Cylindromatosis (CYLD) is a deubiquitinase that was initially identified as a tumor suppressor and has recently been implicated in diverse normal physiologic processes. In this study, we have investigated the involvement of CYLD in angiogenesis, the formation of new blood vessels from preexisting ones. We find that knockdown of CYLD expression significantly impairs angiogenesis in vitro in both matrigel-based tube formation assay and collagen-based 3-dimensional capillary sprouting assay. Disruption of CYLD also remarkably inhibits angiogenic response in vivo, as evidenced by diminished blood vessel growth into the angioreactors implanted in mice. Mechanistic studies show that CYLD regulates angiogenesis by mediating the spreading and migration of vascular endothelial cells. Silencing of CYLD dramatically decreases microtubule dynamics in endothelial cells and inhibits endothelial cell migration by blocking the polarization process. Furthermore, we identify Rac1 activation as an important factor contributing to the action of CYLD in regulating endothelial cell migration and angiogenesis. Our findings thus uncover a previously unrecognized role for CYLD in the angiogenic process and provide a novel mechanism for Rac1 activation during endothelial cell migration and angiogenesis.
Introduction
Angiogenesis, which refers to the formation of new capillary blood vessels in the body, is a fundamental event required for a variety of physiologic and pathologic processes, such as embryonic development, wound healing, myocardial ischemia, and tumor growth.1,2 Angiogenesis is tightly regulated by proangiogenic and antiangiogenic factors and requires the migration of vascular endothelial cells from preexisting blood vessels.1,–3 Vascular endothelial cell migration is a multistep process that involves leading-edge protrusion, cycles of adhesion formation at the front and detachment at the cell rear, and contraction of the cell body.3
Migrating endothelial cells have a highly polarized structure, with the appearance of membrane ruffles at the leading edge and asymmetrical localization of signaling molecules and the cytoskeleton.3,4 There is a growing body of evidence that coordinated action of microtubules and actin filaments is critical for cell polarization and migration.4,5 In migrating cells, microtubule plus ends radiate primarily toward the leading edge and minus ends are concentrated at the centrosome, which is positioned between the nucleus and the leading edge.6,7 Microtubule dynamics are regulated exquisitely in migrating cells by a number of microtubule-binding proteins and by the Rho family guanosine triphosphatases (GTPases) Rac1, RhoA, and Cdc42.6,8 Microtubules are also able to modulate the activities of these Rho GTPases, and the interplay between the cytoskeleton and Rho GTPases is required for efficient cell polarization and migration.6,8
Recently, the tumor suppressor cylindromatosis (CYLD) has been identified as a novel microtubule-binding protein and has been implicated in cell migration.9,10 CYLD contains 3 cytoskeleton-associated protein glycine-rich (CAP-Gly) domains in its amino terminus, and both the microtubule-binding and migration-regulatory activities of CYLD are mediated by its first CAP-Gly domain,9 suggesting that this protein might exert its effect on cell migration through modulation of microtubule dynamics. The involvement of CYLD in cell migration also suggests that this protein might play a role in angiogenesis, of which vascular endothelial cell migration is an essential component. This study was undertaken to test these hypotheses directly.
Methods
Cell culture and transfection
Human umbilical vein endothelial cells (HUVECs) were obtained from the ATCC and maintained as described previously.11 Plasmid transfections were performed by electroporation with the use of the ECM830 system (BTX). CYLD and control small interfering RNAs (siRNAs) were synthesized by Ribobio and transfected into cells with the Lipofectamine 2000 reagent (Invitrogen).
Tube formation and cell spreading assays
HUVECs were plated onto 35-mm culture dishes precoated with matrigel. To examine tube formation, photographs were taken 4 and 16 hours later with an Axio Observer A1 fluorescence microscope (Carl Zeiss Inc), and the degree of tube formation was quantified by measuring the cumulative tube length with the use of the ImageJ software (National Institutes of Health). To examine cell spreading, cells were fixed with 4% paraformaldehyde 25 or 60 minutes after plating. Photographs were then taken, and the degree of cell spreading was quantified.
3-Dimensional capillary sprouting assay
Cell spheroids were generated by culturing HUVECs overnight in medium containing 0.25% carboxymethylcellulose in round-bottom 96-well plates. The spheroids were embedded into collagen gels and transferred to 24-well plates. Culture medium was then added on top of the gel. Capillary-like sprout formation was examined by microscopy, and angiogenic activity was quantified by measuring the cumulative sprout length per spheroid.
Measurement of angiogenic response in vivo
Angiogenic response in vivo was examined with the directed in vivo angiogenesis assay kit (Trevigen) as described.12 In brief, semiclosed angioreactors were filled with extracellular matrix containing angiogenic factors (heparin and fibroblast growth factor 2) and CYLD or control siRNAs. In another set of experiments, the angioreactors were filled with extracellular matrix containing angiogenic factors and anti-CYLD antibody or immunoglobulin G (IgG) control. The angioreactors were incubated at 37°C for 1 hour to allow gel formation and then implanted subcutaneously into the dorsal flank of 6- to 8-week-old athymic nude mice. After 11 days, angioreactors were removed and photographed. To quantify the angiogenic response, cell pellets were recovered from the angioreactors by dispase digestion and stained with fluorescein-conjugated lectin-1 (FITC-lectin). The use of mice was approved by the Animal Care and Use Committee of Nankai University.
