Maintenance of membrane integrity and organization in the metazoan cell is accomplished through intracellular tethering of membrane proteins to an extensive, flexible protein network. Spectrin, the principal component of this network, is anchored to membrane proteins through the adaptor protein ankyrin. To elucidate the atomic basis for this interaction, we determined a crystal structure of human βI-spectrin repeats 13 to 15 in complex with the ZU5-ANK domain of human ankyrin R. The structure reveals the role of repeats 14 to 15 in binding, the electrostatic and hydrophobic contributions along the interface, and the necessity for a particular orientation of the spectrin repeats. Using structural and biochemical data as a guide, we characterized the individual proteins and their interactions by binding and thermal stability analyses. In addition to validating the structural model, these data provide insight into the nature of some mutations associated with cell morphology defects, including those found in human diseases such as hereditary spherocytosis and elliptocytosis. Finally, analysis of the ZU5 domain suggests it is a versatile protein-protein interaction module with distinct interaction surfaces. The structure represents not only the first of a spectrin fragment in complex with its binding partner, but also that of an intermolecular complex involving a ZU5 domain.
Introduction
Organization and integrity of the metazoan cell membrane are maintained through intracellular tethering to an extensive, flexible protein network. The principal component of this membrane skeleton scaffold, spectrin, is attached to the membrane via the adaptor protein ankyrin, which directly bridges the interaction between β-spectrin and various membrane proteins.1 Both of these proteins are capable of interacting with multiple binding partners. Depending on the cell type and isoform, ankyrins can bind many different membrane proteins: E-cadherin in epithelial cells,2 the voltage-gated Na+ channel in neurons,3 the inositol 1,4,5-trisphosphate receptor in cardiomyocytes,4 and the anion-exchange transporter band 3 in erythrocytes.5,6 In the case of human erythrocytes, particularly during passage through capillaries, mechanical stress on the membrane is transmitted through these interactions and relieved by deformations in the spectrin network. Remarkably, these reversible distortions occur while the erythroid spectrin network remains attached to the membrane, most likely conserving the spectrin/ankyrin interaction.7 In addition, mutations in both spectrin ankyrin have been associated with a variety of human diseases,8 including hereditary spherocytosis and elliptocytosis,9,10 exemplifying the deleterious effects of network disruption and the importance of the integrity of the spectrin cytoskeleton to the cell.
Members of the spectrin superfamily—which includes not only multiple isoforms of spectrin but also α-actinin, dystrophin (reviewed in Broderick and Winder11 ), and the plakins12 —are characterized by tandem spectrin repeats. Although these proteins show modest sequence identity, spectrin repeats display marked structural similarities: each consists of roughly 106 amino acids, adopts a 3-helix bundle fold, and is linked to adjacent repeats by an ordered, α-helical linker. Despite these structural constraints, high-affinity interactions of specific spectrin repeats with particular binding partners demonstrate that the domain has been adapted to interact with an array of ligands.13 In the case of the human erythroid spectrin heterotetramer, which is assembled as a head-to-head dimer of α/β dimers,14 ankyrin specifically recognizes β-spectrin repeats 14 and 15 of the 37 spectrin repeats present in the heterodimer.15,–17
In humans, there are 3 major ankyrin isoforms (1, or R, in erythrocytes; 2, or B, in the brain; and 3, or G, which is more widely expressed) in addition to several tissue-specific alternative-splice isoforms.1,18 Ankyrins have a modular primary structure with 3 regions: an N-terminal domain composed of tandem ankyrin repeats that mediates attachment to different membrane proteins, a central spectrin-binding domain, and a C-terminal regulatory domain that harbors a death domain and modulates the affinities of the molecule.19 Within the spectrin-binding domain, a small subdomain termed ZU5-ANK (named for its homology to ZO-1, a tight junction-associated protein,20 and Unc5, the netrin receptor21 ) is responsible for specific recognition of repeats 14 and 15 of β-spectrin.16,22,23 Interactions with this domain alone fully account for the binding specificity of the 2 molecules.16,22,24 When compared to all other known structures,25,26 ZU5-ANK shows no structural similarity to any other known protein module.
Given the critical role of spectrin and ankyrin in physically linking the membrane skeleton to the cell membrane, it is important to understand the atomic basis for the interactions between them. To address this, we have crystallized and solved the atomic structure of a complex of human βI-spectrin repeats 13 to 15 with the ZU5-ANK domain of human ankyrin R to 2.75 Å resolution. The structural data elucidate the roles of both repeats 14 and 15 in ankyrin binding, reveal the role of electrostatic and hydrophobic interactions along the binding interface, and illustrate that the relative orientation of the 2 spectrin repeats is crucial for stable complex formation. Using the structure as a guide, we characterized the roles of different regions and amino acids in binding. In addition to validating the structural model, these data provide insight into the nature of mutations associated with cell morphology defects. Analysis of the ZU5 domain suggests that, like spectrin repeats, this domain has distinct binding interfaces that allow it to function as a versatile protein-protein interaction module. The structure represents not only the first of a spectrin fragment in complex with its binding partner but also the first of an intermolecular complex with a ZU5 domain.
Methods
Expression and purification of spectrin and ankyrin fragments
Constructs encompassing the ankyrin-binding region of human erythroid β-spectrin (repeats 13-15; residues 1583-1906) and the ZU5-containing spectrin-binding domain of human erythroid ankyrin (residues 911-1026) were separately expressed and purified as described previously.27 Both proteins included an extra 3 amino acids at the N-terminus from the expression vector. Once purified, the spectrin and ankyrin fragments were concentrated to 9.3 mg/mL (250μM), and 5.4 mg/mL (300μM) respectively, aliquoted, flash frozen in liquid nitrogen, and stored at −80°C.
Mutagenesis
All site-directed mutagenesis was conducted with the use of the QuikChange (Stratagene) method. Successful incorporation of the desired mutation(s) was validated by DNA sequencing (Northwestern University Genomics Core) and by mass spectrometry of the purified proteins.
For experimental phase determination, a panel of 6 cysteine mutants in repeat 13 of spectrin was generated for directed labeling with ethylmercury thiosalicylate (EMTS). The resulting mutant proteins were expressed and purified as described elsewhere.27 Ultimately, one of these mutants led to a satisfactory experimental map (see “Data collection, crystal parameters, structure determination, and refinement”).
