To the editor:
In a recent issue of Blood, Murga Penas et al identified a novel cluster region on MALT1 from t(14;18)(q32;q21) IGH-MALT1 translocations in MALT lymphomas, which bears many similarities to the bcl-1 major translocation cluster (MTC) from mantle cell lymphomas and the bcl-2 major breakpoint region (MBR) from follicular and diffuse large B-cell lymphomas.1
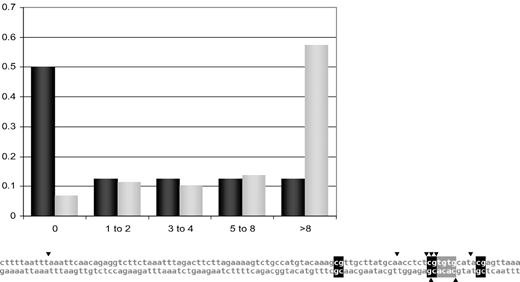
We have previously reported that pro-B/pre-B stage translocation breakpoints are highly clustered to the dinucleotide sequence CpG, including those at the bcl-1 MTC, bcl-2 MBR, bcl-2 intermediate cluster region (icr), bcl-2 minor cluster region (mcr), and E2A cluster region from E2A-PBX1 translocations in pre-B ALLs.2 After plotting the reported breakpoints, we find that CpG proximity is also a feature of the MALT1 cluster region (see figure). Four of the 8 breakpoints are directly at CpGs (P < .002) and breakpoints are much closer to CpGs than expected by random chance (P < .005). All but 1 are within 8 bp of CpG (dark gray bars in the histogram), far more than if breakpoints were randomly distributed throughout the region (light gray bars in the histogram). Methods have been described previously.2
Analysis of MALT lymphoma breakpoints. (Top) Frequency of MALT1 cluster region breakpoints at various distance intervals from CpG. Proportions of MALT1 cluster region breakpoints at distances of 0 bp, 1 to 2 bp, 3 to 4 bp, 5 to 8 bp, and > 8 bp from CpG are graphed. The distribution for actual lymphoma or leukemia breakpoints is in dark gray, while that for a random distribution between the farthest breakpoints is in light gray. If the dark gray and light gray bars parallel one another, then the patient breakpoints appear random in their distribution relative to the specified motif. However, when they follow opposite trends, that is, the light gray bars rise with increasing distance from the specified motif while the dark gray bars fall, then the breakage process appears to concentrate around the motif. (Bottom) Breakpoint distribution on the MALT1 cluster region. MALT lymphoma breakpoints in the MALT1 cluster region identified by Murga Penas et al are plotted. Each breakpoint is represented as a triangle adjoining the breakpoint site, with the top strand sequences running telomeric to centromeric, der(14) breakpoints above, and der(18) breakpoints below. CpGs are highlighted in black, and the lone CAC(A) highlighted in gray.
Analysis of MALT lymphoma breakpoints. (Top) Frequency of MALT1 cluster region breakpoints at various distance intervals from CpG. Proportions of MALT1 cluster region breakpoints at distances of 0 bp, 1 to 2 bp, 3 to 4 bp, 5 to 8 bp, and > 8 bp from CpG are graphed. The distribution for actual lymphoma or leukemia breakpoints is in dark gray, while that for a random distribution between the farthest breakpoints is in light gray. If the dark gray and light gray bars parallel one another, then the patient breakpoints appear random in their distribution relative to the specified motif. However, when they follow opposite trends, that is, the light gray bars rise with increasing distance from the specified motif while the dark gray bars fall, then the breakage process appears to concentrate around the motif. (Bottom) Breakpoint distribution on the MALT1 cluster region. MALT lymphoma breakpoints in the MALT1 cluster region identified by Murga Penas et al are plotted. Each breakpoint is represented as a triangle adjoining the breakpoint site, with the top strand sequences running telomeric to centromeric, der(14) breakpoints above, and der(18) breakpoints below. CpGs are highlighted in black, and the lone CAC(A) highlighted in gray.
While the IGH-MALT1 translocation is found in approximately 10% of MALT lymphomas, the t(11;18)(q21;q21) API2-MALT1 translocation is more common, found in approximately 30% of MALT lymphomas, but also involves the MALT1 locus. The IGH-MALT1 translocation involves a very focused (86 bp) region upstream of the MALT1 promoter, joins to the immunoglobulin heavy chain RSS, involves a CpG-type mechanism, and contains N- and T-nucleotides in its junctions. By contrast, the API2-MALT1 translocation involves a very wide (29 kb) region covering several exons and introns of the MALT1 gene, joins to a 4.5-kb region of the API2 intron, and its junctions mainly use microhomology, without N- or T-nucleotides.3-5 In our analysis of API2-MALT1 translocations, none of the 40 breakpoints on API2 and none of the 41 breakpoints on MALT1 occur at CpGs (p = 1), and breakpoints are no closer to CpGs than expected by random chance (P > .2).
We concur with the observation of Murga Penas et al that none of the MALT1 breakpoints is compatible with a standard V(D)J recombination-type mechanism. The presence of N-nucleotides likely added by TdT, T-nucleotides, and the involvement of the V(D)J recombination signal sequences (RSS) at the immunoglobulin heavy chain locus side of the translocation are all features that are virtually pathognomonic for the pro-B/pre-B stage of development, though the lymphoma itself may originate from a later stage of development than the translocation. By contrast, the API2-MALT1 translocation, which also involves the MALT1 locus, has none of these features and likely occurs either before the pro-B/pre-B stage (ie, at the lymphoid-myeloid hematopoietic stem cell stage) or after the pro-B/pre-B stage (ie, at the mature B-cell stage). It is especially interesting that a single clinical entity—MALT lymphomas—would feature translocations from 2 different stages of B-cell development, but involve the same MALT1 locus. Our analysis affirms the authors' deduction that the bcl-1, bcl-2, and IGH-MALT1 translocations all share a common mechanism, but additionally shows that, like the bcl-1 and bcl-2 translocations, the IGH-MALT1 is a member of the CpG-type translocation family, which appears unique to the B-cell lineage and the pro-B/pre-B stage of development.
Authorship
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael R. Lieber, USC Norris Cancer Ctr, Rm 5428, 1441 Eastlake Ave, MC9176, Los Angeles, CA 90089-9176; e-mail: lieber@usc.edu.