In vitro wound healing assay
HUVECs grown in 24-well plates as confluent monolayers were mechanically scratched with a 20-μL pipette tip to create the wound. Cells were washed with phosphate-buffered saline (PBS) to remove the debris, and complete culture media were then added to allow wound healing. Phase contrast images of the wound were taken 0, 12, and 24 hours later at 3 random locations to examine the extent of wound closure.
Immunofluorescence microscopy
Cells grown on glass coverslips were fixed with methanol for 5 minutes at −20°C or fixed with 4% paraformaldehyde for 30 minutes at room temperature. Cells were blocked with 2% bovine serum albumin in PBS, incubated with antibodies against α-tubulin (Sigma-Aldrich) and pericentrin (Santa Cruz Biotechnology), and then with FITC- and rhodamine-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) followed by staining with 4′,6-diamidino-2-phenylindole (DAPI) for 5 minutes. Coverslips were mounted with 90% glycerol in PBS and examined with the Axio Observer A1 fluorescence microscope (Carl Zeiss). For immunofluorescent staining of blood vessels in angioreactors, frozen sections of the vessel-containing matrigel were mounted onto slides precoated with 3-aminopropyltriethoxysilane. The sections were fixed with 4% paraformaldehyde, stained with antibodies against CD31 (BioLegend) and desmin (Abcam), and then examined by fluorescence microscopy.
Live cell imaging
HUVECs were cultured in a 37°C chamber on a TCS SP5 confocal microscope (Leica) equipped with a live-cell imaging workstation. To examine membrane ruffle dynamics, the fluorescence of GFP at the leading edge of cells was recorded at 20-second intervals with the use of the LAS AF software (Leica). The acquired image sequences were analyzed by ImageJ (National Institutes of Health), and membrane ruffle dynamics were presented as 3-dimensional surface plots. To examine microtubule dynamics, individual GFP-labeled microtubules in migrating cells were recorded at 2-second intervals, and microtubule life history plots representing changes in microtubule length over time were generated.
Immunoblot analysis
Cells were lysed in a buffer containing 1% Triton X-100, 150mM NaCl, and 50mM Tris (pH7.5). Cell membrane was isolated with the plasma membrane protein extraction kit (BioVision) according to the manufacturer's instruction. Proteins were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were blocked in Tris-buffered saline containing 0.2% Tween 20 and 5% fat-free dry milk and incubated with antibodies against CYLD (Lifespan), α-tubulin (Sigma-Aldrich), or GFP (Roche). The membranes were then incubated with horseradish peroxidase–conjugated secondary antibodies (Amersham Biosciences). Specific proteins were visualized with enhanced chemiluminescence detection reagent according to the manufacturer's instructions (Pierce Biotechnologies).
Measurement of Rac1 activity
The GST-PBD (p21-binding domain of Pak1 fused to glutathione-S-transferase) pulldown assays were used to detect cellular activated (GTP-bound) Rac1, as described previously.13 In brief, cell lysates were incubated with GST-PBD–coupled glutathione sepharose beads. The beads were washed extensively, and proteins bound on beads were examined by immunoblot analysis with anti-Rac1 antibody (Cell Signaling Technology).
Results
Knockdown of CYLD expression impairs angiogenesis in vitro
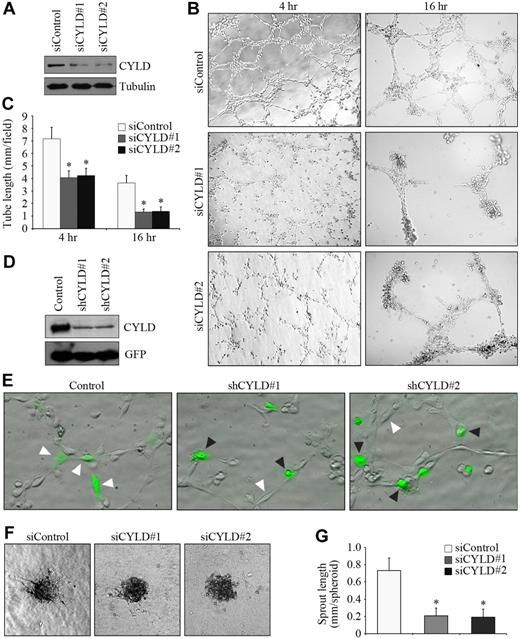
To investigate the potential role of CYLD in angiogenesis, we first examined the effect of CYLD siRNAs on vascular endothelial tube formation in vitro. Two different siRNAs were used to target CYLD. Both of them were able to knockdown CYLD expression effectively in HUVECs (Figure 1A). We observed capillary/tubelike structures as early as 2 hours after plating cells onto matrigel, and the structures were more evident 4 hours after plating. Transfection of the endothelial cells with CYLD siRNAs remarkably impaired tube formation compared with the control group (Figure 1B). By measuring the cumulative tube length, we found that CYLD siRNAs inhibited tube formation from HUVECs by 42% and 63%, respectively, 4 and 16 hours after plating (Figure 1C).