For biophysical studies of both spectrin and ankyrin mutants, a panel of 20 mutants was generated on the basis of clinical mutations in the regions of interest, previous experimental data, and structure-based design. Each mutant protein was expressed and purified as described previously.27
Mass spectrometry
Each purified protein was diluted in water to 1μM and applied to a Phenomenex Jupiter Proteo Column (1.0 mm × 150 mm, 4-μm particle size) at 50 μL/min. In total, approximately 1 pmol (1 μL) of material was injected. A mobile phase gradient from 0.1% formic acid in water to 0.08% formic acid in acetonitrile was used to chromatograph each sample over 6 minutes, which was then taken for in-line electrospray ionization mass spectrometry. Mass spectra were obtained on an Agilent 6510 Accurate-Mass quadrupole time-of flight liquid chromatography/mass spectrometer with an accelerating voltage of 4100 V. Deconvolution was performed with the included software, Agilent MassHunter Qualitative Analysis Version B.01.02. Masses of all proteins were within 1.0 Da of those predicted.
Complex formation, purification, and crystallization
Purified HEβ1315 and ZU5-ANK were mixed in a 1:1 molar ratio and incubated on ice for 10 minutes. The complex was purified with the use of a P-60 (Bio-Rad) size-exclusion column equilibrated with 10mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), pH 7.5. The purified complex was concentrated to 5 to 20 mg/mL for crystallization screening and initial crystal optimization. Later crystallization optimization demonstrated that crystals of the complex could be generated simply by mixing the proteins in a 1:1 molar ratio immediately before the crystallization experiment. Crystals were grown by hanging drop vapor diffusion in 10% PEG-4000; 0.2M NH4OAc; 0.01M CaCl2; 0.05M MES, pH 6.5; and 5mM dithiothreitol (DTT) on siliconized glass coverslips (Hampton). Two microliters of purified protein solution (∼ 5 mg/mL) were mixed with an equal volume of reservoir solution and suspended over 0.75 mL of reservoir solution. Rectangular prismatic crystals (∼ 100 μm × 20 μm × 20 μm) grew at 10°C within 2 weeks and diffracted to approximately 10 Å. Improved crystals, which diffracted to approximately 6.0 Å, could be generated within 16 hours by streak seeding and yielded larger crystals (∼ 300 μm × 50 μm × 50 μm) with better-defined edges. The diffraction quality of the crystals was further improved to 2.75 Å by overnight dehydration of the crystals, which was conducted by replacing the reservoir solution with 2× reservoir solution (20% PEG-4000; 0.4M NH4OAc; 0.02M CaCl2; 0.1M MES, pH 6.5; and 10 mM DTT). Crystals were harvested and frozen in liquid nitrogen after direct addition of glycerol to a final concentration of 10% to the crystal drop.
Data collection, crystal parameters, structure determination, and refinement
Data were collected at 100 K by the use of synchrotron radiation at the Advanced Photon Source (APS) LS-CAT beamline at Argonne National Laboratory. Data were processed with XDS28 and scaled with SCALA.29 Additional processing was performed with programs from the CCP4 suite.29
The crystals belong to space group P212121 with a = 90.5 Å, b = 95.7 Å, c = 137.9 Å, α = β = γ = 90° with 2 complexes per asymmetric unit (∼ 55.1% solvent content). An experimental map was obtained with the use of 2.75 Å data collected from EMTS-derivatized crystals of a cysteine-mutagenized construct (HEβ1315 C1892S, E1680C) at 0.979 Å. Crystals grown for the purpose of derivatization were grown in the absence of DTT; after the crystals formed, EMTS was added to the crystallization drop at a final concentration of 5mM 16 hours before harvesting. Single-wavelength anomalous dispersion phasing was performed with the program autoSHARP,30 which correctly located 8 labeled cysteines in the asymmetric unit. The initial solvent flattened map was of excellent quality and 3-helix spectrin repeats were recognized easily. An initial partial model was constructed by the use of previously solved structures of the component proteins (β-spectrin repeats 14 and 15 PDB ID 3F57; ZU5-ANK PDB ID 3F59).27 Subsequent manual model building and inspection were conducted with the program Coot.31 Model refinement was performed with REFMAC5.32 Even after refinement, not all regions of the molecules were visible, particularly the distal portion of spectrin repeat 15, which has been observed to be disordered in previous structures.27,33 Proper placement of side chains was validated with EMTS-derivatized crystals of seleno-methione–incorporated proteins, which provided peaks in the anomalous difference map that were used as guides to register the sequence with both methionines and cysteines.
As part of the refinement, TLS parameters were refined for each spectrin repeat as well as each ZU5-ANK molecule. The 2 complexes were restrained by noncrystallographic symmetry during the refinement, although the restraints were not too tight because the 2 complexes are slightly different in orientation. The final model consisted of residues 1585 to 1819 and 1841 to 1889 for HEβ1315 chain A; 1587 to 1827 and 1840 to 1892 for HEβ1315 chain B; 910 to 1022 and 1025 to 1068 for ZU5-ANK chain C; and 910 to 997, 1003 to 1021, and 1025 to 1068 for ZU5-ANK chain D. The final model had an Rfactor and Rfree of 24.5% and 29.2%, respectively, with a root mean square deviation (RMSD) of 0.014 Å and 1.475° for bond lengths and bond angles, respectively, and with all residues within the favored regions of the Ramachandran plot. Refinement statistics are listed in Table 1.30,34
Circular dichroism
Circular dichroism measurements were obtained with a Jasco J715 spectropolarimeter equipped with a Peltier device and routinely calibrated with d-10-camphorsulfonic acid (Keck Facility, Northwestern University). Wavelength scans were carried out at room temperature at a final protein concentration of 15 to 20μM in 10mM sodium phosphate, pH 8.0, and 0.14M NaF (0.2-mm path length demountable cuvette). The data pitch was 1 nm, the scan speed 20 nm/min, the response time 8 seconds, and the band width 1 nm. Data for wavelength scans were given after buffer subtraction and presented in units of molar ellipticity ([θ]), deg·cm2·dmol−1. Thermal denaturation scans for each spectrin mutant were measured from 10 to 85°C at 222 nm at a protein concentration of approximately 1μM in a 1-cm cuvette. Samples were heated at a rate of 1°C/min with stirring. Instrumentation and methods for the ankyrin mutants are described in the supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Surface plasmon resonance
Binding affinities and kinetic parameters of ankyrin fragments to biotinylated spectrin fragments were measured with a Biacore 3000 equipped with a streptavidin biosensor. Before immobilization, each purified spectrin fragment was individually biotinylated. With the exception of the R1684C mutant, each spectrin fragment contains only a single cysteine residue (C1892), which was successfully used previously for measuring the affinities of the wild-type fragments. The biotinylated spectrins (ligands) were diluted in HEPES buffered saline with surfactant P20 (HBS-P: 10mM HEPES, pH 7.4; 0.15M NaCl; 0.005% (vol/vol) surfactant P20; Biacore Life Sciences) to a final concentration of 1nM and applied to individual flow cells of streptavidin-modified biosensors (sensor chip SA; GE Healthcare) at a rate of 20 μL/min at 25°C. Between 100 and 150 response units were immobilized for each biotinylated ligand. Immobilization was judged to be stable with no noticeable decay in response. Purified ZU5-ANK fragments (analytes) in HBS-P were diluted serially to the concentrations indicated and maintained at 10°C until the time of injection. Injections were performed in parallel, with the ankyrin fragments being applied over all flow cells simultaneously at a flow rate of 20 μL/min and a sensor temperature of 25°C. This step allowed for binding to be measured for 3 spectrin mutants simultaneously. Injections were performed in triplicate with the use of HBS-P as the system buffer; association and dissociation phases were 60 seconds each.
