Abstract
Transferrin receptor 2 (TFR2) is a transmembrane protein that is mutated in hemochromatosis type 3. The TFR2 gene is transcribed in 2 main isoforms: the full-length (α) and a shorter form (β). α-Tfr2 is the sensor of diferric transferrin, implicated in the modulation of hepcidin, the main regulator of iron homeostasis. The function of the putative β-Tfr2 protein is unknown. We have developed a new mouse model (KI) lacking β-Tfr2 compared with Tfr2 knockout mice (KO). Adult Tfr2 KO mice show liver iron overload and inadequate hepcidin levels relative to body iron stores, even though they increase Bmp6 production. KI mice have normal transferrin saturation, liver iron concentration, hepcidin and Bmp6 levels but show a transient anemia at young age and severe spleen iron accumulation in adult animals. Fpn1 is strikingly decreased in the spleen of these animals. These findings and the expression of β-Tfr2 in wild-type mice spleen suggest a role for β-Tfr2 in Fpn1 transcriptional control. Selective inactivation of liver α-Tfr2 in KI mice (LCKO-KI) returned the phenotype to liver iron overload. Our results strengthen the function of hepatic α-Tfr2 in hepcidin activation, suggest a role for extrahepatic Tfr2 and indicate that β-Tfr2 may specifically control spleen iron efflux.
Introduction
The maintenance of iron homeostasis is essential for all living organisms because iron is indispensable for cell metabolism but toxic when present in excess. Therefore, dietary iron absorption and iron recycling from macrophages, which destroy senescent erythrocytes, are tightly regulated processes.1 This regulation is mainly achieved by the liver peptide hepcidin (Hepc), which controls the expression of the iron exporter ferroportin (Fpn1) on the surface of duodenal and reticuloendothelial cells. After hepcidin binding, the hepcidin/Fpn1 complex is internalized, thus impairing iron export to the circulation.2
Dysregulation of this mechanism leads to iron overload that characterizes hereditary hemochromatosis (HH). HH is a heterogeneous genetic disorder caused by mutations in different genes encoding for proteins of the hepcidin activator (HFE, TFR2, and HJV) or effector (HEPC and SLC40A1) pathway. Mutational inactivation of HEPC or HJV causes a juvenile and severe HH form (type 2 HH, Online Mendelian Inheritance in Man [OMIM] no. 602390) because Hjv regulates hepcidin production through the Smad pathway, acting as a coreceptor that enhances Bmp signaling.3 HFE HH (type 1 HH, OMIM no. 235200) is an adult-onset, low penetrant disease, whereas transferrin receptor 2 (TFR2) hemochromatosis (type 3 HH, OMIM no. 604250)4 is less severe than the juvenile form but has a similar early onset.5,6 In addition, the phenotype produced by HFE and TFR2 inactivation suggests that the 2 proteins might play distinct roles in different periods of life. Mutations of SLC40A1, which encodes Fpn1, produce a different phenotype known as “ferroportin disease” (OMIM no. 604653).7
Although several data suggest a function for the complex Hfe-Tfr2 in iron sensing,8,9 the role of the 2 proteins in hepcidin activation remains elusive. Recent in vivo data show that Bmp6 is involved in the iron-regulatory pathway because its inactivation leads to iron overload in mice.10-12 Furthermore, Hfe is directly involved in the Bmp6-Smad pathway because Hfe-deficient mice show defective Smad signaling.13,14
TFR2 is a member of the transferrin receptor family, has a moderate homology to the transferrin receptor (TFRC), and is capable of binding transferrin, although with lower affinity.15 TFR2 is expressed at high levels in hepatocytes and at low levels in peripheral blood mononuclear cells and spleen15 and is not posttranscriptionally regulated by iron. Diferric transferrin (Fe2-Tf) is a strong modulator of Tfr2 trafficking because it increases the Tfr2 half-life16 by favoring both recycling and surface stabilization of the receptor17 and by inhibiting its lysosomal degradation. It has also been demonstrated that Tfr2 may interact simultaneously with Hfe and Fe2-Tf.8 The current model of Tfr2 function proposes that the Tfr2-Hfe complex, in association with Fe2-Tf, triggers signals that activate hepcidin transcription.9 However, the role of Tfr2 in Hepc activation and its potential relationship with the Hjv/Bmp6/Smad-dependent pathway remain unknown.
The TFR2 gene has 2 main transcripts: a longer tissue-specific form (α), mainly expressed in the liver, and a shorter one (β), with a putative start codon methionine at position 172 in exon 4, which is in frame with the major transcript. The β isoform lacks the intracellular and the transmembrane domains15 and is predicted to produce an intracellular/secreted protein of unknown function. Interestingly, although most of the TFR2 mutations in HH3 affect both isoforms, the M172K variant, occurring in 2 families,18,19 affects the putative translation start site of the β form, suggesting a specific role for this protein in iron regulation.
