Abstract
Fanconi anemia (FA) is a genetically heterogeneous, autosomal recessive disorder characterized by pediatric bone marrow failure and congenital anomalies. The effect of FA gene deficiency on hematopoietic development in utero remains poorly described as mouse models of FA do not develop hematopoietic failure and such studies cannot be performed on patients. We have created a human-specific in vitro system to study early hematopoietic development in FA using a lentiviral RNA interference (RNAi) strategy in human embryonic stem cells (hESCs). We show that knockdown of FANCA and FANCD2 in hESCs leads to a reduction in hematopoietic fates and progenitor numbers that can be rescued by FA gene complementation. Our data indicate that hematopoiesis is impaired in FA from the earliest stages of development, suggesting that deficiencies in embryonic hematopoiesis may underlie the progression to bone marrow failure in FA. This work illustrates how hESCs can provide unique insights into human development and further our understanding of genetic disease.
Introduction
Fanconi anemia (FA) is a complex autosomal recessive disorder characterized by marrow aplasia, congenital abnormalities, and a predisposition to malignancy. The major source of mortality in FA is complications associated with bone marrow failure.1 The median age of onset for pancytopenia in FA is 7 years, with thrombocytopenia and anemia generally preceding neutropenia.2 Blood counts at birth are typically normal, although there is evidence that patient marrow is hypoplastic and deficient in CD34+ hematopoietic progenitors well before hematologic complications arise.3,4 In addition, FA patients have an approximately 15 000-fold increased risk of developing acute myelogenous leukemia compared with the healthy population, as well as an elevated risk for other solid tumors, most notably squamous cell cancers of the head and neck.2 Finally, a spectrum of associated dysmorphologies is frequently found at birth including skeletal abnormalities such as radial ray defects and scoliosis, abnormal skin pigmentation, and aberrant kidney and urinary tract development.5
FA demonstrates genetic heterogeneity; biallelic mutations in any one of at least 13 different genes leads to the same condition. Patients with mutations in the same gene comprise members of the same complementation group, reflecting the initial gene identification strategies using cell-fusion techniques (genetic complementation).1 The genes for the 13 known complementation groups have been cloned and all appear to function in a common pathway regulating DNA repair. The key diagnostic criterion for FA is hypersensitivity to DNA cross-linking agents, such as mitomycin C (MMC), suggesting that FA cells fail to appropriately sense and/or resolve interstrand cross-links.6 Eight of the FA proteins (FANCA, B, C, E, F, G, L, M) form a nuclear complex that functions as an E3 ubiquitin ligase for the FANCI-FANCD2 (ID) heterodimer.7 Upon monoubiquitination, the ID heterodimer is targeted to nuclear foci that contain BRCA1, RAD51, and BRCA2/FANCD1, where it is thought to participate in homology-directed DNA repair.8 In addition, FANCD2, FANCI, and FANCD1, components of the pathway that act “downstream” from the core complex, appear to function within a broader set of interactions aimed at maintaining genomic integrity, intersecting with several pathways that are mutated in other chromosome instability syndromes including ataxia-telangiectasia, Nijmegen breakage syndrome, and Bloom syndrome.1 Although the disease is similar clinically among complementation groups, recent studies have suggested that patients in rare complementation groups that are downstream of the core complex such as FA-D2 (3%-6% of all cases) and FA-D1 (< 1%) have more severe disease than patients with more common mutations in the core complex components such as FA-A (65% of all cases),9,10 perhaps reflecting intrinsic developmental differences among complementation groups.
Although there has been extensive biochemical characterization of the FA pathway and its role in maintaining genomic integrity, the connection between cellular deficiencies in DNA repair and the specific clinical phenotypes of marrow aplasia and skeletal malformation remains poorly understood. Previous studies have described DNA damage–induced apoptosis and aberrant cellular signaling, especially in the STAT1 (signal transducer and activator of transcription 1) pathway, as possible mechanisms of hematopoietic cell loss in FA,11,12 although their importance to the pathophysiology of marrow failure in patients remains uncertain. Classically, patients with FA have normal blood counts at birth, but subsequently undergo progressive loss of hematopoietic progenitors and stem cells, resulting in a median age at presentation of 7 years.2,4,13 Neonatal aplastic anemia in FA has been described,14,15 although hematopoietic dysfunction may not be recognized in childhood because of the significant compensatory mechanisms present in the bone marrow. The observation that marrow hypocellularity often precedes discrete clinical symptoms coupled with the presence of developmental abnormalities at birth has led to speculation that the FA pathway may have an important role during embryonic development, especially within the hematopoietic system.4,16
Studies of the developmental aspects of FA have been lacking because of the absence of hematopoietic dysfunction in mouse models of the disease. Individual gene knockouts of Fanca, Fancc, Fancg, and Fancd2 have been generated, and although all knockout mice display an increased sensitivity to DNA cross-linking agents, none develop marrow hypoplasia, bone marrow failure, leukemia, or skeletal abnormalities (reviewed in Parmar et al17 ). Subtle hematopoietic differences do exist: notably, decreased long-term repopulating activity18 and hypersensitivity to oxidative stress19 and inhibitory cytokines such as interferon gamma.20 However, the absence of marrow aplasia, the central pathology of the human disease, strongly suggests that knockout mouse models are inadequate for the study of hematopoietic failure in FA.
Direct analysis of bone marrow from FA patients is restricted by the limited proliferative potential of primary hematopoietic cells, as well as the specific problem of obtaining tissue samples from aplastic FA marrow. Limited studies with human FA hematopoietic progenitors have described a similar hypersensitivity to inhibitory cytokines and reactive oxygen species,11,21 suggesting that cellular stress–induced apoptosis may play an important role in the development of marrow aplasia in FA.13 However, studies of FA patient marrow are incapable of addressing the effect of FA gene deficiency on the genesis of hematopoietic tissue that occurs during embryonic development. Because such developmental studies cannot be performed in humans for practical and ethical reasons, the effect of FA gene deficiency on early hematopoietic development remains uncharacterized.
Human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) have the potential to serve as a powerful platform for the study of human development and its dysfunction in genetic disease.16 Previous reports have demonstrated that hESC lines carrying mutations for Lesch-Nyhan and fragile X syndrome phenocopy their respective diseases,22,23 and reprogramming of somatic cells from patients has enabled the generation of disease-specific iPSCs.24 Directed differentiation of hESCs/iPSCs into specific tissues from any of the 3 embryonic germ layers through manipulation of culture conditions and/or genetic modification enables detailed study of the cell fate decisions that occur during development. Although hematopoietic differentiation from iPSCs remains to be fully characterized, many groups have reported embryoid body (EB) or stromal coculture differentiation schemes from hESCs that generate CD45+ hematopoietic cells containing mature myeloid and lymphoid elements, erythromyeloid progenitors with methylcellulose colony-forming activity, and low numbers of repopulating cells displaying limited chimerism upon transplantation into immunodeficient mice.25-27 In vitro hematopoietic differentiation from hESCs recapitulates many important aspects of early embryonic hematopoietic development, including specification from a Brachyury-positive mesodermal progenitor,28 the presence of a bipotential hemangioblast,29 and temporal globin switching from embryonic to fetal hemoglobins reflecting the transition from extraembryonic to intraembryonic hematopoiesis.30 Thus, hESCs provide a scalable in vitro model for obtaining unique and valuable insight into early human hematopoietic development.