The ankyrin fragments completely dissociated after approximately 180 seconds; nevertheless, a 300-second postdissociation phase wash was included in the run parameters to ensure complete removal of ZU5-ANK fragments from previous injections. Data were analyzed by use of the included BIAevaluation software (Version 4.1). Sensorgrams for each set of injections were processed globally and simultaneously for ka and kd after reference cell and buffer subtraction. Fits were assessed by correspondence between the data and the calculated fit, randomness of residuals, and nearness of global reduced χ2 values to 1.0. Values presented for the ka, kd, and KD (calculated by ka/kd) represent the average plus or minus SE for the 3 replicates for each ligand. Because several of the mutations led to severely defective binding, double-reciprocal analysis of the equilibrium values of the response was used to validate the values of KD obtained by fitting.
Data deposition
Results
Overall structure and molecular details of the spectrin-ankyrin complex
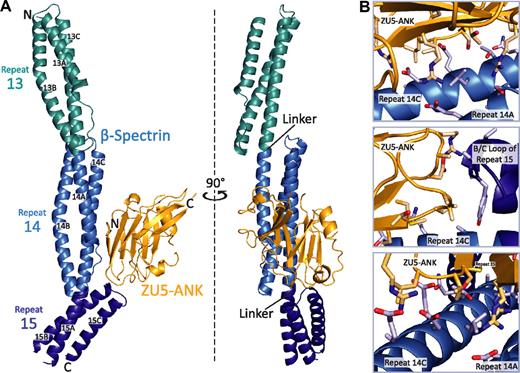
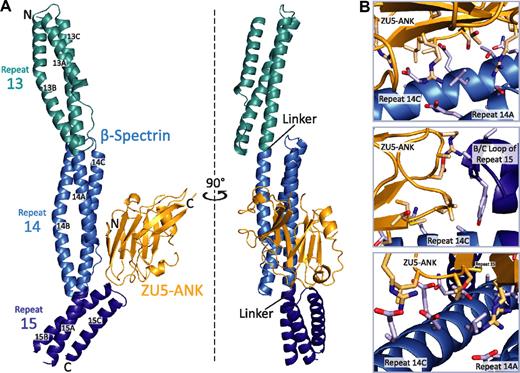
In the complex of βI-spectrin repeats 13 to 15 with the ZU5-ANK domain of human ankyrin R, the individual binding partners adopt largely the same tertiary structure as in their unbound states (Figure 1A).27,33,37 The spectrin repeats fold into well-characterized triple-helical bundles joined by ordered, helical linkers. The ZU5-ANK fragment maintains a compact, well-folded structure with a β-sheet rich core and a number of loops. In the complex, the 2 β-sheets that form the core of ZU5-ANK are oriented parallel to the long axis of spectrin. ZU5-ANK nestles against the B/C loop in repeat 15 of spectrin and sits atop much of repeat 14, creating an extended but narrow contact area of approximately 630 Å2. Because there are 2 copies of the complex in the crystallographic asymmetric unit, it was possible to assess its structural variability (supplemental Figure 1). The 2 copies of the complex superpose well, with an RMSD of 1.4 Å for main chain atoms. The distal portion of repeat 13, which is not involved in the binding interface, shows greater variability. A superposition that uses only ZU5-ANK and β-spectrin repeats 14 to 15 results in an RMSD of 0.9 Å for the main chain atoms, indicating that this region of the complex is largely unchanged.
Structure of the human β I-spectrin/ZU5-ankyrin R complex. (A) HEβ −1315 (blue) comprises 3 tandem canonical spectrin repeats (teal, blue, and dark blue from N- to C-terminus), each folded into a 3-helix bundle with the repeats connected by α-helical linkers. ZU5-ANK (gold), the spectrin binding domain of ankyrin R, maintains a compact β-sandwich fold connected by extended loops. The main interacting surfaces of the complex are formed by helices A and C of repeat 14 and the B/C loop of repeat 15 in β-spectrin and by the 2 strands from the β-core as well as 2 loops of ZU5-ANK. The 2 views are related by a 90° rotation. (B) Three different views of the spectrin/ankyrin interaction, that is, along the C-terminus of helix C of repeat 14 of spectrin (top), near the B/C loop of repeat 15 (middle), and near the N-terminus of helix C of repeat 14 (bottom).
Structure of the human β I-spectrin/ZU5-ankyrin R complex. (A) HEβ −1315 (blue) comprises 3 tandem canonical spectrin repeats (teal, blue, and dark blue from N- to C-terminus), each folded into a 3-helix bundle with the repeats connected by α-helical linkers. ZU5-ANK (gold), the spectrin binding domain of ankyrin R, maintains a compact β-sandwich fold connected by extended loops. The main interacting surfaces of the complex are formed by helices A and C of repeat 14 and the B/C loop of repeat 15 in β-spectrin and by the 2 strands from the β-core as well as 2 loops of ZU5-ANK. The 2 views are related by a 90° rotation. (B) Three different views of the spectrin/ankyrin interaction, that is, along the C-terminus of helix C of repeat 14 of spectrin (top), near the B/C loop of repeat 15 (middle), and near the N-terminus of helix C of repeat 14 (bottom).