Currently, several HH type 3 murine models are available: mouse Y245X/Y245X is the product of targeted mutagenesis harboring the ortholog of human Y250X variant.20 Tfr2 knockout (KO; Tfr2−/−) mouse has been generated by germinal inactivation of the gene21 and tissue-specific Tfr2 KO mice have selective hepatic inactivation of Tfr2.22 All these mice develop iron overload in adult life, with features comparable with the human disease, including an abnormally low hepcidin production, and are thus valuable models of the disease. However, their contribution to a better understanding of the role of Tfr2 in iron metabolism has been limited. Recently, a Hfe/Tfr2 double KO mouse showed that the 2 proteins may act in parallel in vivo because the double null mice have an iron overload more severe than the single KO animals.23
In this paper, we report the generation and phenotype characterization of a new mouse model characterized by the specific inactivation of Tfr2 β isoform (KI mice). This was obtained by introducing into exon 4 of Tfr2 gene the M167K mutation, which corresponds to the human 172 variant found in some HH3 patients. KI mice were compared with Tfr2 germinally inactivated (KO) animals obtained in the identical genetic background. Furthermore, we selectively inactivated α-Tfr2 in the liver of KI animals (LCKO-KI). The study of iron metabolism of these mice at different ages suggests a role in tissues different from the liver for Tfr2 and distinct roles for the α and β isoforms.
Methods
Gene targeting construct
Tfr2-deficient mice were created using the Cre/lox recombination system. A modified version of Tfr2 exon 4 was generated by polymerase chain reaction (PCR) mutagenizing the 167 ATG codon to AAG and inserting a LoxP sequence 107 bp upstream of intron 3 splice acceptor site. This modified fragment was inserted into the 4.3-kb left homology arm; the 4.6-kb right arm encompassed Tfr2 intron 6-exon 10. Finally, the 2 arms were cloned in a Bluescript plasmid containing a Neo-TK double selection cassette flanked by 2 LoxP sites. The result was a multifunctional construct carrying the M167K mutation, 3 codirectional LoxP sequences, and a Neo-TK selection cassette (Figure 1A). After linearization, the construct was electroporated in C14 ES cells. Homologous recombination was verified by Southern blot after DNA digestion with EcoRI restriction enzyme. Heterozygous clones were selected based on the band size (of 10 and 7 kb, respectively; Figure 1B). Two of these clones were amplified and transiently transfected with Cre-expressing plasmid. In the presence of Cre recombinase, 2 different recombination events may occur (Figure 1A): recombination between LoxP 2 and 3 ablates the inactivation cassette and produces a knock-in M167K allele (KI mice) characterized by inactivation of the initiation codon of the β form and by 2 LoxP sites flanking exons 4 and 6, respectively. Recombination between LoxP 1 and 3 produces a constitutively inactive Tfr2 allele (KO mice).
Tfr2 genomic targeting construct. (A) The Tfr2 locus, the targeting construct, and the 2 mutagenized alleles are illustrated. Exons 3 to 7 of the murine Tfr2 gene are shown as black boxes. The inactivation cassette is in gray. LoxP sites are shown as arrows. M167K mutation is highlighted. Recombination between LoxP 2 and 3 generates Tfr2 KI allele. Recombination between LoxP 1 and 3 produces Tfr2 KO allele. E indicates EcoRI restriction sites used for construct cloning and Southern blot analysis. Barred line represents Southern blot probe. (B) Southern blot analysis showing first electroporation results to obtain the full targeted clones (7.1-kb band) versus the WT cell DNA (10-kb band). (C) Southern blot showing KI (3.9-kb band) and KO clones (2.4-kb band) obtained from Cre electroporation.
Tfr2 genomic targeting construct. (A) The Tfr2 locus, the targeting construct, and the 2 mutagenized alleles are illustrated. Exons 3 to 7 of the murine Tfr2 gene are shown as black boxes. The inactivation cassette is in gray. LoxP sites are shown as arrows. M167K mutation is highlighted. Recombination between LoxP 2 and 3 generates Tfr2 KI allele. Recombination between LoxP 1 and 3 produces Tfr2 KO allele. E indicates EcoRI restriction sites used for construct cloning and Southern blot analysis. Barred line represents Southern blot probe. (B) Southern blot analysis showing first electroporation results to obtain the full targeted clones (7.1-kb band) versus the WT cell DNA (10-kb band). (C) Southern blot showing KI (3.9-kb band) and KO clones (2.4-kb band) obtained from Cre electroporation.
Three days after transfection and gancyclovir selection, DNA from resistant clones was digested with EcoRI and analyzed by Southern blotting. Clones heterozygous for KI or KO alleles were identified (3.9- and 2.4-kb bands in Southern blot, respectively; Figure 1C).
Tfr2 KI and KO clones were microinjected in C57BL/6 strain blastocysts, and chimera were obtained. Male chimera were crossed with C57BL/6 females and the agouti descendants were genotyped using primers encompassing the floxed/deleted regions (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). KI alleles were identified using 2 different PCR protocols, using primers F1/R1 and F2/R2 (supplemental Figure 1B); KO alleles were identified using primers F3/R2 and F1/R2 (supplemental Figure 1C). Homozygous KI mice were intercrossed with the 129/sv Alb-Cre line (Alb-Cre)24 (kindly provided by Valeria Poli, University of Turin, Turin, Italy) to obtain a liver-specific inactivation of α-Tfr2 within a KI context (LCKO-KI mice). This genotype was confirmed by PCR using primers F1/R1 (supplemental Figure 1D).