Knockdown of CYLD expression impairs endothelial tube formation and sprouting. (A) Immunoblot analysis of CYLD and tubulin expression in HUVECs transfected with CYLD or control siRNAs for 72 hours. (B) HUVECs transfected with CYLD or control siRNAs were plated onto matrigel, and photographs were taken 4 and 16 hours later. Objective lens used was A-Plan 10×/0.25 NA dry (Carl Zeiss Inc). (C) Experiments were performed as in panel B, and the cumulative tube length was measured. Data are the mean and standard error from 3 experiments with each performed in triplicate (*P < .05 vs control). (D) Immunoblot analysis of CYLD and GFP expression in HUVECs transfected with pEGFPC1 and pSUPER or pSUPER-CYLD plasmids for 72 hours. (E) HUVECs transfected with pEGFPC1 and pSUPER or pSUPER-CYLD plasmids were plated onto matrigel, and photographs were taken 2 hours later. GFP was used to mark transfected cells. White arrowheads indicate cells that align end-to-end, and black arrowheads indicate cells without such a property. Objective lens used was A-Plan 20×/0.3 NA dry (Carl Zeiss Inc). (F) Capillary-like sprout formation from spheroids generated from HUVECs transfected with CYLD or control siRNAs. Objective lens used was A-Plan 10×/0.25 NA dry (Carl Zeiss Inc). (G) Experiments were performed as in panel F, and the cumulative sprout length was measured. Data are the mean and SE from 2 experiments with each performed in triplicate (*P < .05 vs control). All images in this figure and in the following figures (except Figure 3E and Figure 4A) were taken with the Axio Observer A1 fluorescence microscope (Carl Zeiss Inc), equipped with the AxioCam MRm Rev.3 CCD camera and the AxioVision Rel. 4.1 software. All images were processed with the Adobe Photoshop CS8.0 software (Adobe Systems).
Knockdown of CYLD expression impairs endothelial tube formation and sprouting. (A) Immunoblot analysis of CYLD and tubulin expression in HUVECs transfected with CYLD or control siRNAs for 72 hours. (B) HUVECs transfected with CYLD or control siRNAs were plated onto matrigel, and photographs were taken 4 and 16 hours later. Objective lens used was A-Plan 10×/0.25 NA dry (Carl Zeiss Inc). (C) Experiments were performed as in panel B, and the cumulative tube length was measured. Data are the mean and standard error from 3 experiments with each performed in triplicate (*P < .05 vs control). (D) Immunoblot analysis of CYLD and GFP expression in HUVECs transfected with pEGFPC1 and pSUPER or pSUPER-CYLD plasmids for 72 hours. (E) HUVECs transfected with pEGFPC1 and pSUPER or pSUPER-CYLD plasmids were plated onto matrigel, and photographs were taken 2 hours later. GFP was used to mark transfected cells. White arrowheads indicate cells that align end-to-end, and black arrowheads indicate cells without such a property. Objective lens used was A-Plan 20×/0.3 NA dry (Carl Zeiss Inc). (F) Capillary-like sprout formation from spheroids generated from HUVECs transfected with CYLD or control siRNAs. Objective lens used was A-Plan 10×/0.25 NA dry (Carl Zeiss Inc). (G) Experiments were performed as in panel F, and the cumulative sprout length was measured. Data are the mean and SE from 2 experiments with each performed in triplicate (*P < .05 vs control). All images in this figure and in the following figures (except Figure 3E and Figure 4A) were taken with the Axio Observer A1 fluorescence microscope (Carl Zeiss Inc), equipped with the AxioCam MRm Rev.3 CCD camera and the AxioVision Rel. 4.1 software. All images were processed with the Adobe Photoshop CS8.0 software (Adobe Systems).
We then analyzed the behavior of CYLD-knockdown cells during the tube formation process. HUVECs were transfected with pSUPER-CYLD plasmids, which express small hairpin RNAs (shRNAs) targeting CYLD, together with pEGFPC1, which expresses GFP to mark transfected cells. Both of the 2 CYLD shRNAs could efficiently inhibit CYLD expression in HUVECs (Figure 1D). Cells aligned end-to-end 2 hours after plating on matrigel, whereas most CYLD shRNA-transfected cells lost such a property (Figure 1E).
To further investigate the function of CYLD in angiogenesis, we examined the effect of CYLD siRNAs on endothelial sprouting in a collagen-based 3-dimensional angiogenesis assay. We found that CYLD siRNAs significantly blocked capillary-like endothelial sprouting from the spheroids compared with the control group (Figure 1F-G). Together, these data show that CYLD is critically involved in endothelial tube formation and sprouting.
CYLD is critical for angiogenic response in vivo
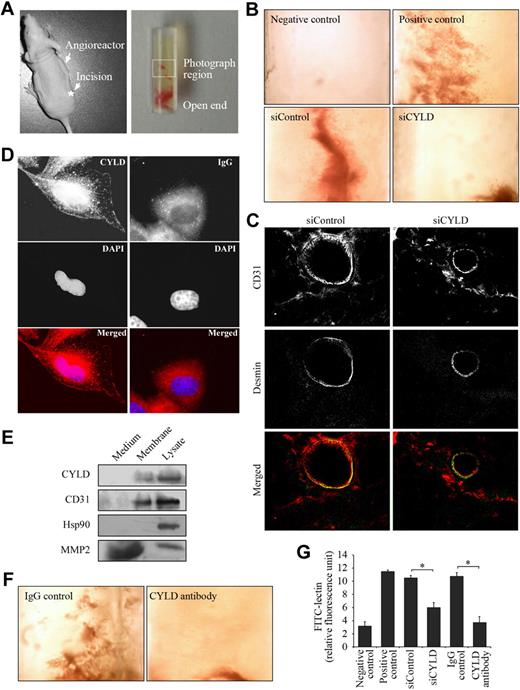
To substantiate the in vitro data indicating the importance of CYLD in angiogenesis, we studied the effect of CYLD on angiogenic response in mice. Semiclosed angioreactors were filled with extracellular matrix containing angiogenic factors (heparin and fibroblast growth factor 2) and implanted subcutaneously into the dorsal flank of athymic nude mice (Figure 2A). We found apparent vascular structures in the angioreactors 11 days after implantation (Figure 2B positive control), whereas no vascular structures were observed in angioreactors that were filled with extracellular matrix not containing angiogenic factors (Figure 2B negative control).