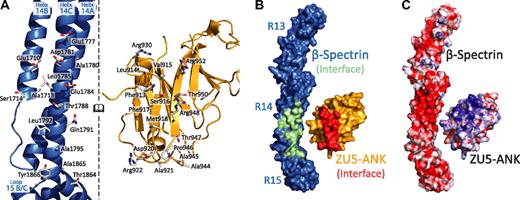
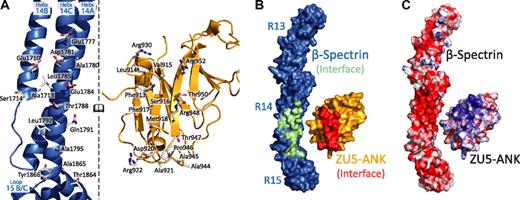
Interactions between the 2 proteins (Figure 1B; supplemental Figure 2) can be classified into 2 groups: charge interactions involving complementary patches between repeat 14 of spectrin and ankyrin and contacts with the B/C loop of spectrin repeat 15 (Figure 2). Most contacts with repeat 14 involve helix C, including Arg930ank-Asp1781spec/Glu1777spec. In contrast, only a few amino acids from helix A interact with ZU5-ANK (Arg952ank-Glu1710spec). Ankyrin sits close to the B/C loop and the linker region, which is ordered and helical. Tight contact between the 2 proteins in this region is facilitated by the presence of several small amino acids in which larger side chains are unlikely to be easily accommodated. For instance, Ala1865spec and Ala1867spec in the B/C loop of spectrin repeat 15 allow the close packing of the 2 molecules, whereas Ala944ank and Ala945ank on the ankyrin side of the interaction limit steric clashes. Interactions in the B/C loop region include few hydrogen bonds between the 2 molecules, mostly between main chain atoms. An exception is the side chain of Arg941ank, which contacts the main chain of Tyr1866spec. The spectrin and ankyrin fragments are closely packed in this region, enabling van der Waals interactions between ankyrin residues Asp920, Ala921, Arg922, Ala944, Ala945, and Pro946 and the spectrin residues in its B/C loop. In addition, a tyrosine (Tyr1866) in the spectrin B/C loop, which is conserved in all human spectrin isoforms that bind ankyrin, points toward the linker region with its side chain against Pro946 in ankyrin. The structure of the complex clearly elucidates the necessity for both repeats 14 and 15 in tandem to form a stable complex with ankyrin.
Binding surfaces of the spectrin-ankyrin interaction reveal a bimodal interaction. In all panels the molecules of the complex are opened and rotated such that the interacting regions of both face the reader. (A) The spectrin residues involved in ankyrin binding are found principally along repeat 14. The interacting surfaces present many charged residues along the N-terminal portion of repeat 14, whereas more hydrophobic residues are localized near the spectrin B/C loop. (B) Surface footprints of spectrin (green/blue) and ankyrin (red/gold) illustrate the shape complementarity. The interacting residues in spectrin and ankyrin are shown in green and red, respectively. (C) The electrostatic surface of the molecules show significant charge interactions involving a negatively charged region along repeat 14 of spectrin and in a positively charged patch on the ankyrin fragment. The molecular surface of the molecules is shown with the equipotential electrostatic surface mapped onto them at ±15 kbT/ec with red corresponding to negative and blue corresponding to positive charges.
Binding surfaces of the spectrin-ankyrin interaction reveal a bimodal interaction. In all panels the molecules of the complex are opened and rotated such that the interacting regions of both face the reader. (A) The spectrin residues involved in ankyrin binding are found principally along repeat 14. The interacting surfaces present many charged residues along the N-terminal portion of repeat 14, whereas more hydrophobic residues are localized near the spectrin B/C loop. (B) Surface footprints of spectrin (green/blue) and ankyrin (red/gold) illustrate the shape complementarity. The interacting residues in spectrin and ankyrin are shown in green and red, respectively. (C) The electrostatic surface of the molecules show significant charge interactions involving a negatively charged region along repeat 14 of spectrin and in a positively charged patch on the ankyrin fragment. The molecular surface of the molecules is shown with the equipotential electrostatic surface mapped onto them at ±15 kbT/ec with red corresponding to negative and blue corresponding to positive charges.
Binding and stability of clinical, loss-of-function, and structure-based mutants
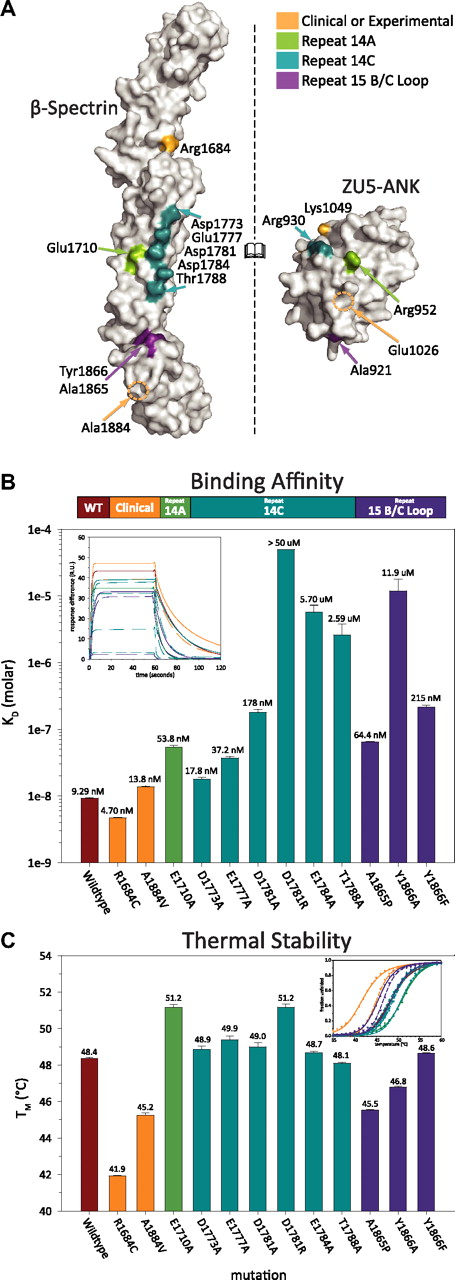
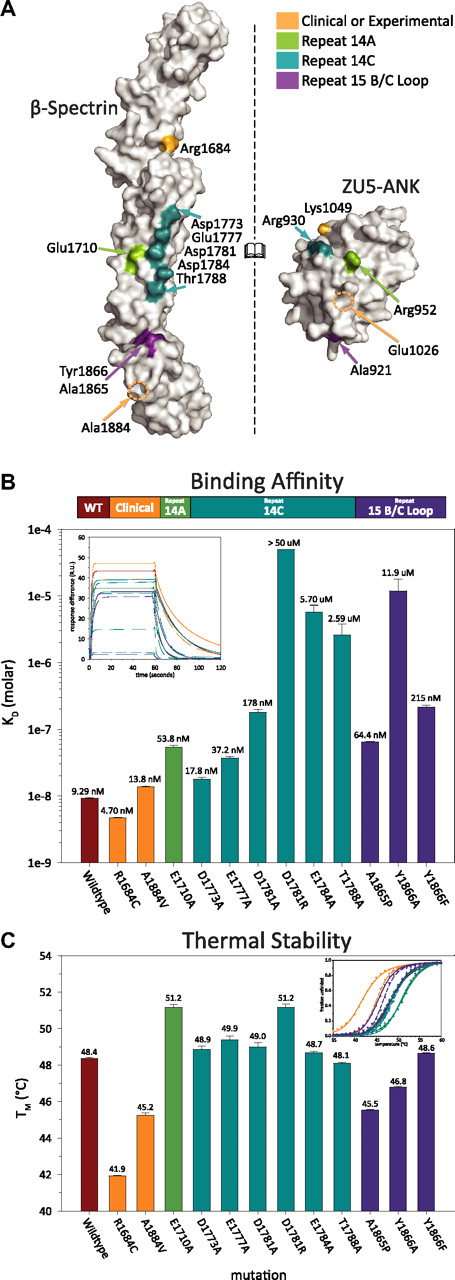
To explore the role of residues at the interface, as well as to provide insights into the nature of mutations associated with human diseases,38 a number of spectrin and ankyrin mutants were prepared (supplemental Figure 3). All of the 11 spectrin mutants were soluble, stable, and could be purified with high yields (Figure 3C; supplemental Figure 3B).38,39 In contrast, only 7 of the 11 ankyrin mutants were found to be soluble, whereas the remaining 4 mutants were highly insoluble upon expression. The insolubility of these 4 mutants suggests that they were misfolded, although this was not directly evaluated. Each of the 7 soluble ankyrin mutants was successfully purified (supplemental Figure 3A), although their yields were consistently lower than that of the wild-type protein (supplemental Figure 3B). Surface plasmon resonance was used to measure the binding affinity and kinetics of each of the mutants to their corresponding wild-type binding partner. The measured binding affinities (Figure 3B; supplemental Figure 5A) ranged from the low nanomolar regime (comparable with wild-type binding of 10-15nM) to very weak binding (>1μM).