Animal care
Tfr2-targeted mice were housed in the barrier facility at the Department of Clinical and Biological Sciences, University of Turin and maintained on a standard diet. All procedures were carried out with approval from the Institutional Animal Care and Use Committee of University of Turin. Experiments were performed on animals from 2 to 10 weeks on. Before any experimental procedures, the 3 Tfr2 lines were back-crossed to 129/sv mice for at least 10 generations, to have the mutations in a pure 129/sv background. The 129/sv mice of the same age were used as controls.
Hematologic and iron parameters
Blood (0.5 mL) was extracted by a single retro-orbital puncture from anesthetized mice. Blood cell counts and erythrocyte parameters were determined using an ADVIA120 Hematology System (Siemens Diagnostics). Transferrin saturation was calculated as a ratio of serum iron and total iron-binding capacity levels (Randox Laboratories Ltd).
To measure iron concentration, tissue samples were dried at 110°C overnight, weighed, and digested in 1 mL of acid solution (3M HCl, 0.6M trichloroacetic acid) for 20 hours at 65°C. The clear acid extract was added to 1 mL of working chromogen reagent (1 volume of 0.1% bathophenanthroline sulfate and 1% thioglycolic acid solution, 5 volumes of water, and 5 volumes of saturated sodium acetate). The solutions were then incubated for 30 minutes at room temperature until color development and the absorbance measured at 535 nm. A standard curve was plotted using an acid solution containing increasing amounts of iron diluted from a stock solution of Titrisol iron standard (Merck). All the hematologic analyses were performed in at least 8 animals.
Histology and Perl staining
For histologic studies and iron staining, tissues were fixed in aqueous formaldehyde solution (4% vol/vol) and embedded in paraffin (PARAPLAST Tissue Embedding medium; Tyco Healthcare). For histologic assessment of nonheme iron, slides of liver and spleen sections were stained with Perl Prussian blue. Hematoxylin and eosin counterstaining was used to mark nucleus and cytoplasm by standard procedures.
Macrophage isolation
Splenic macrophages were isolated from whole spleens of Tfr2-targeted mice and control sib pairs, using MACS CD11b MicroBeads (Miltenyi Biotec).
Quantitative RT-PCR
For reverse transcription, 1 μg of total RNA, 25μM random hexamers, and 100 U of reverse transcriptase (Applera) were used. Gene expression levels were measured by quantitative real-time reverse-transcription (RT)–PCR in an iCycler (Bio-Rad) using a quantitative RT-PCR assay (Assays-on-Demand; Applied Biosystems). For each PCR reaction, 1 μL of cDNA was added to 15 μL of PCR reaction mix containing 10 μL of TaqMan Universal PCR Master Mix (Applera). All analyses were carried out in triplicate; results showing a discrepancy greater than 1 threshold cycle in 1 of the wells were excluded.
Real-time PCR of Bmp6 gene was performed according to published protocols11 using SYBR Green–based methodology.
Gus (β-glucuronidase) gene was used as housekeeping control. The results were analyzed using the ΔΔthreshold cycle (Ct) method. Transcriptional data of targeted mice were evaluated normalizing their ΔΔCt values to wild-type (WT) ΔΔCt mean.
The statistical significance of the differences of mRNA expression levels between controls and targeted mice was evaluated using a nonparametric Student t test (unpaired, 2 tailed).
Western blot
Between 50 and 100 μg of proteins from cell lysates or homogenized liver and spleen tissues was separated in 8% to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotted according to standard protocols.
Antibodies were the following: anti-TFR2 (to detect the α isoform), anti-Fpn1 MTP1 (Alpha Diagnostics International), anti-Tfr2 S-20 (to detect the β isoform), and antiactin (Santa Cruz Biotechnology); the anti–ferritin L antibody was kindly provided by Paolo Arosio (University of Brescia, Brescia, Italy).
Tfr2 transfection
To develop a positive control for α- and β-Tfr2 proteins, full-length cDNAs were reverse transcribed from murine liver and amplified through PCR. The PCR products were cloned into pcDNA3.1 expression vector (Clontech) and transiently transfected in HEK293T cells using Gene Jammer Transfection Reagent (Stratagene), according to the manufacturer's protocols. Transfection efficiency was checked by transfection with pEGFP-N3 plasmid (Clontech).
Results
Generation of targeted animals
A multifunctional construct able to produce 3 different Tfr2-deficient murine models was prepared. Methionine at codon 167 (ATG) of α-Tfr2 transcript was mutagenized, substituting T with A. This mutation should not affect significantly the function of the α isoform because a secondary structure analysis (Network Protein Sequence Analysis, http://npsa-pbil.ibcp.fr; CLC Protein Workbench, http://www.clcbio.com) predicts that the lysine would correctly localize in the extracellular domain. On the other hand, the same mutation is predicted to abolish the translation start site of the β isoform. Using the Cre/lox recombination system, we generated Tfr2 KO mouse, in which both α- and β-Tfr2 isoforms were germinally inactivated, and Tfr2 KI mouse, in which only the β one was silenced. Moreover, crossing KI animals with mice selectively expressing Cre recombinase in the liver (Alb-cre), we generated Tfr2 LCKO-KI mice, in which both the α- and the β-Tfr2 isoforms are inactivated in the liver, whereas only the β-Tfr2 is inactivated in all other tissues (Table 1).
