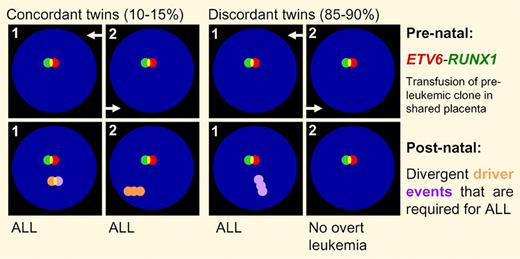
In this issue of Blood, Bateman and colleagues identify the ETV6-RUNX1 fusion as a common prenatal event at the origin of t(12;21)(p13;q22) ALL.1 Additional genetic lesions in the preleukemic clone are required but follow divergent evolutionary pathways toward overt leukemia.
When paleontologists backtrack the sequence of genetic variations in the evolution2 of a species of their interest, they can use radiometric dating methods in the analysis of their fossil samples.3 In the natural history of human acute lymphoblastic leukemia (ALL), relevant (driver) and neutral (passenger) genetic events can be identified, but only as the cumulative outcome of a complex clonal evolution process. The sequence of the individual events and their specific role in branching of the evolutionary tree in the transformation process remain elusive. In analogy to radiometric dating methods, Mel Greaves and colleagues have developed over the past 2 decades a similarly powerful technology, which allows them to resolve the natural history of childhood ALL carrying the t(12;21)(p13;q22). This method is based on the comparative analysis of monozygotic twins and “fossil” evidence from blot spot Guthrie cards and cord bloods.
Acquisition of prenatal and postnatal lesions during the evolution of the preleukemic clone.
Acquisition of prenatal and postnatal lesions during the evolution of the preleukemic clone.
The first case of monozygotic twins concordantly developing acute leukemia was described in 1882 and a large number of additional cases were subsequently identified.4 In approximately 10% to 15% of cases, childhood ALL develops concordantly in monozygotic twins,5 most of whom share a single placenta. In these cases, concordant development of ALL can be explained by prenatal transfusion of a preleukemic clone from one twin to the other. Because even in these cases, the majority (85%-90%) of co-twins do not develop leukemia, and it is likely that multiple genetic events are required for full transformation. Here, Bateman et al analyzed SNP array data from t(12;21)(p13;q22) ALL, which developed concordantly in 5 pairs of monozygotic twins. The central question of their analysis was whether the ETV6-RUNX1 fusion, resulting from the t(12;21)(p13;q22) translocation, indeed represents the initial genetic lesion at the origin of this ALL subtype. Bateman et al identified 32 previously recognized “driver” lesions, which (1) are recurrently found in ALL and (2) are mechanistically implicated in leukemogenesis. Strikingly, not a single one of these 32 lesions was concordant among twin pairs. These findings identify the ETV6-RUNX1 fusion as the only unifying event at the origin of ALL and demonstrates that acquisition of secondary genetic lesions follows divergent pathways of clonal evolution. Bateman et al conclude that most if not all of the secondary “driver” lesions are of postnatal origin. If they were acquired before birth, these lesions were likely shared between both co-twins because of feto-fetal transfusion through vascular anastomoses in one common placenta. In addition to 5 monozygotic twin pairs with concordant leukemia, 1 discordant twin pair was studied. Whereas the ETV6-RUNX1 fusion was found in both twins, the predicted “driver” lesions present in the twin with leukemia were absent in the healthy twin. This supports the concept that the t(12;21)(p13;q22) translocation, resulting in expression of the ETV6-RUNX1 oncogene, occurs prenatally. Whereas ETV6-RUNX1 alone is not sufficient, full leukemic transformation depends on accumulation of secondary “driver” lesions, which are acquired most likely after birth. The SNP analysis used in their study typically does not detect point mutations and small deletions, one or more of which could in theory, represent initiating events at the onset of leukemia that were missed in this analysis. The fact, however, that all of the 32 “driver” lesions that were actually detected in this analysis were indeed acquired subsequent to the ETV6-RUNX1 fusion argues against this possibility.
The finding that ETV6-RUNX1 alone is not sufficient for leukemic transformation is consistent with previous findings that only about 1% of newborns carrying a significant level of preleukemic clones with ETV6-RUNX1 actually develop leukemia.6 This low frequency of leukemic development on the basis of ETV6-RUNX1 seems to contrast the 10- to 15-fold higher frequency of concordant twins with ETV6-RUNX1 ALL (10%-15%). One likely explanation for this seeming discrepancy is based on the fact that monozygotic twins share any genetic risk factors that may predispose to leukemia. Three predisposing genetic variants at 7p12.2, 10q21.2, and 14q11.2 were recently identified in genome-wide association studies7 ; however, whether and how such predisposing factors cooperate with ETV6-RUNX1 remains to be elucidated.
In addition, the mechanism(s) that drive the evolution of preleukemic clones are currently not known. Recent work implicated the B cell–specific immunoglobulin hypermutation enzyme AID in clonal evolution of certain ALL subtypes.8 In addition, the RAG1 and RAG2 enzymes not only mediate V(D)J recombination of immunoglobulin gene segments but also cooperate with AID in the acquisition of chromosomal rearrangements, which may also drive clonal evolution of ALL.9 This raises the intriguing possibility that key mechanisms of genetic diversification in adaptive immune responses, namely RAG1/2-dependent V(D)J recombination and AID-dependent hypermutation, drive clonal evolution of ALL. Fifty years ago, Frank Macfarlane Burnet and others first applied Darwin's selection theory to the clonal evolution of adaptive immune responses.10 The study by Bateman et al adds to the emerging scenario that the same mechanisms may be active in the natural history of ALL.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■