Recently, the Izpisúa-Belmonte group reported that FA patient fibroblasts could not be reprogrammed to FA iPSCs in the absence of genetic complementation (Raya et al),31 which only serves to highlight the need for alternate methods to study early hematopoietic development in FA. We have adopted a lentiviral RNA interference (RNAi) strategy in hESCs to test whether embryonic hematopoiesis is dysfunctional in FA. In this report, we knocked down 2 FA genes: FANCA, the most commonly mutated gene in FA, and FANCD2, a gene mutated in a rare complementation group. We chose FANCA as a representative member of the FA core complex and FANCD2 for its role as a central downstream actor in the FA pathway.8 After generation of stable knockdown hESC lines (referred to as FANCAi and FANCD2i), we demonstrate that FANCAi and FANCD2i hESCs display the characteristic DNA repair defects found in FA. Using directed in vitro differentiation to model embryonic hematopoietic development, we then show that FANCD2i hESCs demonstrate significant reductions in hematopoietic gene expression and progenitor numbers upon differentiation, whereas FANCAi hESCs display a milder hematopoietic phenotype with intermediate reductions in hematopoietic gene expression and progenitor numbers. FA gene complementation of FANCAi and FANCD2i hESCs rescues the hematopoietic phenotype, substantiating that the observed hematopoietic deficits are specifically related to the FA gene knockdowns. These results establish that not only are the earliest stages of hematopoietic development impaired in FA, but that suggested differences in the severity of disease between complementation groups may have a developmental basis. Our work demonstrates the value of hESCs in enabling novel studies of early hematopoietic development in FA and has implications for our understanding of the pathogenesis of bone marrow failure in FA patients.
Methods
hESC cell culture and maintenance
hESC lines H9 and BGO1 were maintained as undifferentiated cells on inactivated primary mouse embryo fibroblasts (MEFs; Specialty Media). hESC lines were cultured in 80% knockout Dulbecco modified Eagle medium or Dulbecco modified Eagle medium: Nutrient Mix F-12 supplemented with 20% knockout serum replacement, 1% nonessential amino acids, 1mM l-glutamine (all from Invitrogen), 0.1mM mercaptoethanol (Sigma-Aldrich), and 10 ng/mL human recombinant basic fibroblast growth factor (Invitrogen). hESC lines were passaged weekly by treatment with collagenase IV (Gibco) for 10 minutes followed by cell scraping and mild trituration.
siRNA vector design
Three oligonucleotides encoding stem-loop structures targeting the FANCA and FANCD2 genes were cloned under the control of the human U6 promoter in the pLentilox vector.32 The targeting sequences within FANCA are AAGCTGTCTTCCCTGTTAGAGTT, AAGCATAACATGGAGCTCTTGTT, and AAGAAGGCCCTGGTCTTCCTGTT and within FANCD2 are AAGGGAGAAGTCATCGAAGTATT, AAGCAGCTCTCTAGCACCGTATT, and AAGCTACAGAAGTTGTGCAACTT. A control oligonucleotide targeting the firefly Luciferase gene was directed against GAGCTGTTTCTGAGGAGCC.
Lentiviral vector production and hESC transduction
Lentiviral vectors, pseudotyped with the vesicular stomatitis virus G protein, were produced in 293T cells as described previously.33 hESCs were transduced on Matrigel (BD) in MEF-conditioned media by single-round infections for 24 hours, followed by expansion onto MEFs and enrichment of infected hESCs by a combination of fluorescence-activated cell sorting (FACS) and manual picking of green fluorescent protein (GFP)–positive colonies. Both FANCAi and FANCD2i hESC lines were infected with a mixture of 3 knockdown viruses (each with a different target sequence).
Southern blot analysis
Genomic DNA was obtained using the DNAEasy kit (QIAGEN). GFP probes were obtained by polymerase chain reaction (PCR) amplification from pLentilox vector. Probes were labeled and Southern hybridization was performed according to standard protocols.
Western blot analysis
Western blotting analysis was carried out using a monoclonal antibody against FANCD2 (1:500 dilution; Abcam) followed by a horseradish peroxidase–conjugated rabbit anti–mouse secondary antibody (1:5000; Roche). FANCA Western blotting was performed using a polyclonal antibody (1:400; Abcam) followed by goat anti–rabbit secondary antibody (1:5000; Roche)
Teratoma formation and histologic analysis
Nonobese diabetic/severe combined immunodeficient mice were injected subcutaneously with 1 to 2 × 106 hESCs resuspended in phosphate-buffered saline. Teratomas were excised, paraffin-embedded, and analyzed histologically via hematoxylin and eosin sections in a blinded fashion for tissues from all 3 embryonic germ layers.
FANCD2 immunofluorescence
hESCs were plated on Matrigel with MEF-conditioned media in 6-well plates and treated with 2mM hydroxyurea (Sigma-Aldrich) for 24 hours. Immunofluorescence staining was performed as described previously.8 The anti-FANCD2 monoclonal antibody (E35 rabbit polyclonal antiserum8 courtesy of A.S. and A. D'A., 1:400 dilution blocking buffer) was added for 2 hours at room temperature, followed by secondary Texas Red anti–mouse antibody (1:400; Jackson ImmunoResearch Laboratories) for 2 hours in the dark. The cells were visualized with a Zeiss Axioplan 2 microscope (63×/1.4 NA oil objective), and samples were scored in a blinded fashion for FANCD2 nuclear foci. A threshold of 4 FANCD2 foci within a cell was required for positivity.34
Chromosomal breakage analysis
hESC cultures were treated with 200nM MMC for 24 hours in the dark at 37°C. Cultures were then harvested after a 2.5-hour exposure to 30 ng/mL colcemid (Sigma-Aldrich). After a 30-minute treatment with 75mM KCl, the cells were fixed with a 3:1 mixture of methanol–acetic acid. Nuclear pellets were sent to Cell Line Genetics in a blinded fashion for cytogenetic analysis. At least 50 metaphase cells from each culture were scored for DNA breaks and radial formations. Breakage scores were calculated using the following scoring guide: chromatid gap, 0.25; chromosome gap, 0.5; chromosome break, 1; deletions, rings, and triradials, 2; and quadriradial, 3.
Hematopoietic differentiation of hESCs and analysis
In vitro EB-based hematopoietic differentiation was performed and single-cell suspensions from EB-differentiated hESCs were obtained as described previously.25 FACS staining was performed using CD45-allophycocyanin and CD34-phycoerythrin antibodies (1:40 dilution; R&D Systems) for 30 minutes on ice, followed by 2 washes with phosphate-buffered saline, and analysis using a FACSCalibur (BD Biosciences). Quantification of cell death was done by FACS staining with 7-amino-actinomycin D (BD Biosciences) either on dissociated EBs directly or after placement for 3 days in a liquid hematopoietic culture described in Wang et al.35 Images visualized using Leica DMII L, 10×/0.22 NA dry objective.