Genomic targeting of the three Tfr2 mouse models
| . | KI . | KO . | LCKO-KI . |
|---|---|---|---|
| Liver | |||
| α-Tfr2 | + | − | − |
| β-Tfr2 | − | − | − |
| Other tissues | |||
| α-Tfr2 | + | − | + |
| β-Tfr2 | − | − | − |
| . | KI . | KO . | LCKO-KI . |
|---|---|---|---|
| Liver | |||
| α-Tfr2 | + | − | − |
| β-Tfr2 | − | − | − |
| Other tissues | |||
| α-Tfr2 | + | − | + |
| β-Tfr2 | − | − | − |
KO indicates knockout; KI, knockin; LCKO-KI, hepatic knockout in a knockin background; +, positive; and −, negative.
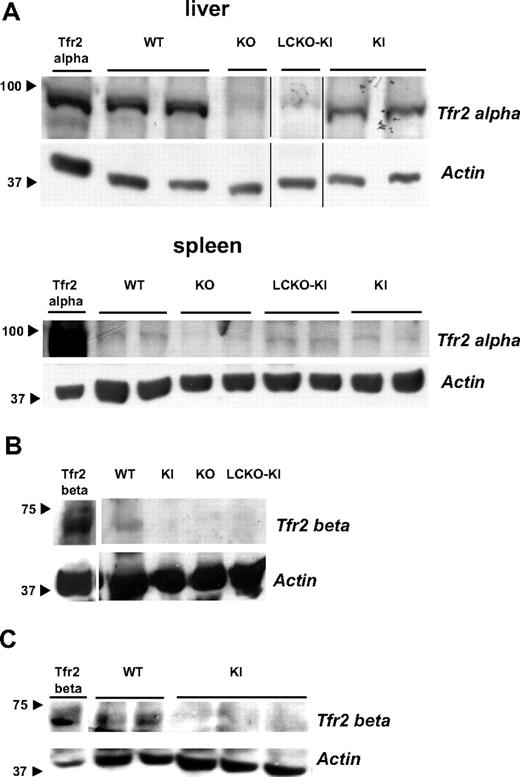
Western blot analysis showed that α-Tfr2 protein is absent in the liver of KO and LCKO-KI but present in tissues other than the liver (spleen) in both KI and LCKO-KI animals (Figure 2A). β-Tfr2 was found to be a 65-kDa protein, as expected based on the amino acid composition. It was detected in the spleen of control animals but in none of the 3 models (Figure 2B). Western Blot analysis demonstrated that β-Tfr2 is produced in WT splenic macrophages (Figure 2C).
Molecular detection of the Tfr2 proteins in the targeted mice. (A) Western blot demonstrating lack of α-Tfr2 in liver and spleen of KO and LCKO-KI and persistence of the protein in KI mice. First lane indicates transfected α-Tfr2 used as positive control. Tissues from different animals per genotype are shown; anti-TFR2 (Alpha Diagnostics International) antibody was used. Black arrows indicate marker size. Vertical lines indicate reposition of the same gel's image. (B) Western blot demonstrating lack of β-Tfr2 in KI, KO, and LCKO-KI spleen. First lane indicates transfected β-Tfr2 used as positive control; anti-Tfr2 S-20 antibody (Santa Cruz Biotechnology) was used. Black arrows indicate marker size. (C) Western blot showing β-Tfr2 in WT and band absence in KI splenic macrophages. First lane indicates transfected β-Tfr2 used as positive control; anti-Tfr2 S-20 antibody (Santa Cruz Biotechnology) was used. Black arrows indicate marker sizes.
Molecular detection of the Tfr2 proteins in the targeted mice. (A) Western blot demonstrating lack of α-Tfr2 in liver and spleen of KO and LCKO-KI and persistence of the protein in KI mice. First lane indicates transfected α-Tfr2 used as positive control. Tissues from different animals per genotype are shown; anti-TFR2 (Alpha Diagnostics International) antibody was used. Black arrows indicate marker size. Vertical lines indicate reposition of the same gel's image. (B) Western blot demonstrating lack of β-Tfr2 in KI, KO, and LCKO-KI spleen. First lane indicates transfected β-Tfr2 used as positive control; anti-Tfr2 S-20 antibody (Santa Cruz Biotechnology) was used. Black arrows indicate marker size. (C) Western blot showing β-Tfr2 in WT and band absence in KI splenic macrophages. First lane indicates transfected β-Tfr2 used as positive control; anti-Tfr2 S-20 antibody (Santa Cruz Biotechnology) was used. Black arrows indicate marker sizes.
Phenotype analysis
The progeny obtained by intercrossing all the heterozygous strains had the expected Mendelian frequencies.
Phenotype analysis of the different Tfr2 models was performed in 10 weeks and in young (14-day-old) mice. The choice of the latter point was based on the low levels of Hepc transcription observed at this age in normal C57BL/6 mice, probably resulting from weaning and body growth (A.R., R.M.P., C.C., unpublished data, 2008).