Clinical mutations in the ankyrin binding region of β-spectrin disrupt protein stability rather than binding affinity. (A) The characterized human β-spectrin mutants can be classified into 4 categories: those from clinical38 sources or functionally based39 experiments (orange), those along the helix A of repeat 14 interface (green), those along helix C of repeat 14 (blue), and those in the B/C loop of repeat 15 (purple). The ankyrin mutations coloring is assigned on the basis of the position of their spectrin binding partner where applicable. (B) Surface plasmon resonance measurements of the binding affinity (color-coded as in panel A) illustrate that structure-guided mutations disrupt binding significantly, whereas many clinical mutations do not. The inset depicts the fit of each mutant's sensorgram upon injection of ZU5-ANK at 200nM. (C) In contrast, a similar analysis of thermal stability demonstrated that clinical mutations have a significant destabilizing effect, whereas structure-based mutations do not. Bars indicate average values with error bars corresponding to the SE. The insert presents the circular dichroism thermal denaturation data (fraction unfolded vs temperature) and 2-state fits for each mutant.
Clinical mutations in the ankyrin binding region of β-spectrin disrupt protein stability rather than binding affinity. (A) The characterized human β-spectrin mutants can be classified into 4 categories: those from clinical38 sources or functionally based39 experiments (orange), those along the helix A of repeat 14 interface (green), those along helix C of repeat 14 (blue), and those in the B/C loop of repeat 15 (purple). The ankyrin mutations coloring is assigned on the basis of the position of their spectrin binding partner where applicable. (B) Surface plasmon resonance measurements of the binding affinity (color-coded as in panel A) illustrate that structure-guided mutations disrupt binding significantly, whereas many clinical mutations do not. The inset depicts the fit of each mutant's sensorgram upon injection of ZU5-ANK at 200nM. (C) In contrast, a similar analysis of thermal stability demonstrated that clinical mutations have a significant destabilizing effect, whereas structure-based mutations do not. Bars indicate average values with error bars corresponding to the SE. The insert presents the circular dichroism thermal denaturation data (fraction unfolded vs temperature) and 2-state fits for each mutant.
As expected, the type of structure-guided mutation affected the degree to which binding was impaired. Disruption of salt bridges between the 2 molecules had a severe effect on binding. Furthermore, equivalent mutations along the C helix of spectrin had a varying degree of effect on binding, which was dependent upon their proximity to the core of the binding interface. For instance, Glu1784Ala reduced the binding affinity to weaker than 1μM, Asp1781Ala to approximately 200nM, Glu1777Ala to approximately 50nM, and finally Asp1773Ala, with an affinity of around 20nM, approached wild-type affinity. Mutations introducing steric clashes at the B/C loop/ankyrin interface weakened binding in proportion to their bulkiness, which is consistent with the more qualitative observations of Stabach et al.33 Thus, ZU5-ANK Ala921Thr had an affinity of approximately 100nM, whereas that of Ala921Tyr was near 300nM. An exception was the Ala1865Pro mutation in the B/C loop, which affected the binding affinity only modestly (KD ∼ 60nM), a less drastic impact than expected given the structural constraints imposed by prolines. Presumably, the flexibility of other residues in the B/C loop compensates for the rigidity imposed by the proline in this region of the molecule. Finally, ankyrin mutations distant from the contact interface (ESY1026AAAank, KR1049AAank) showed modest changes in affinity. This was particularly interesting because equivalent experimental mutations in ankyrin-G have been previously shown to have a devastating effect on cell morphology.39 It is possible that these mutations affect general ankyrin folding or disrupt interactions with other domains or proteins. Overall, however, the measured effects on binding affinity are consistent with the roles of these residues suggested by the structure of the complex.
In addition to structure-based mutagenesis, the effects on binding by 2, naturally occurring, spherocytosis-inducing mutations of β-spectrin—Arg1684Cys (Birmingham) and Ala1884Val (São Paulo)—also were assessed (Figure 3). Neither of these clinical mutations is in the immediate vicinity of the binding interface. Consistent with the qualitative observation that the Ala1884Val mutant (in helix 15C) does not disrupt binding,33 the affinity was measured to be approximately 15nM. The Arg1684Cys mutation (in the linker region between repeats 13 and 14) also displayed nanomolar affinity (∼ 5nM). The finding that these mutations implicated in spherocytosis did not have a robust effect on binding affinity suggests that other attributes must be disrupted to produce erythrocyte membrane skeleton defects.
Because spectrin has been shown to unfold and refold both in direct in vitro experiments40 and in indirect in vivo studies,41 we hypothesized that spectrin stability could be adversely affected by these mutations. Thermal melting temperatures (TM) of the fragments demonstrated striking differences among the spectrin mutants (Figure 3C), spanning almost 10°C (from ∼ 42°C to 51°C), even though all of the fragments were folded to a similar extent at lower temperatures with greater than 90% α-helical content (supplemental Figure 4). The ΔΔGunfolding,RT for each mutant compared with the wild type ranged from roughly −2.9 to 1.5 kcal/mol (supplemental Figure 6). Many of the mutants designed based on the structure of the complex (mostly alanine substitutions of acidic residues) showed increases in TM attributable to a decrease in charge repulsion in the anion-rich ankyrin-binding region. In contrast, both of the clinical mutations led to significant thermal destabilization of the fragments.