Gene expression analysis
Multiple independent biologic samples were obtained for RNA isolations. Total RNA was extracted using the RNAEasy kit (QIAGEN), treated with DNase I (Ambion), and cDNAs were prepared using the Superscript III First Strand Synthesis System (Invitrogen) according to the manufacturer's instructions. Real-time PCR was performed with SYBR green reagent kits (Stratagene) on an iCycler Real-Time machine (Bio-Rad). Primers were obtained from Integrated DNA Technologies. Primer sequences are listed in supplemental Table 6 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Human homeobox and cell-cycle gene quantitative PCR array was performed on a Stratagene Mx3000P real-time machine and data were analyzed according to the manufacturer's instructions (SABiosciences).
Copy number analysis
Quantitative real-time PCR was used to determine the average number of lentiviral integrations in genomic DNA isolated from infected hESCs as previously described.36 In all the real-time PCR analyses, a genomic DNA sample from a cloned induced pluripotent stem cell line with 6 copies of an enhanced GFP lentiviral transgene was included as a reference control.
Colony-forming unit assays
EB-differentiated hESC populations (1 × 104 to 2 × 105) were plated into methylcellulose H4434 (StemCell Technologies) and incubated at 37°C and 5% CO2 for 14 days. When indicated, MMC (Sigma-Aldrich) was added to H4434 at a final concentration of 5 nM. Colony-forming units (CFUs) were scored based on morphologic characteristics.
Retroviral vector infection and selection
Construction of the pMMP-puro, pMMP-puro hFANCA, and pMMP-puro hFANCD2 retroviral vectors and generation of retroviral supernatants was described previously.37 hESCs were transduced on Matrigel (BD Biosciences) in MEF-conditioned media by single-round infections for 24 hours, followed by expansion onto MEFs and 2 rounds of puromycin selection (1 μg/mL) for 72 hours.
Mouse Fancd2−/− ESC derivation and hematopoietic differentiation
Fancd2−/− (and wild-type control) mouse ESCs were derived from timed matings of gene-trap Fancd2 heterozygote mice (K. Parmar, J. Kim, S. Sykes, A.S., P. Stuckert, K. Zhu, A. Hamilton, M. K. DeLoach, J. L. Kutok, K. Akashi, D. G. Gilliland, A.D., submitted manuscript). ESC genotypes were confirmed by genomic PCR. EB-based in vitro hematopoietic differentiation was performed as described previously.38 All animal studies were performed with the approval of the Children's Hospital Boston Institutional Animal Care and Use Committee.
Statistical analysis
Student t test and the χ2 test were used for pairwise statistical analysis. The one-way analysis of variance was used in Figures 4 and 5 to determine significance in multiway comparison, followed by Tukey posthoc test. P values are indicated in supplemental Table 5 and in the figures (***P < .01; **.01 < P < .03; and *.03 < P < .05).
Results
Generation of FA hESCs
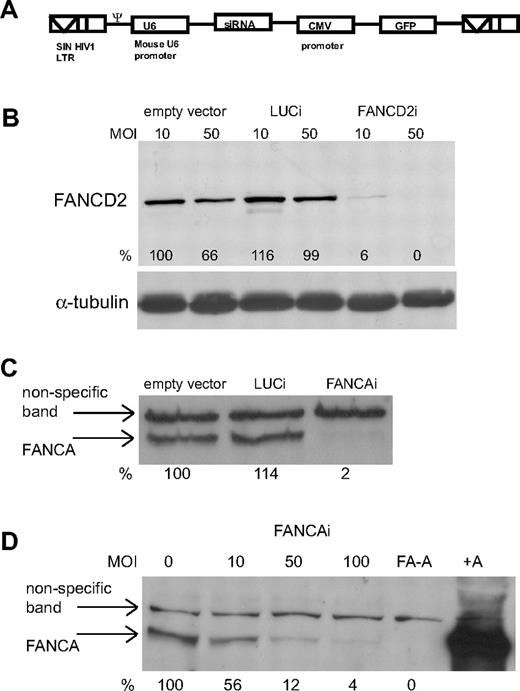
We have previously shown that lentiviral vectors can be used to efficiently knock down gene expression in hESCs.33 A schematic of the lentiviral vector is shown in Figure 1A. We designed short hairpin RNAs (shRNAs) targeting FANCA and FANCD2 and infected hESCs at various multiplicities of infection (MOIs). In all experiments, shRNAs against the Luciferase gene (LUCi) served as a control. Knockdown of FANCD2 and FANCA is highly efficient in hESCs (Figure 1B-C). Furthermore, a titration of MOIs for the FANCA knockdown virus shows that gene knockdowns occur in a dose-dependent manner (Figure 1D).
Efficient knockdown of FA proteins in hESCs by RNAi. (A) Schematic of the lentiviral vector used to deliver small interfering RNAs. (B) FANCD2 Western blot on lysates from GFP+ FACS for hESCs infected at MOI 10 and 50 with empty vector, LUCi, and FANCD2i virus at day 4 after infection. Percentage of knockdown is calculated using ImageJ software (National Institutes of Health) after normalization to α-tubulin levels. (C-D) FANCA Western blot on lysates from GFP+ FACS of hESCs infected either with empty vector, LUCi, and FANCAi virus at MOI 50 (C) or increasing MOIs of FANCAi virus (D) at day 4 after infection. Percentage of knockdown is calculated using ImageJ software after normalization to a nonspecific band. (D lane 5, FA-A) A lysate from a FA-A patient fibroblast; (lane 6, +A) the patient fibroblast line retrovirally corrected with FANCA.
Efficient knockdown of FA proteins in hESCs by RNAi. (A) Schematic of the lentiviral vector used to deliver small interfering RNAs. (B) FANCD2 Western blot on lysates from GFP+ FACS for hESCs infected at MOI 10 and 50 with empty vector, LUCi, and FANCD2i virus at day 4 after infection. Percentage of knockdown is calculated using ImageJ software (National Institutes of Health) after normalization to α-tubulin levels. (C-D) FANCA Western blot on lysates from GFP+ FACS of hESCs infected either with empty vector, LUCi, and FANCAi virus at MOI 50 (C) or increasing MOIs of FANCAi virus (D) at day 4 after infection. Percentage of knockdown is calculated using ImageJ software after normalization to a nonspecific band. (D lane 5, FA-A) A lysate from a FA-A patient fibroblast; (lane 6, +A) the patient fibroblast line retrovirally corrected with FANCA.
To generate stable, uniformly GFP+ cell lines for each knockdown virus and LUCi controls, 3 independent infections of hESCs were subjected to a combination of serial FACS and manual picking of GFP+ colonies. This was especially important as the carrier state in FA has no increased risk for disease, suggesting that a 50% reduction in these proteins is not pathogenic.39 During the enrichment process, hESC lines were always maintained as a population to minimize any nonspecific effects related to the selection of individual aberrant clones. Genomic copy number analysis showed, on average, between 4 and 7 lentiviral integrations per cell and, importantly, Southern blot analysis confirmed that the selection process did not cause the emergence of dominant clones and that all of our hESC lines remain polyclonal (supplemental Figure 1).