Blood cell counts showed that KI mice had moderate macrocytic anemia when evaluated at 14 days (supplemental Table 1). These mice were smaller than age-matched WT sib pairs (8.2 ± 0.1 g vs 11.6 ± 0.4 g; P < .001). However, they reached normal size (21.7 ± 0.6 g vs 21.8 ± 0.2 g) and normal hematologic parameters by 10 weeks of age. Conversely, adult KO mice have hemoglobin and MCH levels higher than controls, whereas LCKO-KI have normal hematologic values at all stages (supplemental Table 1).
At 10 weeks of age, homozygous KO and LCKO-KI mice displayed high transferrin saturation, whereas KI mice had normal transferrin saturation (Figure 3A). Moreover, the transferrin saturation showed a trend toward greater levels in LCKO-KI than in KO, although the difference was not statistically significant.
Iron status of Tfr2-deficient mice. (A) Transferrin saturation values in the 3 mouse models at 10 weeks of age. Transferrin saturation was measured on 5 animals for each strain. ***P < .001 versus WT. Bars represent SD in the analyzed animals group. (B) LIC in Tfr2-targeted mice at different ages. Statistical differences between LIC values of the different Tf2-targeted mice versus WT sib pairs at the same ages: *P < .05, **P < .01, and ***P < .001. Statistical difference between LIC values of Tfr2 KO and LCKO-KI mice at 22 weeks of age is also reported (P < .001). Bars represent SD in the analyzed animals group.
Iron status of Tfr2-deficient mice. (A) Transferrin saturation values in the 3 mouse models at 10 weeks of age. Transferrin saturation was measured on 5 animals for each strain. ***P < .001 versus WT. Bars represent SD in the analyzed animals group. (B) LIC in Tfr2-targeted mice at different ages. Statistical differences between LIC values of the different Tf2-targeted mice versus WT sib pairs at the same ages: *P < .05, **P < .01, and ***P < .001. Statistical difference between LIC values of Tfr2 KO and LCKO-KI mice at 22 weeks of age is also reported (P < .001). Bars represent SD in the analyzed animals group.
The kinetics of liver iron deposition in the 3 models was examined (Figure 3B). Liver iron concentration (LIC) was already increased even in 14-day-old KO and LCKO-KI mice compared with WT, further increasing at 22 and 52 weeks. Statistical analysis between 22-week-old animals revealed higher iron deposits in LCKO-KI than in KO (P < .001). Surprisingly, KI mice at a young age had normal LIC. Liver iron deposition was observed only in older mice (P < .05 at 22 weeks and P < .01 at 52 weeks) when the LIC value was significantly increased compared with control animals of the same age.
Interestingly, the inactivation of liver α-Tfr2 in KI mice (LCKO-KI) reverted the phenotype to liver iron overload (Figure 3). Hepatic ferritin light chain (Ft L) was increased in livers of KO and LCKO-KI from 10 weeks of age, which further confirms iron overload, although it was similar in KI and control mice (not shown).
Molecular characterization of hepatic iron genes
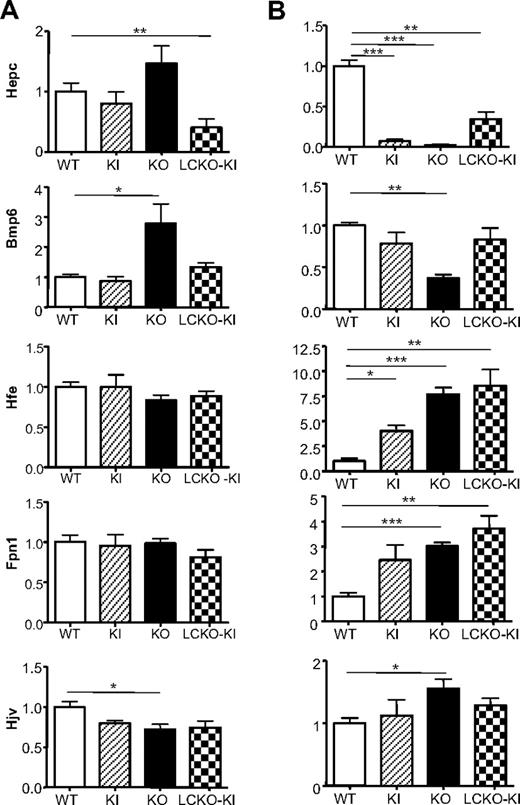
Quantitative analysis of the transcripts of liver Hepc and those of other iron genes provided different results in the 3 models (Figure 4). At 10 weeks of age (Figure 4A), Hepc expression was similar to that of controls in KO and KI mice but significantly decreased in LCKO-KI. Considering the different degree of iron overload, Hepc levels were inappropriate to LIC in KO, severely depressed in LCKO-KI, and normal in KI. Bmp6 transcripts were increased in KO and normal in KI animals as expected, but they were normal also in iron-loaded LCKO-KI. Liver Hfe and Fpn1 RNA levels were similar to controls in the 3 Tfr2-deficient animals, whereas Hjv mRNA was significantly reduced only in KO mice.
Liver iron genes expression in Tfr2-deficient mice. Relative hepatic iron gene expression in (A) 10-week-old and (B) 14-day-old KI, KO, and LCKO-KI mice compared with age-matched WT sib pairs. Gene expression analysis was performed on 18 animals for Hepc transcripts and on at least 5 animals for strain for the other genes. *P < .05, **P < .01, and ***P < .001 versus WT. Bars represent SD in the analyzed animals group.