Discussion
Spectrin recognition by ankyrin
The structure of the complex together with the biochemical data and previous structures of spectrin repeats 14 to 15 and ZU5-ANK in the unbound state provide a number of insights into the overall mechanism of spectrin recognition by ankyrin. In addition, most of the residues involved in the binding interface are highly conserved,27,33 suggesting that the mechanism of recognition also is conserved. Recognition by ankyrin involves 2 important determinants: (1) the presence of complementary charge and hydrophobic surfaces in the proteins, and (2) a large tilt in the relative orientation of repeats 14 to 15 that allows for proper docking of ankyrin. These 2 features provide a mechanism for distinguishing repeats 14 to 15 from all other spectrin family repeats. The structure and mutagenesis data clearly show the importance of the charge interactions, especially in a complex with a relatively small buried surface area. The critical role of charge interactions is affirmed by the fact that all the interacting amino acids are highly conserved and that most have a measurable effect on binding affinity.
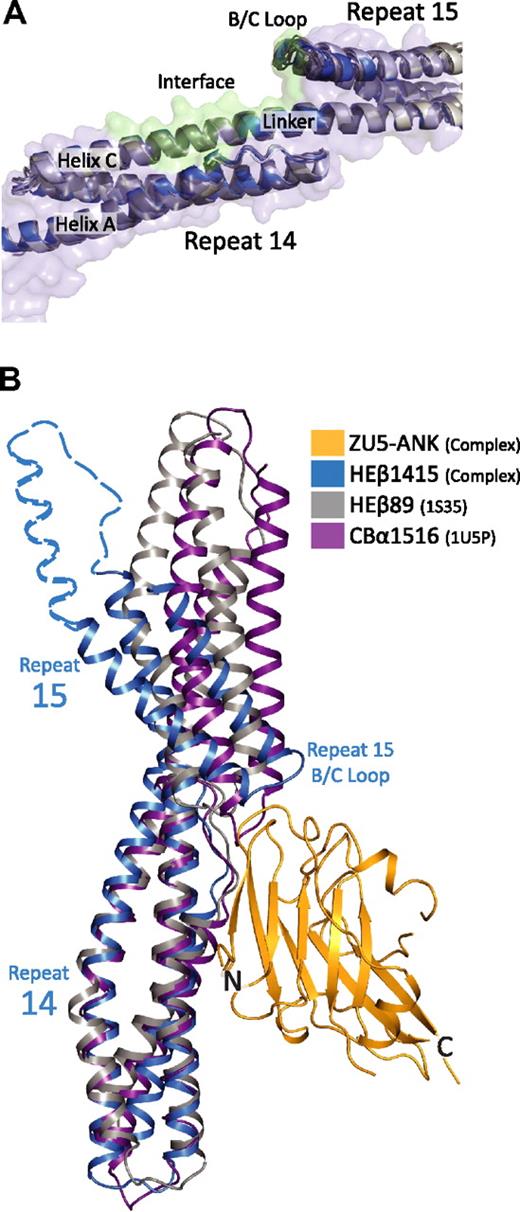
The role of the relative orientation of repeats 14 and 15 can be analyzed by comparing 3 structures of the ankyrin-binding domain of spectrin recently solved independently by x-ray crystallography. Two of these structures correspond to repeats 14 to15 of human erythroid β-spectrin27,33 whereas the third corresponds to a fragment spanning repeats 14 to 16 of human brain β-spectrin.37 Superposition of the repeat 14 to 15 structures onto repeats 14 to 15 from the complex demonstrates that the structural models are markedly similar with RMSD of main chain atoms approximately 1.5 Å (Figure 4A). Moreover, in the 3 structures, repeat 15 is tilted with respect to repeat 14 to a larger extent than in other structures of spectrin repeats (Figure 4B).27,33,37,43,–45 More importantly, the repeats in all 3 structures display the same relative orientations despite their significantly different crystal environments. This occurrence is an unusual one for spectrin repeat structures, which tend to exhibit variability in the relative repeat orientations, even among structures of identical molecules.43,–45 This unusual uniformity in repeat orientation suggests that the recognition of spectrin by ankyrin—although dependent on both charge and hydrophobic interactions—also requires shape complementarity without need for an induced fit. Moreover, the structures suggest that repeats 14 to 15 adopt a kinked conformation even in the absence of ankyrin and that the adoption of this special conformation, together with the presence of a highly negatively charged patch, serves to distinguish these repeats.
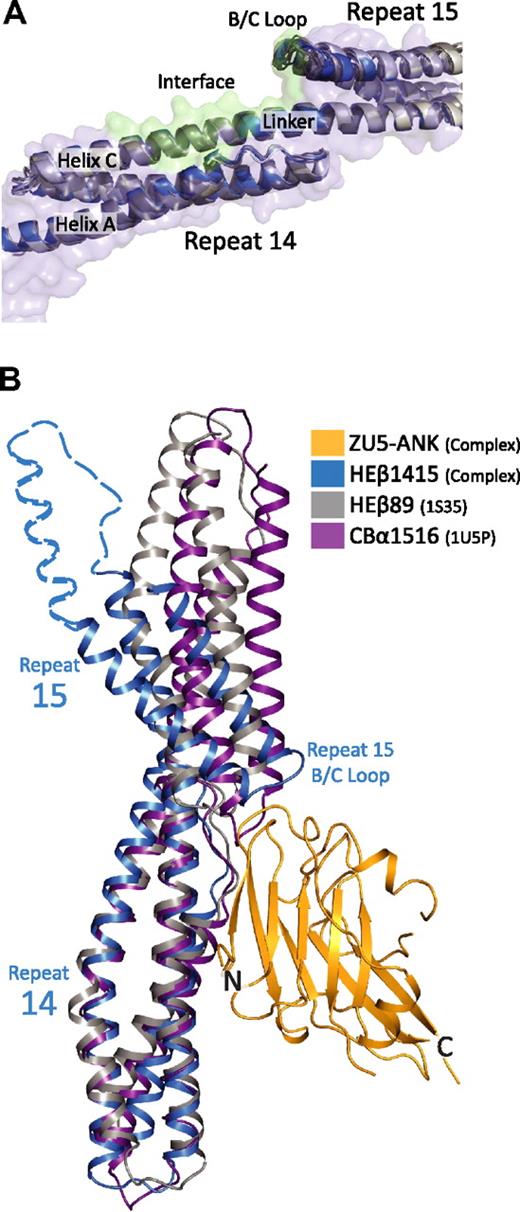
Comparison of structural models of spectrin di-repeats suggest a binding mechanism aided by a specific bend angle between repeats 14 and 15. (A) Superposition of multiple structures of repeats 14 and 15 of β-spectrin27,33,37 in the unbound state (gray) onto repeats 14 and 15 in the complex (blue) show marked structural similarities. In total, 5 models from 3 independent crystal structures of unbound β-spectrin repeats 14 and 15 were superimposed. The structures are virtually identical, showing the same relative orientation of the 2 repeats (RMSD values of ∼ 1.5 Å between all of the structures). The similarity among the structures suggests that the presence of a specific bending angle is important in recognition. (B) Alignment of 3 different spectrin di-repeat structures to repeat 14 of the complex demonstrate how the overall bend angle between β-spectrin repeats 14 and 15 (blue; HEβ1415) generates a close-fitting and matching interaction with ZU5-ANK (gold; ZU5-ANK). In contrast, the shallower bend angles seen in human erythroid β-spectrin repeats 8 and 944 (gray; HEβ89) and chicken brain α-spectrin repeats 15 and 1645 (purple; CBα1516) would reduce the interacting surface or induce steric clashes, respectively. The dashed blue outline completing repeat 15 was modeled from the structure of brain β-spectrin repeats 14 to 16.37
Comparison of structural models of spectrin di-repeats suggest a binding mechanism aided by a specific bend angle between repeats 14 and 15. (A) Superposition of multiple structures of repeats 14 and 15 of β-spectrin27,33,37 in the unbound state (gray) onto repeats 14 and 15 in the complex (blue) show marked structural similarities. In total, 5 models from 3 independent crystal structures of unbound β-spectrin repeats 14 and 15 were superimposed. The structures are virtually identical, showing the same relative orientation of the 2 repeats (RMSD values of ∼ 1.5 Å between all of the structures). The similarity among the structures suggests that the presence of a specific bending angle is important in recognition. (B) Alignment of 3 different spectrin di-repeat structures to repeat 14 of the complex demonstrate how the overall bend angle between β-spectrin repeats 14 and 15 (blue; HEβ1415) generates a close-fitting and matching interaction with ZU5-ANK (gold; ZU5-ANK). In contrast, the shallower bend angles seen in human erythroid β-spectrin repeats 8 and 944 (gray; HEβ89) and chicken brain α-spectrin repeats 15 and 1645 (purple; CBα1516) would reduce the interacting surface or induce steric clashes, respectively. The dashed blue outline completing repeat 15 was modeled from the structure of brain β-spectrin repeats 14 to 16.37
