure 1. In this case AAV-Hfe (■) or AAV-c (▩) was injected into Tfr2245x/245x mice. Mice injected with AAV-Hfe express Hfe as judged by immunoblots and increased Hfe mRNA. No differences in nonheme iron were detected in the livers of AAV-Hfe–injected mice compared with AAV-c–injected Tfr2245x/245x mice. Three mice were used in each set of experiments. The experiment was repeated twice with similar results. Error bars represent SD.
Expression of Hfe in Tfr2245x/245x mice fails to increase hepcidin expression and to reduce iron levels. (A-B) An identical protocol was used as outlined in Figure 1. In this case AAV-Hfe (■) or AAV-c (▩) was injected into Tfr2245x/245x mice. Mice injected with AAV-Hfe express Hfe as judged by immunoblots and increased Hfe mRNA. No differences in nonheme iron were detected in the livers of AAV-Hfe–injected mice compared with AAV-c–injected Tfr2245x/245x mice. Three mice were used in each set of experiments. The experiment was repeated twice with similar results. Error bars represent SD.
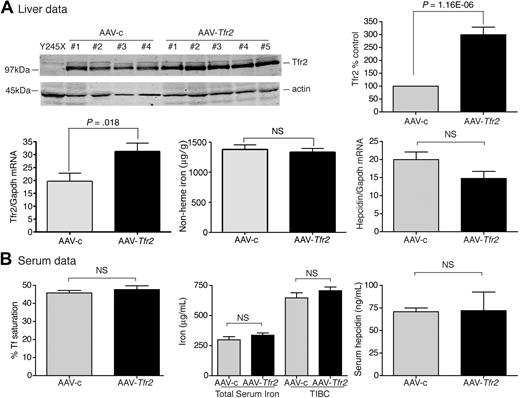
In a complementary set of experiments, we observed that expression of Tfr2 in Hfe-null mice also had no effect on iron homeostasis. In AAV-Tfr2–injected animals, Tfr2 mRNA increased by 2.1-fold and Tfr2 increased 3-fold compared with the AAV-c–injected animals (Figure 4A), but the levels of iron and hepcidin in the livers and serum remained unchanged (Figure 4A-B). A similar level of viral-mediated Tfr2 expression in AAV-Tfr2–injected Tfr2245x/245x mice decreased nonheme iron and increased hepcidin mRNA in the liver, along with a decrease in Tf saturation (Figure 2). The combined results of these experiments indicate that neither overexpression of Tfr2 in Hfe-null mice nor overexpression of Hfe in mice that lack functional Tfr2 is sufficient to correct the iron overload of the mutant strain, supporting a model in which the 2 proteins are both required.
Increased expression of Tfr2 in Hfe−/− mice fails to change hepcidin and iron levels. (A-B) An identical protocol was used as outlined in Figure 1. AAV-Tfr2 was injected into Hfe−/− mice. Increased Tfr2 expression in mice injected with AAV-Tfr2 (■) was detected by immunoblot with the use of an antibody to Tfr2 and not in AAV-c (▩) injected mice. Similarly, increased Tfr2 mRNA was detected in the livers of AAV-Tfr2–injected mice. No significant changes in the iron parameters of the liver or serum were detected despite the increased Tfr2 mRNA levels and Tfr2 protein levels. The experiment was repeated once with 4 mice in each group with similar results. Error bars represent SD.
Increased expression of Tfr2 in Hfe−/− mice fails to change hepcidin and iron levels. (A-B) An identical protocol was used as outlined in Figure 1. AAV-Tfr2 was injected into Hfe−/− mice. Increased Tfr2 expression in mice injected with AAV-Tfr2 (■) was detected by immunoblot with the use of an antibody to Tfr2 and not in AAV-c (▩) injected mice. Similarly, increased Tfr2 mRNA was detected in the livers of AAV-Tfr2–injected mice. No significant changes in the iron parameters of the liver or serum were detected despite the increased Tfr2 mRNA levels and Tfr2 protein levels. The experiment was repeated once with 4 mice in each group with similar results. Error bars represent SD.