FANCAi and FANCD2i hESC lines maintain a stable knockdown for more than 40 passages with high expression of the pluripotency markers OCT4, NANOG, and Tra-1-60, display multilineage differentiation from EBs in vitro, and form teratomas upon injection into immunodeficient mice (data are summarized in supplemental Table 1). No significant differences were found among the FANCAi, FANCD2i, and LUCi hESC lines in terms of pluripotency markers, growth characteristics, or broad differentiation potential. We suggest the inability to generate FA iPSCs31 may relate to a requirement for the FA pathway during the reprogramming process, as it does not appear necessary for maintenance of the pluripotent state.
FA hESCs have defective DNA repair
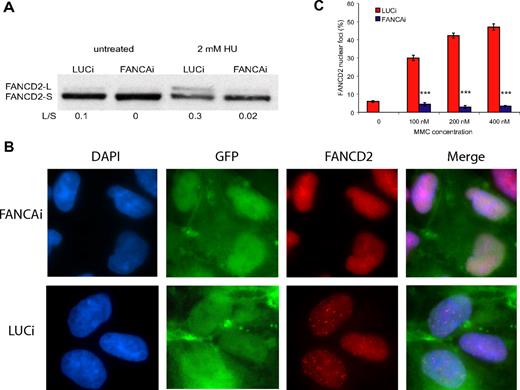
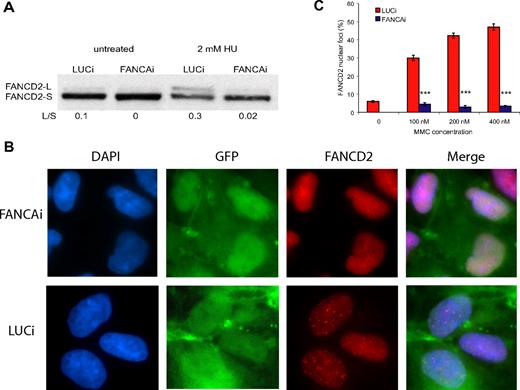
FANCA is a member of the FA core complex, which acts to monoubiquitinate the ID heterodimer during S phase and in response to DNA damage or cellular stress. Upon treatment with 2mM hydroxyurea (HU), LUCi hESCs show a strong up-regulation of FANCD2 monoubiquitination, which is greatly attenuated in FANCAi hESCs (Figure 2A). To further study the effect of FANCA knockdown on FA core complex function, we examined the relocalization of FANCD2 to nuclear foci, the putative sites of DNA repair. FANCD2 monoubiquitination is necessary and sufficient to cause FANCD2 relocalization to nuclear foci8 and thus serves as an indicator of FA core complex function. As shown in Figure 2B, FANCD2 nuclear foci are prominent in control cells treated with 200nM MMC, whereas they are absent in FANCAi hESCs. Quantification of FANCD2 nuclear foci after 100nM, 200nM, and 400nM MMC treatment demonstrates a dramatic reduction in the percentage of foci in the FANCAi hESC line, which never rises above baseline even at the highest MMC dose (Figure 2C). Taken together with the strong attenuation of FANCD2 monoubiquitination, this demonstrates that FANCA knockdown disrupts FA core complex function in hESCs.
FANCA knockdown results in marked reduction of FANCD2 monoubiquitination and nuclear foci formation. (A) FANCAi and LUCi hESCs were treated with 2mM HU for 24 hours, and lysates were subjected to Western blotting for FANCD2. FANCD2-L is the active, monoubiquitinated form of FANCD2-S. The ratio of L/S was calculated using ImageJ software. (B) FANCD2 immunofluorescence of FANCAi and LUCi hESCs treated with 200nM MMC treatment for 24 hours. DAPI (4,6 diamidino-2-phenylindole) stains for DNA content and GFP marks infected cells. (C) Quantification of FANCD2 nuclear foci after 100, 200, and 400 nM MMC treatment for 24 hours. Percentage FANCD2 nuclear foci (y-axis) represents the fraction of cells with 4 or more nuclear foci, as assayed by FANCD2 immunofluorescence. Data are represented as the average ± SEM; each data point represents 3 independent sets of 50 cells. ***P < .01 by Student t test.
FANCA knockdown results in marked reduction of FANCD2 monoubiquitination and nuclear foci formation. (A) FANCAi and LUCi hESCs were treated with 2mM HU for 24 hours, and lysates were subjected to Western blotting for FANCD2. FANCD2-L is the active, monoubiquitinated form of FANCD2-S. The ratio of L/S was calculated using ImageJ software. (B) FANCD2 immunofluorescence of FANCAi and LUCi hESCs treated with 200nM MMC treatment for 24 hours. DAPI (4,6 diamidino-2-phenylindole) stains for DNA content and GFP marks infected cells. (C) Quantification of FANCD2 nuclear foci after 100, 200, and 400 nM MMC treatment for 24 hours. Percentage FANCD2 nuclear foci (y-axis) represents the fraction of cells with 4 or more nuclear foci, as assayed by FANCD2 immunofluorescence. Data are represented as the average ± SEM; each data point represents 3 independent sets of 50 cells. ***P < .01 by Student t test.
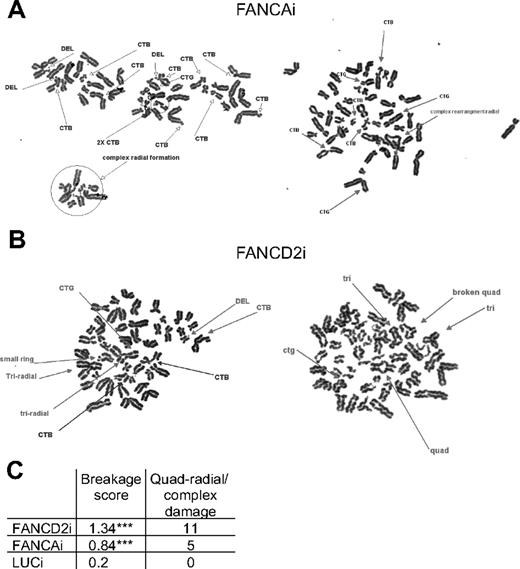
Clinically, the “gold standard” for FA diagnosis is increased levels of chromosomal breakage after treatment with DNA cross-linking agents, such as MMC.1 This diagnostic test reflects the underlying defect in the DNA damage response seen across all FA complementation groups. We treated the FANCAi, FANCD2i, and LUCi hESC lines with 200 nM MMC for 24 hours and harvested nuclear pellets for cytogenetic analysis.40 FANCAi and FANCD2i hESCs display extensive chromosomal breakage and radial formation upon MMC treatment (Figure 3A-B). Quantification of total damage per cell reveals significant differences between the 2 FA hESC lines and the LUCi control line in 50 scored metaphases (P < .001 by χ2 test). Quadriradials and other significant damage (ring structures, extensive chromosomal breakage/rearrangement precluding precise quantitation) were found only in the 2 FA hESC lines (Figure 3C). These data demonstrate that both the FANCAi and FANCD2i hESC lines exhibit the characteristic defects in DNA repair observed in cells from FA patients and gene knockout mice.
FANCD2i and FANCAi hESCs exhibit increased chromosomal breakage and radial formation upon MMC treatment. (A-B) Representative metaphase spreads from FANCAi and FANCD2i hESCs after treatment with 200nM MMC for 24 hours. DEL indicates deletion; CTB, chromatid break; CTG, chromatid gap; and CSB, chromosome break. (C) Quantification of total damage/cell (additional details on breakage score calculation in “Chromosomal breakage analysis”) and quadriradials/complex damage (ring structures, extensive chromosomal breakage/rearrangement precluding precise quantitation) for 50 scored metaphases. ***P < .01 by χ2 test.
FANCD2i and FANCAi hESCs exhibit increased chromosomal breakage and radial formation upon MMC treatment. (A-B) Representative metaphase spreads from FANCAi and FANCD2i hESCs after treatment with 200nM MMC for 24 hours. DEL indicates deletion; CTB, chromatid break; CTG, chromatid gap; and CSB, chromosome break. (C) Quantification of total damage/cell (additional details on breakage score calculation in “Chromosomal breakage analysis”) and quadriradials/complex damage (ring structures, extensive chromosomal breakage/rearrangement precluding precise quantitation) for 50 scored metaphases. ***P < .01 by χ2 test.
Impaired hematopoietic development from FA hESCs
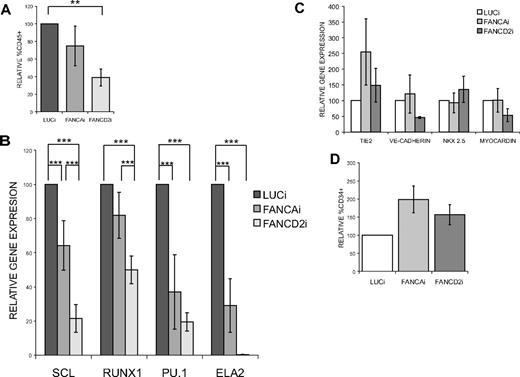
Having established that our hESC system faithfully replicates the cellular phenotype in FA, we next tested the hypothesis that hematopoietic development is dysfunctional in FA using in vitro hematopoietic differentiation from hESCs as a surrogate assay for early developmental events. An EB-based differentiation system supplemented with a cocktail of hematopoietic cytokines was used as previously described.25 FANCAi, FANCD2i, and LUCi hESCs were allowed to differentiate as EBs, disaggregated at various time points, and subjected to quantitative reverse-transcription (qRT)–PCR, FACS analysis, and methylcellulose colony-forming assays. Temporal studies from day 10 onward identified days 18 to 22 as a plateau of maximal hematopoietic activity in all lines tested (data not shown). Day 20 FACS analysis revealed that FANCD2i EB-derived cells show a significantly reduced commitment to the hematopoietic lineage as assayed by the percentage of CD45+ cells (Figure 4A). FANCAi EB-derived cells display a trend toward reduced CD45+ cells, although the difference was not statistically significant. qRT-PCR analysis from day 20 EBs revealed dramatic reductions in the expression of characteristic hematopoietic genes in FANCD2i EBs compared with the LUCi controls. FANCAi EB-derived cells consistently displayed an intermediate hematopoietic phenotype, with significant reductions in hematopoietic gene expression relative to LUCi (SCL, PU.1, neutrophil elastase [ELA2]), but not as severe an impairment as in the FANCD2i cells (Figure 4B). To exclude the possibility that this effect was due to a general defect in mesodermal lineage specification, we interrogated a set of cardiac (NKX 2.5, myocardin) and endothelial (TIE2, VE-CADHERIN, CD34) markers via qRT-PCR and FACS analysis and found no significant differences among the FANCAi, FANCD2i, and LUCi hESC lines (Figure 4C-D). Moreover, analysis of hESC-derived teratomas showed no generalized loss of mesodermal structures from the FA hESC lines (supplemental Figure 1, data not shown). Thus, we observed a specific deficit in hematopoietic commitment for both FA hESC lines, with a more severe reduction in hematopoietic fates for loss of FANCD2 than for loss of FANCA.
FANCAi and FANCD2i hESCs display reduced hematopoietic gene expression during in vitro differentiation. All data are presented as mean ± SEM. The number of biologic replicates (n) is reported for each panel. ***P < .01, **.01 < P < .03. (A) FACS analysis of day 20 EB-derived cells for the percentage of CD45+ cells. n = 8. (B-C) qRT-PCR analysis of representative hematopoietic (B), endothelial, and cardiac (C) markers in day 20 EBs. Relative gene expression (y-axis) is expressed as percentage and was calculated relative to LUCi expression levels after normalization with GAPDH. n = 6. (D) FACS analysis of day 20 EB-derived cells for the percentage of CD34+ cells. n = 6.
FANCAi and FANCD2i hESCs display reduced hematopoietic gene expression during in vitro differentiation. All data are presented as mean ± SEM. The number of biologic replicates (n) is reported for each panel. ***P < .01, **.01 < P < .03. (A) FACS analysis of day 20 EB-derived cells for the percentage of CD45+ cells. n = 8. (B-C) qRT-PCR analysis of representative hematopoietic (B), endothelial, and cardiac (C) markers in day 20 EBs. Relative gene expression (y-axis) is expressed as percentage and was calculated relative to LUCi expression levels after normalization with GAPDH. n = 6. (D) FACS analysis of day 20 EB-derived cells for the percentage of CD34+ cells. n = 6.
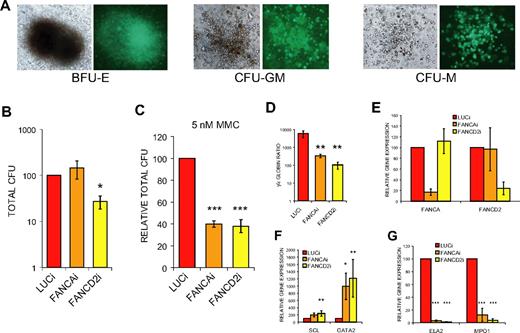
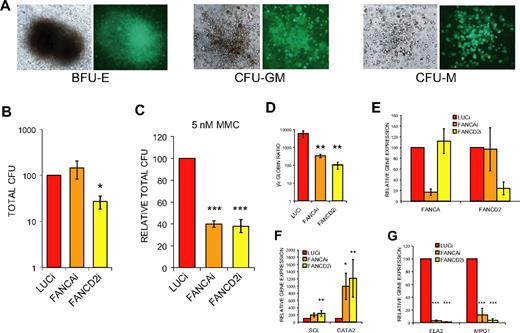
To further characterize the day 20 EB-derived populations, we evaluated the potential to form hematopoietic colonies in methylcellulose media. FANCD2i EB-derived cells displayed a highly significant, 4-fold reduction in total colony-forming units (CFUs; Figure 5B). The majority of the colonies were myeloid (CFU-granulocyte, CFU-macrophage, CFU-granulocyte/macrophage) in all 3 hESC populations (Figure 5A), and there was no significant skewing of the ratios between colony types or significant differences in colony size between FA hESCs and controls (data not shown). Moreover, no significant differences in rates of apoptosis within the EB or in EB-derived liquid hematopoietic cultures were observed among the 3 lines (supplemental Table 2), suggesting primary loss of cells may not play a large role in the observed phenotypes. Interestingly, FANCAi EB-derived cells did not display a reduction in total CFU numbers under baseline conditions (Figure 5B), suggesting that the milder hematopoietic impairment observed in terms of hematopoietic gene expression was not sufficient to affect total CFU numbers in this progenitor assay. Upon treatment with 5 nM MMC, both FANCAi and FANCD2i EB-derived cells demonstrate a comparable and marked reduction in total CFUs relative to treated LUCi controls (Figure 5C), establishing that FA EB-derived hematopoietic progenitors, just like the parental FA hESCs, are hypersensitive to MMC. To clarify these findings, qRT-PCR analysis was performed on the CD45+ population from day 20 EBs, which contains all of the hematopoietic activity (data not shown). Both FANCAi and FANCD2i EB-derived CD45+ populations show a robust knockdown at the RNA level that is comparable with the RNA knockdown levels observed in undifferentiated hESCs (Figure 5E). Importantly, the FANCAi CD45+ population has a greater degree of knockdown than the FANCD2i CD45+ population, demonstrating that the milder hematopoietic phenotype observed for FANCAi hESCs is not the result of a less efficient knockdown and instead perhaps reflects differing severities of the clinical course between the 2 complementation groups.
FANCD2i hESCs demonstrate significant reductions in hematopoietic CFUs. (A) Representative light and fluorescence micrographs of hematopoietic colonies. BFU-E indicates blast-forming unit erythroid; CFU-GM, colony-forming unit granulocyte/monocyte; and CFU-M, colony-forming unit macrophage. (B-C) Total hematopoietic CFUs from day 20 EB-derived cells plated in H4434 media (StemCell Technologies) without (B) or with (C) 5nM MMC and scored at day 14 after plating. Total CFUs are normalized directly to LUCi (B) or first normalized to baseline untreated total CFUs to assess MMC hypersensitivity of hematopoietic progenitors (C). Data are presented as mean ± SEM. n = 6, 3. (D) Normalized ratio of gamma-globin expression to epsilon-globin expression by qRT-PCR from CD45+ population of day 20 EB-derived cells. Data are presented as mean ± SEM. n = 6. (E-G) qRT-PCR analysis of gene expression from CD45+ population of day 20 EBs. Expression is relative to LUCi expression levels after normalization with GAPDH. Data are presented as mean ± SEM and n = 6. ***P < .01; **.01 < P < .03; *.03 < P < .05.
FANCD2i hESCs demonstrate significant reductions in hematopoietic CFUs. (A) Representative light and fluorescence micrographs of hematopoietic colonies. BFU-E indicates blast-forming unit erythroid; CFU-GM, colony-forming unit granulocyte/monocyte; and CFU-M, colony-forming unit macrophage. (B-C) Total hematopoietic CFUs from day 20 EB-derived cells plated in H4434 media (StemCell Technologies) without (B) or with (C) 5nM MMC and scored at day 14 after plating. Total CFUs are normalized directly to LUCi (B) or first normalized to baseline untreated total CFUs to assess MMC hypersensitivity of hematopoietic progenitors (C). Data are presented as mean ± SEM. n = 6, 3. (D) Normalized ratio of gamma-globin expression to epsilon-globin expression by qRT-PCR from CD45+ population of day 20 EB-derived cells. Data are presented as mean ± SEM. n = 6. (E-G) qRT-PCR analysis of gene expression from CD45+ population of day 20 EBs. Expression is relative to LUCi expression levels after normalization with GAPDH. Data are presented as mean ± SEM and n = 6. ***P < .01; **.01 < P < .03; *.03 < P < .05.
Further analysis of the CD45+ hematopoietic populations of FANCAi and FANCD2i EBs revealed remarkably similar gene expression profiles. Both populations showed a dramatic reduction in the ratio of gamma- to epsilon-globin compared with controls, suggesting a failure of the normal transition from epsilon-globin to gamma-globin–positive erythroid progenitors that occurs in a time-dependent manner within EBs (Figure 5D). Beta-globin was not expressed in any EB population, although it was expressed in erythroid methylcellulose colonies derived from all hESC lines studied, a finding that is consistent with other reports.30 Analogous to the globin profile, the expression of other genes marking primitive hematopoietic populations such as SCL and GATA2 was greatly increased in the CD45+ population of both FANCAi and FANCD2i EBs, whereas expression of definitive markers in the CD45+ population such as neutrophil elastase (ELA2) and myeloperoxidase (MPO1) was strongly reduced (Figure 5F-G). This suggests that both FANCAi and FANCD2i hESCs display similar profiles of impaired in vitro hematopoietic development with a bias toward primitive populations.
FA gene complementation rescues hematopoietic deficits
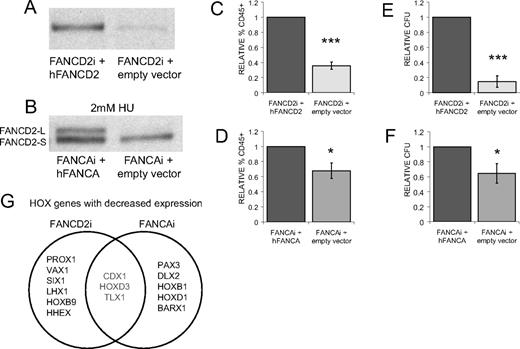
To demonstrate that the observed hematopoietic deficits are the direct result of FA gene knockdown and not due to selection artifacts or off-target effects of shRNAs, FANCAi and FANCD2i hESCs were infected with retroviral vectors overexpressing human FANCA and FANCD2 cDNAs, respectively. Infection with an empty puromycin selectable vector served as a control in both cases. After selection for stably infected populations, “FANCD2i + hFANCD2” hESCs show restoration of FANCD2 protein and “FANCAi + hFANCA” hESCs reestablish FANCD2 monoubiquitination (Figure 6A-B), thereby confirming gene complementation in both FA hESC lines. Overexpression of FANCD2 fully corrects the hematopoietic deficits seen with FANCD2i hESCs upon in vitro hematopoietic differentiation. FANCD2i + hFANCD2 day 20 EB-derived cells display an almost 3-fold increase in percentage of CD45+ cells and an almost 7-fold increase in CFUs compared with the uncorrected FANCD2i EB-derived controls (Figure 6C,E). Genetic complementation of FANCAi hESCs yielded smaller, but still significant, increases in CD45 percentage and CFU numbers, providing further support that loss of FANCA results in a milder hematopoietic phenotype than loss of FANCD2 (Figure 6D,F). Taken together, these data conclusively demonstrate that reductions in hematopoiesis during in vitro hESC differentiation are a primary result of FA gene knockdown.