CYLD is critical for angiogenic response in vivo. (A left) An athymic nude mouse with a semiclosed angioreactor implanted into the dorsal flank. The asterisk indicates the site for surgical incision. (Right) A semiclosed angioreactor. The boxed area indicates the region photographed. (B) Vascular growth in the angioreactors implanted in mice for 11 days. Angioreactors filled with extracellular matrix containing angiogenic factors serve as the positive control, and angioreactors filled with extracellular matrix not containing angiogenic factors serve as the negative control. In siRNA experiments, angioreactors were filled with extracellular matrix containing angiogenic factors and CYLD or control siRNAs. Objective lens used was A-Plan 4×/0.1 NA dry (Carl Zeiss Inc). (C) Immunofluorescence microscopy of blood vessels in angioreactors. Frozen sections of the vessel-containing matrigel were stained with anti-CD31 and anti–desmin antibodies to detect endothelial cells and mural cells, respectively. Objective lens used was A-Plan 40×/0.5 NA dry (Carl Zeiss Inc). (D) Immunofluorescent images of HUVECs stained with anti-CYLD or IgG control antibodies and the DNA dye DAPI. Objective lens used was Plan-Apochromat 63×/1.25 NA oil (Carl Zeiss Inc). (E) Immunoblot analysis of CYLD, CD31, Hsp90, and MMP2 in the culture medium, plasma membrane, and lysate of HUVECs. (F) Vascular growth in the angioreactors filled with extracellular matrix containing angiogenic factors and anti-CYLD antibody or IgG control. Objective lens used was A-Plan 4×/0.1 NA dry (Carl Zeiss Inc). (G) Experiments were performed as in panels B and F, and angiogenic responses were quantified by FITC-lectin staining of cell pellets recovered from the angioreactors. Data are the mean and standard error from 2 experiments with each performed in octaplicate (*P < .05).
CYLD is critical for angiogenic response in vivo. (A left) An athymic nude mouse with a semiclosed angioreactor implanted into the dorsal flank. The asterisk indicates the site for surgical incision. (Right) A semiclosed angioreactor. The boxed area indicates the region photographed. (B) Vascular growth in the angioreactors implanted in mice for 11 days. Angioreactors filled with extracellular matrix containing angiogenic factors serve as the positive control, and angioreactors filled with extracellular matrix not containing angiogenic factors serve as the negative control. In siRNA experiments, angioreactors were filled with extracellular matrix containing angiogenic factors and CYLD or control siRNAs. Objective lens used was A-Plan 4×/0.1 NA dry (Carl Zeiss Inc). (C) Immunofluorescence microscopy of blood vessels in angioreactors. Frozen sections of the vessel-containing matrigel were stained with anti-CD31 and anti–desmin antibodies to detect endothelial cells and mural cells, respectively. Objective lens used was A-Plan 40×/0.5 NA dry (Carl Zeiss Inc). (D) Immunofluorescent images of HUVECs stained with anti-CYLD or IgG control antibodies and the DNA dye DAPI. Objective lens used was Plan-Apochromat 63×/1.25 NA oil (Carl Zeiss Inc). (E) Immunoblot analysis of CYLD, CD31, Hsp90, and MMP2 in the culture medium, plasma membrane, and lysate of HUVECs. (F) Vascular growth in the angioreactors filled with extracellular matrix containing angiogenic factors and anti-CYLD antibody or IgG control. Objective lens used was A-Plan 4×/0.1 NA dry (Carl Zeiss Inc). (G) Experiments were performed as in panels B and F, and angiogenic responses were quantified by FITC-lectin staining of cell pellets recovered from the angioreactors. Data are the mean and standard error from 2 experiments with each performed in octaplicate (*P < .05).
Importantly, addition of CYLD siRNA in the angioreactors largely blocked vascular growth induced by the angiogenic factors; in contrast, control siRNA did not affect vascular growth into the angioreactors (Figure 2B). To examine the vessel architecture in angioreactors, frozen sections of the vessel-containing matrigel were double-stained with anti-CD31 antibody to visualize endothelial cells and anti–desmin antibody to visualize mural cells (vascular smooth muscle cells/pericytes). A number of blood vessels were found in the control siRNA group, which exhibited a layer of endothelial cells surrounded by mural cells (Figure 2C). In contrast, blood vessels were difficult to detect in the CYLD siRNA group, and only a few thinner vessels were observed; interestingly, these thinner vessels also exhibited a layer of endothelial cells covered by mural cells (Figure 2C).
By immunofluorescence microscopy, we found in HUVECs the localization of CYLD on the cell membrane in addition to its intracellular distribution (Figure 2D). Immunoblot analysis confirmed the association of CYLD with the cell membrane (Figure 2E). The membrane-localization pattern of CYLD suggested that blocking its function by a specific antibody might be able to inhibit vascular growth into the angioreactors. Indeed, we found that the addition of anti-CYLD antibody in the angioreactors significantly blocked vascular growth induced by the angiogenic factors (Figure 2F).
To quantify the angiogenic response in vivo, cell pellets were recovered from the angioreactors by dispase digestion and stained with FITC-lectin, a specific marker of endothelial cells.12 As shown in Figure 2G, the fluorescence intensity of cells recovered from CYLD siRNA or antibody-treated angioreactors was dramatically decreased compared with the control siRNA and IgG groups, indicating that disruption of CYLD in vivo could significantly inhibit blood vessel growth into the angioreactors. These results thus show a critical role for CYLD in angiogenesis in vivo.