Recognition of distinct spectrin repeats by specific proteins is not only important for the integrity of the metazoan cytoskeleton but also has been implicated in many different functions such as cell polarization46 and T-cell differentiation.47 With roughly 30% sequence identity and 90% amino acid similarity among repeats in the spectrin superfamily, however, spectrin fragments must use highly specific interactions to generate binding surfaces while maintaining their canonical 3 helix bundle fold.13 This structure of the spectrin/ankyrin complex demonstrates that spectrin repeats 14 to 15 use shape and charge complementarity to generate a binding region of high specificity without altering the overall structure of the spectrin repeats. A similar strategy may be used by other spectrin repeats to bind different substrates with high affinities. Particular arrangements of adjacent repeats exploiting unique charge and hydrophobic regions along different faces of the spectrin fold could present a diverse set of surfaces, allowing for a variety of distinct ligands to bind.
Clinical mutations greatly destabilize spectrin while only marginally affecting ankyrin binding in vitro
Strikingly, the Arg1684Cys (Birmingham) mutation, which had the strongest binding affinity (∼ 5nM) and was most distant from the binding interface, had the most dramatic destabilization (ΔTM approximately −7°C and ΔΔGunfolding,RT ∼ 1.5 kcal/mol). Interestingly, this residue resides in the linker between repeats 13 and 14. Simulations of tandem spectrin repeat unfolding have revealed the importance of the α-helical linkers on multirepeat stability,42 which could explain the dramatic thermal destabilization observed for this mutant. Structurally, the basis of the Ala1884Val mutation (São Paulo) is not immediately obvious. It is possible that the introduction of the bulkier, more hydrophobic valine side chain in this somewhat solvent-exposed location destabilizes the packing of the repeat 15 3-helix bundle. Regardless of the precise nature of the disruptions, these data suggest that some disease-associated mutants of erythroid spectrin near the ankyrin binding site do not disrupt the spectrin-ankyrin complex.
In addition, a construct incorporating a clinical mutation in the ZU5-ANK region of ankyrin (A945Pank) was not soluble upon expression. Unlike the case of the spectrin mutants, thermal denaturation of the ankyrin mutants studied (supplemental Figure 5), although demonstrating differences from the melting behavior of the wild-type protein, did not show a clear trend in changes in stability. Of the mutations that have been found to cause cell morphology defects in cell culture, the KR1049AAank mutation did lead to a decrease in thermal stability, whereas the ESY1026AAAank mutation actually stabilized the protein, although it is possible that such a change could disrupt intramolecular binding interfaces in ankyrin R. These observations suggest that for the clinical mutations found near the ankyrin binding site, folding instability of spectrin, rather than binding affinity, may lead to a clinical loss of membrane skeleton integrity in a manner yet to be determined.
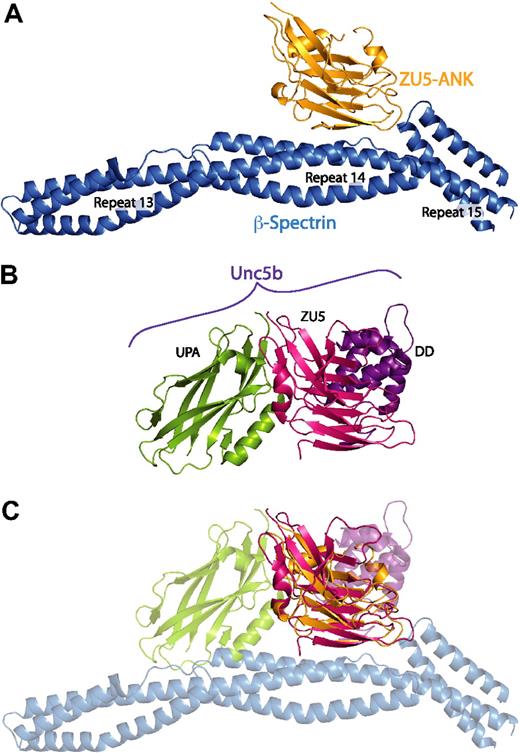
ZU5 domains use multiple interaction surfaces
Recent structural work, which has provided atomic models of ZU5-ANK in the unbound state (supplemental Figure 8),27 and a cytoplasmic portion of the ZU5-containing netrin receptor UNC5b (Figure 5B),48 also suggests that the ZU5-ANK subdomain may be capable of binding multiple ligands. The UNC5b structure, corresponding to an auto-inhibited molecular conformation formed by an intramolecular complex involving the UPA, ZU5, and Death domains of UNC5b,48 provided the first example of intramolecular interactions among a ZU5 domain. The UNC5b structure suggests disparate roles for 2 tandem ZU5 domains present in ankyrin: the first one corresponding to ZU5-ANK and interacting with spectrin, and the second (not ZU5-ANK) interacting with the putative UPA and Death domains of ankyrin. Interestingly, superposition of the ZU5 domain in the UNC5b structure onto the ZU5 domain in the spectrin-ankyrin complex reveals that ZU5 domains utilize 3 separate and different binding interfaces (Figure 5). This finding suggests that ZU5 domains may have evolved into versatile protein-protein binding modules with multiple surfaces for binding different ligands. Moreover, ZU5-ANK may be capable of interacting with other domains of ankyrin or even other proteins to serve as an organizer of proteins near the cell membrane. The alternative interaction surfaces of ZU5-ANK may be those disrupted by some of the mutations, which lead to catastrophic disruption of normal cell morphology in cell culture or to disease states but maintain wild type binding affinity in our in vitro spectrin/ZU5-ANK assays.
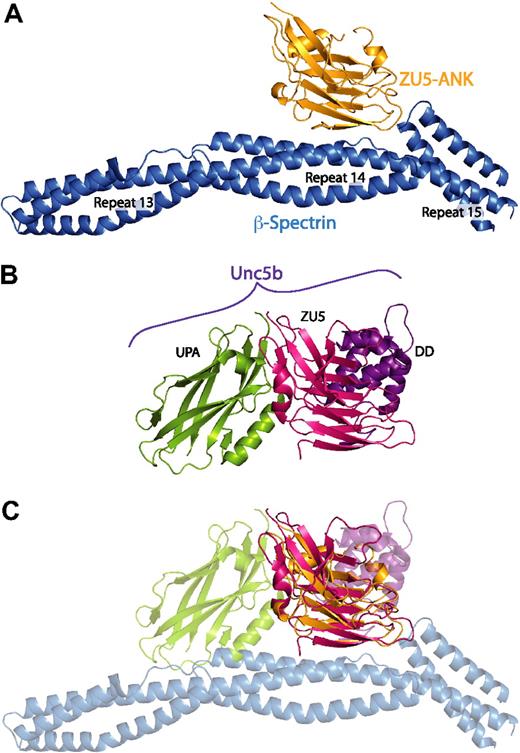
The ZU5 subdomain is capable of forming multiple, specific interactions with distinct surfaces. (A) Structure of the complex between spectrin (blue) and the ZU5-ANK domain of ankyrin (gold). (B) Structure of the cytoplasmic portion of the netrin receptor UNC5b (ZU5-2 domain in magenta, UPA domain in green, DD domain in purple) in the same orientation as panel A. (C) Superposition of the ZU5 domains of the 2 structures demonstrates that ZU5 domains have been adapted to bind multiple ligands with the use of different molecular surfaces. Note that recent sequence analysis of ankyrin R has revealed 2 tandem ZU5 domains48 : ZU5-ANK, which binds to spectrin, and a second ZU5 domain (ZU5-2). On the basis of this alignment, the ZU5 domain in the UNC5b structure correlates with the ZU5-2 domain of ankyrin. The superimposition of the models shown in panel C therefore does not reflect a predicted macromolecular assembly; it serves only to demonstrate that distinct interaction surfaces of ZU5 domains have now been observed.
The ZU5 subdomain is capable of forming multiple, specific interactions with distinct surfaces. (A) Structure of the complex between spectrin (blue) and the ZU5-ANK domain of ankyrin (gold). (B) Structure of the cytoplasmic portion of the netrin receptor UNC5b (ZU5-2 domain in magenta, UPA domain in green, DD domain in purple) in the same orientation as panel A. (C) Superposition of the ZU5 domains of the 2 structures demonstrates that ZU5 domains have been adapted to bind multiple ligands with the use of different molecular surfaces. Note that recent sequence analysis of ankyrin R has revealed 2 tandem ZU5 domains48 : ZU5-ANK, which binds to spectrin, and a second ZU5 domain (ZU5-2). On the basis of this alignment, the ZU5 domain in the UNC5b structure correlates with the ZU5-2 domain of ankyrin. The superimposition of the models shown in panel C therefore does not reflect a predicted macromolecular assembly; it serves only to demonstrate that distinct interaction surfaces of ZU5 domains have now been observed.
Conclusions
Structural and biochemical studies to date have considerably advanced our understanding of spectrin/ankyrin recognition. Whereas the fold of all spectrin repeats is highly conserved, the structure of the spectrin/ankyrin complex illustrates how different binding partners could distinguish specific molecular epitopes. In addition, structural and biochemical data reveal the versatility of ZU5 domains as a protein binding module. Finally, the possible molecular basis for some human blood diseases has been revealed. In conclusion, structural studies have begun to illuminate the relationship between the metazoan membrane and the underlying cytoskeleton at the atomic level, paving the way for further studies of this vital cellular connection.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank B. Forget and P. Gallagher for human erythroid β-spectrin and human erythroid ankyrin cDNA, respectively, as well as Lei Huang for initial help in crystallization screening. We thank the Chemistry Core at the Institute for BioNanotechnology in Medicine at Northwestern University, which is supported by National Institutes of Health award 5P50NS054287. We acknowledge staff and instrumentation support from the University of Chicago Biophysics Core Facility, the Keck Biophysics Facility at Northwestern University, the Center for Structural Biology at Northwestern University, and LS-CAT at APS at Argonne National Labs. Use of the APS is supported by the Department of Energy (DOE). Support from the R.H. Lurie Comprehensive Cancer Center of Northwestern University to the Structural Biology Facility is also acknowledged. We thank C.-Y. Koh and E. Solomaha for instrument access and assistance with electrospray ionization mass spectrometry and surface plasmon resonance experiments, respectively. We also thank N. M. Baker, R. Rajan, N. J. Reiter, and R. I. MacDonald for comments and suggestions.
This work was supported by National Institutes of Health grant (GM057692) to A.M.
National Institutes of Health
Authorship
Contribution: J.J.I. designed and performed research, analyzed and interpreted data, and drafted the manuscript; and A.M. directed research, assisted with data interpretation, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alfonso Mondragón, Department of BMBCB, Northwestern University, 2205 Tech Dr, 2-100 Hogan Bldg, Evanston, IL 60208; e-mail: a-mondragon@northwestern.edu.