Overexpression of Hfe, but not Tfr2, in wild-type mice increases hepcidin expression and lowers iron levels
To determine whether Hfe or Tfr2 was limiting in the regulation of hepcidin expression and iron homeostasis, wild-type mice, which already express normal levels of both Tfr2 and Hfe, were injected with AAV-Hfe, AAV-TfR2, or control (AAV-c) virus. When either of 129/S or FVB mouse strains were injected with AAV-Hfe, Hfe mRNA increased. Nonheme iron levels decreased and hepcidin mRNA levels increased (Figure 5A). Serum hepcidin levels similarly increased, but serum Tf saturation and iron levels in the blood remained unchanged (Figure 5B). In contrast, injection of AAV-Tfr2, produced a 2- to 3-fold increases in the levels of Tfr2 protein and mRNA in the liver, but it did not generate any significant changes in levels of hepcidin or iron in either the liver or serum (Figure 6). These results indicate that Hfe is limiting for hepcidin production in wild-type mice but Tfr2 is not.
Increased expression of Hfe in wild-type 129/S and FVB mice increases hepcidin expression and lowers iron levels. (A-B) AAV-Hfe (■) or AAV-c (▩) was injected into wild-type 129/S mice or wild-type FVB mice according to the protocol listed in Figure 1. Mice injected with AAV-Hfe (■) exhibit significantly reduced nonheme iron in liver compared with AAV-c (▩) injected 129/S and FVB mice. No detectable changes in Tf saturation, total serum iron (TSI), or transferrin iron-binding capacity (TIBC) were detected. In each strain of mice, expression of HFE was detectable by immunoblot analysis, and increased Hfe mRNA could be detected in the AAV-Hfe– but not the AAV-c–injected animals. Three to 4 mice were used in each group. The results are shown for 1 group of mice. The experiment was repeated twice with similar results.
Increased expression of Hfe in wild-type 129/S and FVB mice increases hepcidin expression and lowers iron levels. (A-B) AAV-Hfe (■) or AAV-c (▩) was injected into wild-type 129/S mice or wild-type FVB mice according to the protocol listed in Figure 1. Mice injected with AAV-Hfe (■) exhibit significantly reduced nonheme iron in liver compared with AAV-c (▩) injected 129/S and FVB mice. No detectable changes in Tf saturation, total serum iron (TSI), or transferrin iron-binding capacity (TIBC) were detected. In each strain of mice, expression of HFE was detectable by immunoblot analysis, and increased Hfe mRNA could be detected in the AAV-Hfe– but not the AAV-c–injected animals. Three to 4 mice were used in each group. The results are shown for 1 group of mice. The experiment was repeated twice with similar results.
Increased expression of Tfr2 in 129/S and FVB mice fails to increase hepcidin levels. (A-B) The same protocols were used and measurements were made as in Figure 5, except AAV-Tfr2 was injected into wild-type FVB or wild-type 129/S mice. No significant changes in the iron parameters of the liver or serum, including Tf saturation, total serum iron (TSI), transferrin iron-binding capacity (TIBC), or hepcidin, were detected despite the higher Tfr2 mRNA levels and TfR2 protein levels in the AAV-Tfr2–injected mice (■) compared with the AAV-c–injected mice (▩). Four mice were used in each set of measurements. The experiment was repeated with 4 more mice in each group with similar results. Error bars represent SD.
Increased expression of Tfr2 in 129/S and FVB mice fails to increase hepcidin levels. (A-B) The same protocols were used and measurements were made as in Figure 5, except AAV-Tfr2 was injected into wild-type FVB or wild-type 129/S mice. No significant changes in the iron parameters of the liver or serum, including Tf saturation, total serum iron (TSI), transferrin iron-binding capacity (TIBC), or hepcidin, were detected despite the higher Tfr2 mRNA levels and TfR2 protein levels in the AAV-Tfr2–injected mice (■) compared with the AAV-c–injected mice (▩). Four mice were used in each set of measurements. The experiment was repeated with 4 more mice in each group with similar results. Error bars represent SD.
Discussion
The liver plays a major role in the maintenance of iron homeostasis in both humans and mice by controlling the transcription of hepcidin, which is primarily expressed in this organ. Both direct and indirect evidence points to the importance of the HFE/TfR2/Tf complex in the regulation of hepcidin.37 Mice lacking functional Hfe or Tfr2 have a blunted hepcidin response in comparison to wild-type mice with a comparable iron load.25,26 In the present study, we used an AAV2/8 vector to express either Hfe or Tfr2 under the control of a hepatocyte-specific promoter. Expression of Hfe in Hfe-null mice alleviated iron overload in Hfe-null mice but not in mice deficient in Tfr2. Similarly, expression of Tfr2 corrected the iron overload of mice lacking functional Tfr2 but not in Hfe-null mice. These results are consistent with previous findings that hepatocyte-specific ablation of Hfe or Tfr2 in mice is sufficient to result in low hepcidin levels and increased iron accumulation.34,46 Importantly, we determined that, in wild-type mice, Hfe mRNA is a limiting factor in hepcidin signaling and that overexpression of Hfe increases hepcidin levels, which in turn decreases iron levels. This result is consistent with the observation that the molar amount of HFE protein is lower than that of TfR1 and TfR2 in solubilized human liver extracts.47 Superficially, these results appear to be in conflict with the idea that type 1 HH is a recessive disease. Both mouse studies and, to a lesser extent, human studies indicate that, although heterozygous persons do show detectably higher levels of iron loading, their iron loading is not close to what would be expected for a haplotype insufficiency.10,11 The protein levels of Hfe in Hfe+/− mice as well as in the AAV-Hfe–transduced mice remain to be determined.
HFE and TfR2 form a complex when expressed in tissue culture cells, leading to the hypothesis that they interact to form a sensor to detect the degree of Tf saturation.35,36 HFE also interacts with TfR1, but in contrast to TfR2, which forms a complex with Tf and HFE, Tf competes with HFE for binding to TfR1. Both crystallographic and extensive mutational analyses indicate that TfR1 interacts with the α1α2 domains of HFE.16,48,49 In contrast, TfR2 interacts with the α3 domain of HFE.36 Recently, Schmidt et al50 demonstrated that the 2 different types of interaction are physiologically important, by showing that mice that express a mutant Tfr1, which fails to interact with Hfe, have higher hepcidin mRNA levels and lower iron levels. These results suggest that TfR1 serves to sequester HFE and prevents its interaction with TfR2. Our results show that HFE mRNA is indeed limiting and, combined with those of Schimdt et al,50 imply 2 functions for iron-bound Tf beyond just delivering iron to cells: the release of HFE from TfR1 and the formation of an HFE/TfR2/Tf signaling complex.
In addition, we demonstrated that the AAV2/8 vector allows the controlled expression of proteins. Thus, this virus provides another tool to study the physiologic consequences of proteins that interact with each other or to study the disruption of protein interactions in vivo.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Nancy Andrews, Duke University, for providing the Hfe−/− mice; Christopher Corless, Oregon Health & Science University, for help in analyzing the liver sections; and Maria Chloupkova, Julia Maxson, Kia Nicholson, and An-Sheng Zhang, Oregon Health & Science University, for careful reading of the manuscript.
This work was supported by National Institutes of Health (grants R01-DK054408 and R01-DK072166; C.A.E), in part by (grant 5T32-HD049309; J.C.) and by the Center of Excellence in Molecular Hematology (grant SP30-DK072437; I.D.D.)
National Institutes of Health
Authorship
Contribution: J.G. and J.C. performed the experiments, analyzed data, and wrote the manuscript; I.D.D. performed experiments and analyzed data; D.M.K., C.O.H., and D.D.K. provided the viral vectors and assisted with experimental design and experiments; R.E.F. provided the Tfr2245x/245x mice and analyzed the data; and C.A.E. supervised the project, designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Caroline Enns, Department of Cell and Developmental Biology L215, 3181 SW Sam Jackson Park Rd, Portland, OR 97239; e-mail: ennsca@ohsu.edu.
References
Author notes
J.G. and J.C. contributed equally to this study.