CYLD regulates angiogenesis by mediating cell spreading and migration
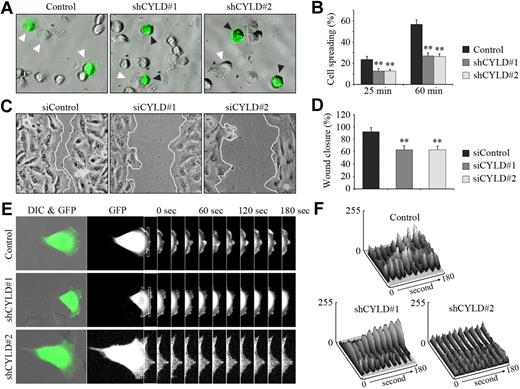
We next sought to investigate the molecular mechanisms underlying the function of CYLD in angiogenesis. We first examined the effect of CYLD knockdown on vascular endothelial cell spreading, an important early step in angiogenesis.3 We found that shRNA-mediated knockdown of CYLD expression significantly impaired the spreading of HUVECs on matrigel compared with the control group (Figure 3A). Quantitative analysis showed that CYLD shRNAs decreased cell spreading by 47% and 53%, respectively, 25 and 60 minutes after plating (Figure 3B). In vitro wound healing assay further showed that knockdown of CYLD expression decreased the migration of HUVECs, as evidenced by delayed wound closure (Figure 3C-D). These data suggest that CYLD regulates angiogenesis by mediating the spreading and migration of vascular endothelial cells.
CYLD regulates angiogenesis by mediating cell spreading and migration. (A) HUVECs transfected with pEGFPC1 and pSUPER or pSUPER-CYLD plasmids for 72 hours were plated onto matrigel, and photographs were taken 25 minutes later. White arrowheads indicate cells spreading on the plates, and black arrowheads indicate cells without such a property. Objective lens used was A-Plan 20×/0.3 NA dry (Carl Zeiss Inc). (B) Experiments were performed as in panel A, and the degree of cell spreading was quantified 25 and 60 minutes after plating. Data are the mean and SE from 2 experiments (600 transfected cells were measured for each experiment; **P < .01 vs control). (C) HUVECs transfected with CYLD or control siRNAs for 72 hours were scratched, and wound margins were imaged 12 hours later. Objective lens used was A-Plan 20×/0.3 NA dry (Carl Zeiss Inc). (D) Experiments were performed as in panel C, and the extent of wound closure was quantified by measuring the wound area compared with the initial wound area. Data are the mean and SE from 2 experiments with each performed in triplicate (**P < .01 vs control). (E) HUVECs were transfected with pEGFPC1 and pSUPER or pSUPER-CYLD plasmids, and the fluorescence of GFP at the leading edge of cells was recorded at 20-second intervals with the use of the TCS SP5 confocal microscope (Leica), equipped with a live-cell imaging workstation and the LAS AF software. Rectangular regions were selected as indicated to analyze membrane ruffle dynamics. Objective lens used was HCX Plan-Apochromat 20×/0.7 NA dry (Leica). (F) Experiments were performed as in panel E, and membrane ruffle dynamics were presented as 3-dimensional surface plots.
CYLD regulates angiogenesis by mediating cell spreading and migration. (A) HUVECs transfected with pEGFPC1 and pSUPER or pSUPER-CYLD plasmids for 72 hours were plated onto matrigel, and photographs were taken 25 minutes later. White arrowheads indicate cells spreading on the plates, and black arrowheads indicate cells without such a property. Objective lens used was A-Plan 20×/0.3 NA dry (Carl Zeiss Inc). (B) Experiments were performed as in panel A, and the degree of cell spreading was quantified 25 and 60 minutes after plating. Data are the mean and SE from 2 experiments (600 transfected cells were measured for each experiment; **P < .01 vs control). (C) HUVECs transfected with CYLD or control siRNAs for 72 hours were scratched, and wound margins were imaged 12 hours later. Objective lens used was A-Plan 20×/0.3 NA dry (Carl Zeiss Inc). (D) Experiments were performed as in panel C, and the extent of wound closure was quantified by measuring the wound area compared with the initial wound area. Data are the mean and SE from 2 experiments with each performed in triplicate (**P < .01 vs control). (E) HUVECs were transfected with pEGFPC1 and pSUPER or pSUPER-CYLD plasmids, and the fluorescence of GFP at the leading edge of cells was recorded at 20-second intervals with the use of the TCS SP5 confocal microscope (Leica), equipped with a live-cell imaging workstation and the LAS AF software. Rectangular regions were selected as indicated to analyze membrane ruffle dynamics. Objective lens used was HCX Plan-Apochromat 20×/0.7 NA dry (Leica). (F) Experiments were performed as in panel E, and membrane ruffle dynamics were presented as 3-dimensional surface plots.
To better understand the action of CYLD in cell migration, we examined membrane ruffling at the leading edge of cells, which reflects an active state of cell protrusion during migration.14 HUVECs were transfected with pEGFPC1 and pSUPER-CYLD plasmids, and GFP fluorescence at the leading edge was recorded at 20-second intervals as a measure of membrane ruffle dynamics (Figure 3E). Three-dimensional surface plots depicting dynamic changes in GFP fluorescence were generated from the sequential images. As shown in Figure 3F, knockdown of CYLD expression significantly inhibited the dynamic changes in GFP fluorescence at the leading edge of migrating HUVECs, indicating diminished membrane ruffling. These results provide further evidence for the involvement of CYLD in vascular endothelial cell migration.