Liver iron genes expression in Tfr2-deficient mice. Relative hepatic iron gene expression in (A) 10-week-old and (B) 14-day-old KI, KO, and LCKO-KI mice compared with age-matched WT sib pairs. Gene expression analysis was performed on 18 animals for Hepc transcripts and on at least 5 animals for strain for the other genes. *P < .05, **P < .01, and ***P < .001 versus WT. Bars represent SD in the analyzed animals group.
The expression of liver Hepc and of the other iron genes was analyzed also in 14-day-old animals (Figure 4B). Although Hepc levels showed high variability at this age, they were significantly decreased in all 3 recombinant models. Bmp6 RNA levels were decreased in KO but normal in LCKO-KI and KI. Hfe transcripts were significantly increased in all models, although to a lower extent in KI mice. A statistically significant increase of hepatic Fpn1 RNA and protein (not shown) was found in KO and LCKO-KI, but not in KI animals. The Hjv transcripts were significantly increased only in KO animals.
Spleen iron overload of adult KI mice
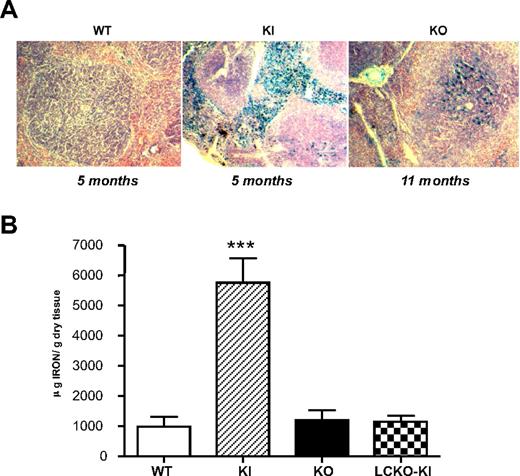
The spleens of KI adult mice compared with age-matched WT animals on Perl staining were strongly positive (Figure 5A). Iron overload was evident mainly in the red pulp, where iron-recycling macrophages are localized. In KO animals, spleen iron deposition was evident, mainly in the white pulp, only from 11 months of age (Figure 5A).
Spleen histology and iron staining. (A) Spleen histology and Perl staining of 5-month-old KI mice compared with aged-matched controls (WT) and 11-month-old KO animals. A Motic BA300 microscope (Mikron Instrument; original magnification ×200) was used with DPX mounting medium and a DPlanApo 20 UV PL, 20×/0.7 NA dry objective (Olympus), captured with a DFC320 (Leica) camera using Leica application suite Version 2.8.1, and processed using Photoshop Version 7.0 (Adobe Systems). (B) SIC in the 3 mouse models at 10 weeks of age. SIC was measured on at least 5 animals for each strain. ***P < .001 versus WT. Bars represent SD in the analyzed animals group.
Spleen histology and iron staining. (A) Spleen histology and Perl staining of 5-month-old KI mice compared with aged-matched controls (WT) and 11-month-old KO animals. A Motic BA300 microscope (Mikron Instrument; original magnification ×200) was used with DPX mounting medium and a DPlanApo 20 UV PL, 20×/0.7 NA dry objective (Olympus), captured with a DFC320 (Leica) camera using Leica application suite Version 2.8.1, and processed using Photoshop Version 7.0 (Adobe Systems). (B) SIC in the 3 mouse models at 10 weeks of age. SIC was measured on at least 5 animals for each strain. ***P < .001 versus WT. Bars represent SD in the analyzed animals group.
Consistently, spleen iron concentration (SIC) at 10 weeks of age was remarkably increased in KI mice. On the contrary, SIC was comparable with WT in KO and LCKO-KI animals (Figure 5B).
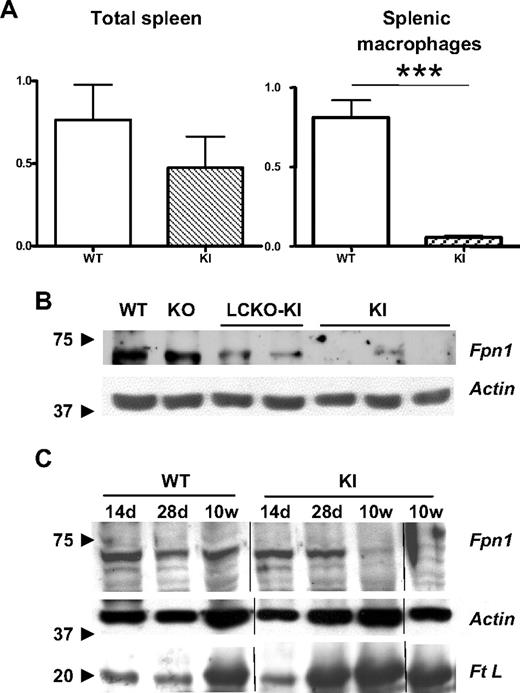
Because of the heavy iron deposition, spleen Fpn1 expression was studied in KI adult mice. Fpn1 transcription was slightly down-modulated in total spleen tissue but significantly impaired in isolated macrophages (Figure 6A). These cells showed a strong decrease also of Fpn1 protein (Figure 6B). The same findings were observed in isolated LCKO-KI but not in KO macrophages (Figure 6B), supporting the hypothesis of a regulatory effect of β-Tfr2 on Fpn1 expression in the spleen.