FA gene complementation rescues hematopoietic deficits. (A) FANCD2 Western blot on lysates from FANCD2i hESCs infected with pMMP-puro hFANCD2 or pMMP-puro (empty vector). (B) FANCAi hESCs infected with pMMP-puro hFANCA or pMMP-puro were treated with 2mM HU for 24 hours and lysates were subjected to Western blotting for FANCD2. FANCD2-L is the active, monoubiquitinated form of FANCD2-S. (C-D) FACS analysis of day 20 EB-derived cells for the percentage of CD45+ cells. Percentage CD45 is normalized to FANCD2i + hFANCD2 (C) or FANCAi + hFANCAi (D) and presented as ± SEM. n = 4, 6. (E-F) Total hematopoietic CFUs from day 20 EB-derived cells plated in H4434 media (StemCell Technologies) scored at day 14 after plating. Total CFUs are normalized to FANCD2i + hFANCD2 (E) or FANCAi + hFANCAi (F) and presented as ± SEM. n = 4, 6. (G) Venn diagram showing HOX genes with decreased expression in day 20 EBs. Eighty-four HOX genes were analyzed by quantitative PCR array (SABiosciences) and the top 8 genes, all with fold reduction greater than −1.5 relative to gene complement control, were included. FANCD2i + empty vector was compared with FANCD2I + hFANCD2 and FANCAi + empty vector was compared with FANCAi + hFANCA to generate the list of genes. Data are provided in supplemental Table 3. ***P < .01; *.03 < P < .05.
FA gene complementation rescues hematopoietic deficits. (A) FANCD2 Western blot on lysates from FANCD2i hESCs infected with pMMP-puro hFANCD2 or pMMP-puro (empty vector). (B) FANCAi hESCs infected with pMMP-puro hFANCA or pMMP-puro were treated with 2mM HU for 24 hours and lysates were subjected to Western blotting for FANCD2. FANCD2-L is the active, monoubiquitinated form of FANCD2-S. (C-D) FACS analysis of day 20 EB-derived cells for the percentage of CD45+ cells. Percentage CD45 is normalized to FANCD2i + hFANCD2 (C) or FANCAi + hFANCAi (D) and presented as ± SEM. n = 4, 6. (E-F) Total hematopoietic CFUs from day 20 EB-derived cells plated in H4434 media (StemCell Technologies) scored at day 14 after plating. Total CFUs are normalized to FANCD2i + hFANCD2 (E) or FANCAi + hFANCAi (F) and presented as ± SEM. n = 4, 6. (G) Venn diagram showing HOX genes with decreased expression in day 20 EBs. Eighty-four HOX genes were analyzed by quantitative PCR array (SABiosciences) and the top 8 genes, all with fold reduction greater than −1.5 relative to gene complement control, were included. FANCD2i + empty vector was compared with FANCD2I + hFANCD2 and FANCAi + empty vector was compared with FANCAi + hFANCA to generate the list of genes. Data are provided in supplemental Table 3. ***P < .01; *.03 < P < .05.
Having established a connection between the FA pathway and early hematopoietic development, we investigated whether loss of the FA pathway had a more general effect on tissue patterning within the EB. Numerous genetic and biochemical studies in mice, zebrafish, and Drosophila have identified key roles for homeodomain-containing transcription factors (HOX and related genes) in directing segmental patterning, organ morphogenesis, and tissue development.41 Early hematopoietic dysfunction coupled with the skeletal and kidney malformations seen in FA patients suggests that dysregulated HOX gene expression may contribute to disease pathogenesis42 ; however, little is known about the specific role of HOX genes in human hematopoietic development. Using FA hESCs as a developmental platform, we profiled the expression of 84 Homeobox (HOX) genes from FANCD2i and FANCAi day 20 EB populations using their gene-corrected cell lines as controls. Comparisons of differentially expressed HOX genes revealed Caudal type homeobox 1 (CDX1), T-cell leukemia homeobox 1 (TLX1), and HOXD3 among the most significantly decreased in both FANCAi and FANCD2i day 20 EBs (Figure 6G). CDX1 and TLX1 have known roles in hematopoietic specification and progenitor proliferation,43,44 whereas HOXD3 has been shown to play a role in patterning the anterior-posterior axis and the embryonic skeleton.45 In addition to their role in tissue specification, HOX genes have been shown to directly influence the cell cycle machinery and cellular proliferation. Expression analysis of cell cycle regulators by qRT-PCR array revealed BRCA1 as the most up-regulated transcript in both FANCAi and FANCD2i EBs compared with gene-corrected controls (supplemental Figure 4), perhaps suggesting compensatory feedback from the cellular network regulating genomic stability. FANCD2i and FANCAi EBs also displayed reduced expression of some key regulators of the G1/S transition, most notably cyclin-dependent kinase inhibitor 2B and 1B (CDKN2B and CDKN1B, respectively). Future experiments will investigate the role of these candidate HOX genes in patterning of the hematopoietic system and appendicular skeleton, as well as alterations in hematopoietic cell cycle regulation in FA.
Finally, we were interested in testing whether our hESC-based system reflected a human-specific effect on hematopoietic development or represented a conserved feature across species. We derived mouse Fancd2 −/− ESCs, subjected them to established in vitro differentiation protocols, and assayed hematopoietic progenitor activity at day 6, a documented time point of peak maximal hematopoietic activity within the mouse EB.38 In contrast to the dramatic reduction in total CFUs from human FANCD2i hESCs (Figure 5B), mouse Fancd2 −/− ESCs do not display reduced progenitor numbers during in vitro differentiation (supplemental Figure 2). These data provide additional evidence that the mouse may be an incomplete model for the hematopoietic failure seen in FA patients and that our hESC-based system uniquely enables studies of early human hematopoietic development and disease. Thus, we conclude that FA hESCs display human-specific hematopoietic deficits during in vitro differentiation.
Discussion
Our limited understanding of human embryonic development comes from ex post facto analysis of clinical cases and experimentation in model organisms. FA manifests congenital skeletal anomalies and early hematopoietic dysfunction that progresses to bone marrow failure in childhood.16 However, an understanding of early hematopoietic development in FA has been lacking, as mouse models do not develop bone marrow failure, and studies using primary hematopoietic cells from FA patients are incapable of addressing the developmental genesis of the hematopoietic system. hESCs and patient-specific iPSCs enable the study of uniquely human aspects of early development and can provide insight into the mechanisms underlying disease. However, in the case of FA, a recent report by the Izpisúa-Belmonte group suggests that it is not feasible to reprogram somatic tissues from FA patients to generate patient-specific iPSCs (Raya et al),31 which serves to highlight the need for alternative approaches to study early development in FA. In this study, we use a lentiviral RNAi strategy in hESCs to study the effect of FA gene knockdown on early hematopoietic development. We show that knockdown of FA genes in hESCs leads to reduced production of CD45+ hematopoietic progenitor populations and that FA gene complementation corrects the hematopoietic deficits. As the process of in vitro hematopoietic differentiation from hESCs closely mirrors early events in embryonic hematopoietic development, our results indicate that hematopoiesis is impaired in FA from the earliest stages of blood cell formation. This is in contrast to the traditional model of hematopoietic failure in FA that describes a progressive loss of stem cell and multipotent progenitor populations during early childhood2,13 ; our results suggest that hematopoietic dysfunction in FA begins much earlier.
Multiple mechanisms have been proposed to explain the development of bone marrow failure in FA including increased apoptosis due to cellular stressors (DNA damage, inhibitory cytokines),11,21 limitations in stem cell and progenitor self-renewal capacities,18 and aberrant cellular signaling,46 although a consensus regarding the underlying pathophysiology of FA remains elusive. Out of necessity, our mechanistic understanding of hematopoietic failure in FA is derived almost exclusively from the study of preformed, postnatal hematopoietic cells from the peripheral blood or bone marrow. We have comparatively little insight into the generation of the earliest stages of hematopoietic tissue that form during embryonic development, a time window that seems especially critical considering that disease manifestations in FA originate in utero. Based on our findings that FA hESCs display decreased in vitro hematopoietic development, we propose that deficits in embryonic hematopoiesis may partially explain the clinical findings of presymptomatic marrow hypocellularity and reduced CD34+ progenitor numbers and may underlie the progression to bone marrow failure in FA. In our model, impaired embryonic and fetal hematopoietic development results in a smaller-than-normal neonatal stem cell and progenitor pool, although the reductions are often masked in infancy by compensatory mechanisms in the bone marrow. The progression to bone marrow failure then results from a still unknown combination of well-studied mechanisms such as DNA damage–induced apoptosis or intrinsic proliferation defects that act to further compromise an already reduced stem cell and progenitor pool. Our hESC-based system provides a unique model of these early developmental events in FA and allows the creation of large numbers of FA hematopoietic progenitors for in vitro and in vivo maturation, thus enabling further mechanistic dissection of the underlying disease pathogenesis from the earliest stages of development onward.
An additional, unique feature of a hESC-based system is the opportunity to compare different subtypes of FA. Genotype-phenotype correlations have been the subject of much debate and remain poorly elucidated, in part because of the complications of mosaicism and somatic reversion.47 Although specific mutations within a complementation group have been associated with earlier onset of bone marrow failure and reduced survival,9 the more general question of whether loss of certain FA genes results in differential outcomes remains unresolved. FANCA is by far the most commonly mutated gene in FA (65%), followed by other core complex components FANCC (15%) and FANCG (10%), whereas mutations in downstream genes such as FANCD2 are relatively rare (3%-6%).9 Interestingly, FA-D2 patients have hypomorphic mutations that typically compromise splicing and all patients retain residual FANCD2 protein.10 In contrast, FA-A patients can have large deletions and biallelic null mutations in FANCA are relatively common.48 Despite the hypomorphic mutations in the FA-D2 complementation group, studies suggest that these patients have a more severe phenotype with an earlier onset of bone marrow failure (median age, 4.5 years) and increased frequency of skeletal abnormalities compared with the FA patient population as a whole.10 The absence of biallelic null mutations in FANCD2 has also led to speculation that a complete loss of FANCD2 function may produce a severe and possibly lethal embryonic phenotype. Given the multifunctionality of downstream components in the FA pathway and that mutations in these components are uncommon in FA, it has been suggested that intrinsic developmental differences may exist between core complex and downstream complementation groups. Understandably, these have been difficult questions to approach experimentally. Here we show that knockdown of FANCD2 results in a greater impairment of hematopoietic development from hESCs than knockdown of FANCA, despite a more efficient extinction of FANCA compared with FANCD2. Furthermore, whereas overexpression of FANCA in FANCAi hESCs resulted in a moderate increase in the CD45 fraction and CFU numbers, overexpression of FANCD2 in FANCD2i hESCs yielded significantly greater increases in hematopoietic populations. Therefore, our data suggest that early hematopoietic development is more affected by loss of FANCD2 than loss of FANCA, and greater deficiencies in embryonic hematopoiesis (leading to an even smaller neonatal stem cell and progenitor pool) may underlie the accelerated progression to bone marrow failure in FA-D2 patients. These data also lend experimental weight to the long-held suspicion that there is a gradient of disease within FA, with the more common core complex complementation groups such as FA-A reflective of a milder form of the disease than the rare downstream complementation groups such as FA-D2.9,10
Our work establishes a new platform for the study of hematopoietic development in FA. The unlimited replicative potential of hESCs enables the production of FA hematopoietic cells en masse for studies that were previously unfeasible. Long-term, large-scale culture of FA hESC-derived hematopoietic progenitors in the presence of inhibitory cytokines can serve to simulate the process of leukemic transformation in vitro, by recreating the selective pressure within aplastic FA marrow for mutations that enable rapid proliferation and suppression of apoptosis leading to the outgrowth of malignant clones. High-throughput screening for compounds that reduce cell stress-mediated apoptosis in FA progenitors or increase the production of fetal hemoglobin in FA red blood cells has potentially significant clinical implications for delaying aplasia and increasing the oxygen-carrying capacity of the blood, respectively. Finally, the scalability of this system facilitates further mechanistic analysis of the role of FA proteins in development. It remains to be fully clarified exactly how knockdown of FANCA and FANCD2 leads to reduced hematopoietic commitment during EB differentiation and whether that effect is related to the well-established defects in DNA repair that FA hESCs and their derivative hematopoietic progenitors also display. It is notable that there was no significant difference in apoptosis rates between FA hematopoietic progenitors and controls, suggesting that the observed hematopoietic deficits may not be a result of direct cell loss, as is seen in DNA damage–induced hematopoietic failure.49 We show that FANCAi and FANCD2i hEB-derived populations exhibit impaired in vitro hematopoietic development with primitive type gene expression and shared reductions in certain key homeotic regulators such as CDX1. CDX1 is known to play an important specification role in embryonic hematopoiesis by establishing a HOX code for appropriate patterning of the ventral mesoderm.38,44,50 Clinically, the presence of congenital skeletal abnormalities and early hematopoietic dysfunction suggests that disruption of the FA pathway has an effect on early development, and future studies will further characterize the possible connection between abnormal signaling in FA and the dysregulated expression of HOX genes that are key to patterning both skeletal and hematopoietic tissue. Our results indicate that hematopoietic failure in FA traces back to the earliest stages of blood development and establish a new model system for further studies of hematopoietic disease.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge Lorraine Meisner from Cell Line Genetics for her assistance with the breakage studies and Jason West for his critical comments.
This work was supported by the National Institutes of Health, National Institutes of Health Director's Pioneer Award, Burroughs Welcome Fund, Leukemia & Lymphoma Society, Howard Hughes Medical Institute, and the Harvard Stem Cell Institute.
Authorship
Contribution: A.T. designed and performed research and wrote the paper; M.W.L. performed research and edited the paper; J.D.M. and K.A. performed research; A.D. contributed vital new reagents; T.M.S. contributed resources and edited the paper; A.S. contributed resources and vital new reagents, and edited the paper; and G.Q.D. helped design research and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: George Daley, Division of Pediatric Hematology/Oncology, Karp bldg, 7th fl, Children's Hospital Boston, Blackfun Cir, Boston, MA 02115; e-mail: george.daley@childrens.harvard.edu.