CYLD modulates microtubule dynamics and cell polarization in migrating cells
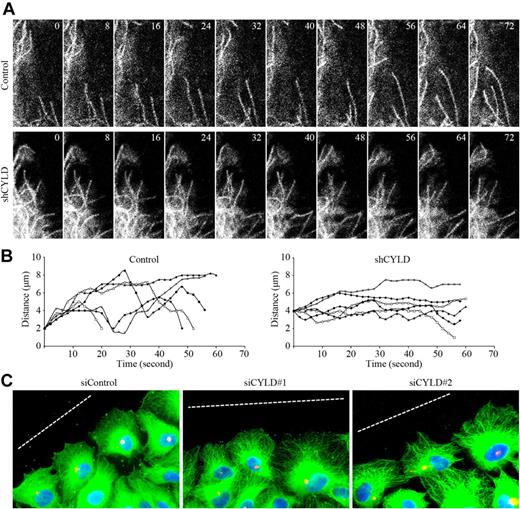
On the basis of the finding that both the microtubule-binding and migration-regulatory activities of CYLD are mediated by its first CAP-Gly domain,9 we hypothesized that CYLD might regulate the migration of vascular endothelial cells through modulation of microtubule dynamics. To test this hypothesis, HUVECs were transfected with pEGFPC1–α-tubulin and pSUPER-CYLD, and the dynamics of individual GFP-labeled microtubules in migrating cells were recorded (Figure 4A). Individual microtubule life history plots representing changes in microtubule length over time were generated. As shown in Figure 4B, knockdown of CYLD expression remarkably decreased the amount of time that microtubules spent in growth and shorting and increased the time that microtubules spent in a pause state.
CYLD modulates microtubule dynamics and cell polarization in migrating cells. (A) HUVECs were transfected with pEGFPC1–α-tubulin and pSUPER or pSUPER-CYLD, and the dynamics of individual GFP-labeled microtubules in migrating cells were then recorded with the TCS SP5 confocal microscope (Leica), equipped with a live-cell imaging workstation and the LAS AF software. Objective lens used was HCX Plan-Apochromat 63×/1.4 NA oil (Leica). (B) Experiments were performed as in panel A, and individual microtubule life history plots representing changes in microtubule length over time were generated. Growth events exhibit an increase in distance from a fixed point (y-axis) over time (x-axis), and shortening events are seen as a decrease in distance over time. (C) HUVECs transfected with CYLD or control siRNAs were scratched, and cells were fixed 2 hours later and stained with anti–α-tubulin antibody, anti–pericentrin antibody, and DAPI to visualize microtubules (green), centrosomes (red), and nuclei (blue), respectively. Broken white lines indicate the wound direction. Objective lens used was Plan-Apochromat 63×/1.25 NA oil (Carl Zeiss Inc).
CYLD modulates microtubule dynamics and cell polarization in migrating cells. (A) HUVECs were transfected with pEGFPC1–α-tubulin and pSUPER or pSUPER-CYLD, and the dynamics of individual GFP-labeled microtubules in migrating cells were then recorded with the TCS SP5 confocal microscope (Leica), equipped with a live-cell imaging workstation and the LAS AF software. Objective lens used was HCX Plan-Apochromat 63×/1.4 NA oil (Leica). (B) Experiments were performed as in panel A, and individual microtubule life history plots representing changes in microtubule length over time were generated. Growth events exhibit an increase in distance from a fixed point (y-axis) over time (x-axis), and shortening events are seen as a decrease in distance over time. (C) HUVECs transfected with CYLD or control siRNAs were scratched, and cells were fixed 2 hours later and stained with anti–α-tubulin antibody, anti–pericentrin antibody, and DAPI to visualize microtubules (green), centrosomes (red), and nuclei (blue), respectively. Broken white lines indicate the wound direction. Objective lens used was Plan-Apochromat 63×/1.25 NA oil (Carl Zeiss Inc).
The finding that CYLD regulates microtubule dynamics in migrating HUVECs suggests that this protein might function in cell polarization, a critical step in cell migration that involves the rearrangement of microtubules and reorientation of the centrosome toward the leading edge of cells.3,4 To investigate this possibility, HUVECs transfected with CYLD or control siRNAs were scratched, and cells were fixed 2 hours later and immunostained to visualize microtubules, centrosomes, and nuclei. As shown in Figure 4C, in the control group, cells at the wound margin exhibited a typical polarized structure with centrosomes localized between the nuclei and the leading edge. In contrast, cells transfected with CYLD siRNAs had polarization defects with centrosomes randomly localized. Collectively, these data show that CYLD modulates microtubule dynamics and cell polarization in migrating vascular endothelial cells.