Spleen Fpn1 gene and protein expression in KI mice. (A) Relative spleen Fpn1 gene expression in 10-week-old KI mice compared with WT sib pairs. The assay was performed on at least 5 animals for each strain. ***P < .001 versus WT. Bars represent SD in the analyzed animal groups. (B) Western blot analysis of Fpn1 protein in splenic macrophages isolated from the 3 Tfr2 models at 10 weeks of age and compared with WT. Arrows indicate the position of the marker proteins. (C) Western blot analysis of spleen Fpn1 and Ft L proteins in KI mice of different ages versus WT. d indicates days; and w, weeks. Arrows indicate the position of the marker proteins. Vertical lines indicate reposition of the same gel's image.
Spleen Fpn1 gene and protein expression in KI mice. (A) Relative spleen Fpn1 gene expression in 10-week-old KI mice compared with WT sib pairs. The assay was performed on at least 5 animals for each strain. ***P < .001 versus WT. Bars represent SD in the analyzed animal groups. (B) Western blot analysis of Fpn1 protein in splenic macrophages isolated from the 3 Tfr2 models at 10 weeks of age and compared with WT. Arrows indicate the position of the marker proteins. (C) Western blot analysis of spleen Fpn1 and Ft L proteins in KI mice of different ages versus WT. d indicates days; and w, weeks. Arrows indicate the position of the marker proteins. Vertical lines indicate reposition of the same gel's image.
To investigate the kinetics of spleen iron deposition in KI animals, Ft L was analyzed by Western blot at different time points compared with Fpn1 expression. Spleen Ft L was strikingly increased in KI mice already at 28 days, whereas in controls the protein increase started at 10 weeks (Figure 6C). Furthermore, Fpn1 decreased with age in KI mice, in agreement with iron accumulation.
To monitor the evolution of the phenotype of KI animals during aging, the main iron parameters in 52-week-old animals were evaluated. Blood values were in the normal range; LIC was lower than KO and LCKO-KI mice even if it were significantly increased compared with age-matched controls (Figure 3B), SIC remained high (4700 μg/g ± 1000), and Hepc transcription was low and comparable with that of normal controls (not shown).
Discussion
Previous models of Tfr2 inactivation in mice recapitulate HH, with liver iron loading, increased transferrin saturation, and decreased hepcidin production. To further investigate the function of α-Tfr2 and to demonstrate a role of the β-Tfr2 isoform, we generated mice in which the M167K mutation was introduced by targeted mutagenesis of murine Tfr2 coding sequence (KI). In this way, the β-Tfr2 protein production was abrogated.
The phenotype of KI mice was then compared with that of KO generated in the same genetic background by measuring blood cell counts, iron parameters, and hepcidin regulation. Because iron metabolism is strongly influenced by growth and development, the comparison was performed between adult (10-week-old) and young (14-day-old) mice. This analysis provides distinct information about hepcidin regulation because in adult animals we explore the ability to activate hepcidin to prevent animals from iron overload (store regulator), whereas in young animals because of the expanding erythroid mass, we explore the ability to inhibit hepcidin (erythroid regulator). The comparison of the different genotypes revealed interesting and surprising findings.
As expected, adult KO mice have increased transferrin saturation and high LIC levels in the presence of normal splenic iron, features that confirm previous findings20-23 and recapitulate the human disease. They have hemoglobin levels in the normal range but slightly higher than controls and macrocytic red cells. In contrast, adult KI mice have normal transferrin saturation and LIC and develop liver iron overload only with aging. Although it has been previously shown that, in overexpressing cells, the mutated Tfr2-M167K α protein is predominantly retained in the endoplasmic reticulum and does not reach the plasma membrane,25 the phenotype of KI mice indicates that in vivo this variant is functional. Furthermore, KI mice show transient macrocytic anemia during weaning but surprisingly accumulate iron in spleen macrophages. Increased splenic macrophage iron could be the result of increased uptake or decreased export. Because macrophages acquire iron mainly from the breakdown of red cells and young KI mice are anemic, an increased destruction of erythroid cells was considered. However, reticulocyte counts similar to controls rule out a hemolytic component. In addition, anemia is transient and adult KI mice have normal hemoglobin levels. Because both Fpn1 RNA and protein are decreased in total spleen of adult KI animals and even more in isolated macrophages, we propose that spleen iron sequestration is the consequence of impaired iron release. This mechanism would be consistent with the normal transferrin saturation and, together with the lack of hepatocyte iron loading, account for normal liver Hepc and Bmp6, whose levels were shown to be related to liver iron stores.26
On this basis, we speculate that β-Tfr2, which is mostly expressed in the spleen15 (and present data), might be a sensor of the iron recycled from erythropoiesis and that inactivation of the sensor might lead to spleen iron retention. β-Tfr2 expression in isolated macrophages is compatible with this hypothesis. Based on the strong reduction of Fpn1 mRNA in isolated macrophages of KI and LCKO-KI mice, we hypothesize that β-Tfr2 exerts a positive transcriptional control on Fpn1. Thus, loss of β-Tfr2 would reduce Fpn1 mRNA, whereas normal hepcidin levels would have the additive effect of decreasing Fpn1 protein in KI mice. When α-Tfr2 is inactivated in LCKO-KI, the strong hepcidin reduction would restore iron export by residual Fpn1, but at the same time, the increased transferrin saturation would favor liver iron deposition. In this model, the effect on hepcidin exerted by α-Tfr2 appears to prevail over the function of the β isoform in the spleen.