Rac1 activation contributes to the effects of CYLD on endothelial cell migration and tube formation
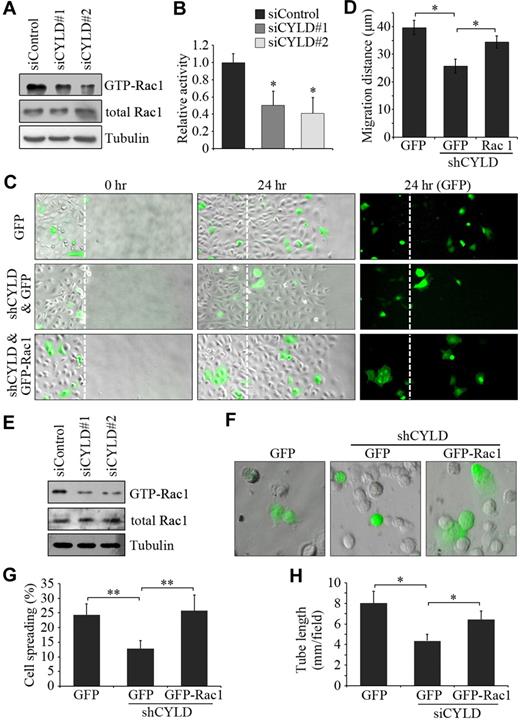
To gain more mechanistic insight into how CYLD mediates endothelial cell migration, we examined the activities of the Rhofamily GTPases Rac1, RhoA, and Cdc42, which are known to participate in cell polarization and migration primarily through interplay with the cytoskeleton.6,8 HUVECs transfected with CYLD and control siRNAs were scratched, and GST-PBD pulldown assays were then performed. CYLD siRNAs were found to significantly reduce Rac1 activity in HUVECs compared with the control group (Figure 5A-B). In contrast, the activities of RhoA and Cdc42 were not obviously affected by CYLD siRNAs (data not shown). To examine whether Rac1 activation contributes to the action of CYLD in endothelial cell migration, HUVECs were transfected with pSUPER-CYLD together with pEGFPC1-Rac1, and wound healing assay was then performed. We found that overexpression of GFP-Rac1 could remarkably rescue CYLD shRNA-induced migration defects (Figure 5C-D), indicating that Rac1 activation underlies the effect of CYLD on endothelial cell migration.
Rac1 activation contributes to the effects of CYLD on vascular endothelial cell migration and tube formation. (A) HUVECs transfected with CYLD or control siRNAs were scratched, and the level of activated (GTP-bound) Rac1 was examined by immunoblot analysis of the GST-PBD pulldown preparation with anti-Rac1 antibody. The levels of total Rac1 and tubulin were examined by immunoblot analysis of cell lysates. (B) Experiments were performed as in panel A, and the relative activity of Rac1 was measured by densitometric analysis of the blots. Data are the mean and SE from 3 experiments (*P < .05 vs control). (C) HUVECs transfected with pSUPER-CYLD and pEGFPC1-Rac1 or pEGFPC1 were scratched, and wound margins were imaged 0 and 24 hours later. Broken white lines indicate initial wound margins. Objective lens used was A-Plan 20×/0.3 NA dry (Carl Zeiss Inc). (D) Experiments were performed as in panel C, and the average migration distance of the transfected cells from the wound margin was measured. Data are the mean and SE from 2 experiments (500 transfected cells were measured for each experiment; *P < .05). (E) HUVECs transfected with CYLD or control siRNAs were plated onto matrigel, and the levels of GTP-Rac1, total Rac1, and tubulin were examined 2 hours later as in panel A. (F) HUVECs transfected with pSUPER-CYLD and pEGFPC1-Rac1 or pEGFPC1 were plated onto matrigel, and photographs were taken 25 minutes later. Objective lens used was A-Plan 20×/0.3 NA dry (Carl Zeiss Inc). (G) Experiments were performed as in panel F, and the degree of cell spreading was quantified. Data are the mean and SE from 2 experiments (600 transfected cells were measured for each experiment; **P < .01). (H) HUVECs transfected with CYLD siRNA and pEGFPC1-Rac1 or pEGFPC1 were plated onto matrigel, and the cumulative tube length was measured 4 hours later. Data are the mean and SE from 2 experiments with each performed in triplicate (*P < .05).
Rac1 activation contributes to the effects of CYLD on vascular endothelial cell migration and tube formation. (A) HUVECs transfected with CYLD or control siRNAs were scratched, and the level of activated (GTP-bound) Rac1 was examined by immunoblot analysis of the GST-PBD pulldown preparation with anti-Rac1 antibody. The levels of total Rac1 and tubulin were examined by immunoblot analysis of cell lysates. (B) Experiments were performed as in panel A, and the relative activity of Rac1 was measured by densitometric analysis of the blots. Data are the mean and SE from 3 experiments (*P < .05 vs control). (C) HUVECs transfected with pSUPER-CYLD and pEGFPC1-Rac1 or pEGFPC1 were scratched, and wound margins were imaged 0 and 24 hours later. Broken white lines indicate initial wound margins. Objective lens used was A-Plan 20×/0.3 NA dry (Carl Zeiss Inc). (D) Experiments were performed as in panel C, and the average migration distance of the transfected cells from the wound margin was measured. Data are the mean and SE from 2 experiments (500 transfected cells were measured for each experiment; *P < .05). (E) HUVECs transfected with CYLD or control siRNAs were plated onto matrigel, and the levels of GTP-Rac1, total Rac1, and tubulin were examined 2 hours later as in panel A. (F) HUVECs transfected with pSUPER-CYLD and pEGFPC1-Rac1 or pEGFPC1 were plated onto matrigel, and photographs were taken 25 minutes later. Objective lens used was A-Plan 20×/0.3 NA dry (Carl Zeiss Inc). (G) Experiments were performed as in panel F, and the degree of cell spreading was quantified. Data are the mean and SE from 2 experiments (600 transfected cells were measured for each experiment; **P < .01). (H) HUVECs transfected with CYLD siRNA and pEGFPC1-Rac1 or pEGFPC1 were plated onto matrigel, and the cumulative tube length was measured 4 hours later. Data are the mean and SE from 2 experiments with each performed in triplicate (*P < .05).
Next, we investigated whether Rac1 activation also contributes to the action of CYLD in angiogenesis. GST-PBD pulldown assay showed that CYLD siRNAs decreased Rac1 activity in HUVECs plated on matrigel (Figure 5E). Importantly, overexpression of GFP-Rac1 significantly rescued the inhibitory effect of CYLD shRNA on endothelial cell spreading on matrigel (Figure 5F-G). In addition, overexpression of GFP-Rac1 was able to rescue the inhibitory effect of CYLD siRNA on tube formation in vitro from HUVECs (Figure 5H). Taken together, these data suggest that Rac1 activation is critically involved in the action of CYLD in endothelial cell migration and angiogenesis.
Discussion
CYLD is best characterized as a tumor suppressor in skin appendage tumors.15,16 It possesses deubiquitinase activity in its carboxyl terminus that negatively regulates the nuclear factor-κB pathway by removing lysine 63–linked polyubiquitin chains from upstream signaling molecules, and this action of CYLD is believed to contribute to its tumor suppressor function.17,–19 In the past years, CYLD has increasingly been implicated in normal physiologic processes, such as immune response and inflammation, osteoclastogenesis, cell cycle progression, and cell migration,9,10,20,,,–24 although the precise molecular mechanisms remain to be elucidated. In this study, we provide the first evidence showing a role for CYLD in angiogenesis. Our results show that CYLD is critical for endothelial tube formation and capillary sprouting in vitro and for angiogenesis in vivo. These findings thus add CYLD to the growing list of proteins that regulate the angiogenic process.
Using gene silencing approaches, we have found that CYLD regulates angiogenesis by mediating vascular endothelial cell migration. It has been shown previously that siRNAs are able to activate the interferon system in certain cell types.25,26 In this study, by quantitative real-time reverse transcription–polymerase chain reaction analyses, we find that the expression of interferon-α, -β, and -γ in HUVECs is not significantly affected by either CYLD or control siRNAs, although a significant increase in interferon-β expression is observed in T98G and SH-SY5Y neuroblastoma cells on treatment with CYLD or control siRNAs (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). These data are consistent with the previous finding that siRNA-induced interferon response is cell type dependent.27 Nevertheless, no significant difference in interferon expression is observed between the control siRNA group and the CYLD siRNA groups in HUVECs, T98G cells, or SH-SY5Y cells (supplemental Figure 1). Collectively, these data suggest that the inhibitory effects of CYLD siRNAs on endothelial cells are not due to an interferon effect.
Note that overexpression of CYLD in HUVECs does not obviously promote cell migration or tube formation in vitro but results in apoptosis (data not shown), implicating that angiogenesis requires an exquisite control of CYLD expression in vascular endothelial cells. Interestingly, CYLD-deficient mice do not show overt phenotype and develop normally,24 suggesting that this protein does not play a significant role in normal embryonic angiogenesis. Instead, given that CYLD-deficient mice are more susceptible to chemical-induced tumors and pathogen-induced inflammation,24,28,–30 it is tempting to speculate that CYLD may function in angiogenesis primarily under pathologic conditions such as wound healing.
The microtubule cytoskeleton is known to undergo dramatic rearrangement in response to signals that stimulate cell migration, and such a property of microtubules relies on precise control of microtubule dynamics in cells.6 In this scenario, it is not difficult to understand that suppression of microtubule dynamics by knockdown of CYLD expression significantly impairs vascular endothelial cell migration. Our previous studies have shown that purified CYLD interacts, through its first CAP-Gly domain, with microtubules assembled from microtubule-associated protein-free tubulin.9 In addition, CYLD affects tubulin polymerization into microtubules in the purified system.9 These findings suggest that CYLD probably regulates microtubule dynamics in migrating cells through direct interaction. However, because CYLD associates with a great number of proteins in cells,16,23 it would not be surprising if an indirect mechanism were identified in the future for its regulation of microtubule dynamics.
Our finding that Rac1 activation contributes to the effects of CYLD on endothelial cell migration and angiogenesis is very intriguing. Given the interplay between Rac1 activity and microtubules in migrating cells,6,8 it is conceivable that the action of CYLD in Rac1 activation might be an event downstream of the modulation of microtubule dynamics by CYLD. Alternatively, CYLD may deubiquitinate Rac1 and thereby promote Rac1 activation to enable efficient cell migration. This notion is supported by recent observations that Rac1 activity is regulated by ubiquitination.31,32 It will be important to investigate in the future whether the deubiquitinase activity of CYLD is required for its function in endothelial cell migration and angiogenesis and whether lysine 63–linked ubiquitination is involved in the regulation of microtubule dynamics and Rac1 activation during cell migration.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs George Mosialos, Rene Bernards, and Xueliang Zhu for providing reagents, Ms Ruming Liu for technical assistance, and members of the Zhou laboratory for stimulating discussions.
This work was supported by the National Natural Science Foundation of China (grants 30600313 and 30825022), the Tianjin Natural Science Foundation (grant 07JCZDJC03000), and the National Basic Research Program of China (grant 2007CB914503).
Authorship
Contribution: J.G. performed research, analyzed data, and wrote the paper; L.S., L.H., M.L., and D.L. performed research; and J.Z. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jun Zhou, Department of Genetics and Cell Biology, College of Life Sciences, Nankai University, 94 Weijin Rd, Tianjin 300071, China; e-mail: junzhou@nankai.edu.cn.