It is puzzling that 10-week-old KI mice show a phenotype different from patients with type 3 HH resulting from the same mutation.18,19 Some differences in iron metabolism between mice and humans are to be expected considering the different dietary iron intake and absorption. Nevertheless, anemia may never have been reported in patients with type 3 HH because they only underwent clinical assessment for hemochromatosis as adults, when anemia is also compensated in the mouse model. Moreover, because splenic iron is usually not measured in patients for technical reasons, its increase would probably remain undetected. In addition, the single M172K patient who had a liver biopsy consistent with HH features had an associated β-thalassemia trait and clinical manifestation could be partially be the result of secondary iron overload.27 On the other hand, phenotypic analysis of 52-week-old KI mice supports the hypothesis that hepatic iron overload could only be delayed. We conclude that the difference between the iron phenotype of mice and humans with β-Tfr2 inactivation requires further investigation.
The HH phenotype of LCKO-KI strengthens the role of hepatic α-Tfr2 and is compatible with the proposed complex Tfr2-Hfe, whose formation is favored by increased diferric transferrin to activate hepcidin,9 a function lost in type 3 HH.28 Iron overload in LCKO-KI is even more severe than in KO mice. Accordingly, Hepc RNA, although inappropriately low relative to body iron stores, was similar in adult controls and KO animals but is significantly down-regulated in LCKO-KI mice. BMP6 was recently identified as the cytokine that orchestrates the Bmp-Hjv-Smad hepcidin-activating pathway.10-12 KO mice retain some ability (∼ 3-fold) to up-regulate Bmp6, as observed in Hfe KO mice,13,14 but this ability is entirely lost in LCKO-KI.
Because both KO and LCKO-KI have an identical hepatic Tfr2 genotype, discrepant Bmp6 and Hepc levels may indicate a role for extrahepatic α-Tfr2. Our hypothesis is that the severity of iron overload in adult KO is partially mitigated by the absence of a Tfr2 inhibitory function in some other tissues.
It has been demonstrated that α-Tfr2 has a remarkable expression in early erythroid precursors.29 We speculate that the absence of α-Tfr2 in bone marrow might result in an impaired inhibitory effect on Bmp6/Hepc pathway and that this effect persists in LCKO-KI. It remains to be established whether this regulatory pathway has to be accounted for by the release of cytokines reported to inhibit hepcidin transcription.30,31 KO animals have macrocytic red cells and have slightly higher hemoglobin values than WT, whereas no major abnormalities are observed in LCKO-KI, except for mild macrocytosis. Macrocytic red cells are observed in iron-loaded HH patients of different types32 and in Hfe KO mice33 and could result from erythropoiesis occurring in conditions of excess iron.
Hepc transcription in young mice is low, with large variations even in controls and is associated with low to normal Bmp6 in all models. Hepcidin levels have been found to be inappropriately impaired also in young Hfe KO mice.13 At 14 days, increased Hfe transcription is observed in all models and increased Fpn1 in KO and LCKO-KI. Considering that in adult life there is no difference between Tfr2-deficient and WT mice, we suggest that, in the absence of Tfr2, the increased expression of Hfe (and Fpn1) that occurs in adulthood is anticipated.
Based on our findings, the absence of β-Tfr2 in the presence of a preserved function of the α form seems to affect erythropoiesis in young mice and iron metabolism in spleen macrophages rather than in hepatocytes, even protecting the latter from iron loading. Our results reveal that α and β-Tfr2 have a distinct function in mouse liver and spleen, respectively. The role of Tfr2 isoforms in other tissues warrants further studies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Emanuela Tolosano and Enrico Bracco, University of Torino, for their precious advice and support in discussing the results.
This work was supported by Regione Piemonte (grant A200) and Telethon Fondazione Onlus, Rome (grant GGP08089; C.C.).
Authorship
Contribution: A.R. designed and performed the research, analyzed the data, and wrote the paper; F.D.C. and E.H. designed the research and reviewed the paper; O.A. performed the ES cell culture and selection; R.M.P. and A.B. analyzed animal phenotype and performed the experiments; S.C. performed real-time PCR experiments; B.M. and F.R. did the hematologic analysis; I.D. made the transfection experiments; L.S., D.C., and F.A. reviewed and provided critical revision to the paper; and C.C. and G.S. analyzed the data, contributed to writing the paper, and reviewed the paper.
Conflict-of-interest disclosure: G.S. is a member of the advisory board and speaker's bureau of Novartis and Bristol-Myers Squibb and received research grants from Novartis. The remaining authors declare no competing financial interests.
Correspondence: Antonella Roetto, University of Torino, Department of Clinical and Biological Sciences, AO San Luigi Gonzaga, 10043, Orbassano, Torino, Italy; e-mail: antonella.roetto@unito.it.
References
Author notes
C.C. and G.S. contributed equally (as senior authors) to this study.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal