Abstract
The 46/1 JAK2 haplotype predisposes to V617F-positive myeloproliferative neoplasms, but the underlying mechanism is obscure. We analyzed essential thrombocythemia patients entered into the PT-1 studies and, as expected, found that 46/1 was overrepresented in V617F-positive cases (n = 404) versus controls (n = 1492, P = 3.9 × 10−11). The 46/1 haplotype was also overrepresented in cases without V617F (n = 347, P = .009), with an excess seen for both MPL exon 10 mutated and V617F, MPL exon 10 nonmutated cases. Analysis of further MPL-positive, V617F-negative cases confirmed an excess of 46/1 (n = 176, P = .002), but no association between MPL mutations and MPL haplotype was seen. An excess of 46/1 was also seen in JAK2 exon 12 mutated cases (n = 69, P = .002), and these mutations preferentially arose on the 46/1 chromosome (P = .029). No association between 46/1 and clinical or laboratory features was seen in the PT-1 cohort either with or without V617F. The excess of 46/1 in JAK2 exon 12 cases is compatible with both the “hypermutability” and “fertile ground” hypotheses, but the excess in MPL-mutated cases argues against the former. No difference in sequence, splicing, or expression of JAK2 was found on 46/1 compared with other haplotypes, suggesting that any functional difference of JAK2 on 46/1, if it exists, must be relatively subtle.
Introduction
Myeloproliferative neoplasms (MPNs) are frequently characterized by deregulated tyrosine kinase signaling, particularly pathways involving JAK2. By far the most common mutation is V617F JAK2, which is seen in more than 95% of cases of polycythemia vera (PV) and 50% to 60% of essential thrombocythemia (ET) and primary myelofibrosis (PMF).1 In addition, diverse mutations involving JAK2 exon 12 have been identified in V617F-negative PV2 and abnormalities affecting W515 or, less commonly, S505 of the thrombopoietin receptor MPL (which signals though JAK2) are found in up to 10% of cases with ET or PMF.3-6
Recently, we and others made the unexpected finding that V617F JAK2 is not acquired randomly but instead arises preferentially on a specific constitutional JAK2 haplotype, at least in white populations.7-9 This haplotype, which we refer to as 46/17 but others have called GGCC,8 is found in approximately 50% of normal persons and thus is a common, very low penetrance predisposition allele of the type that is typically detected by genome-wide association studies. We estimated that 46/1 accounts for 50% of the population attributable risk of developing an MPN described in epidemiologic studies, but it does not account for familial MPNs, which are expected to be caused by rare, highly penetrant variants in genes that remain to be identified.10 Furthermore, 46/1 was seen at comparable frequencies in different MPN subtypes and thus does not explain the phenotypic diversity associated with V617F JAK2.7
It is not clear why an acquired mutation as prevalent as V617F is associated with a particular inherited background, but 2 hypotheses have been suggested.7,8,11 First, 46/1 may be inherently more genetically unstable, acquiring V617F at a faster rate than other haplotypes (hypermutability hypothesis). Second, V617F may arise on all haplotypes at equal rates, but 46/1 may carry an additional factor that either gives a selective advantage to the V617F-positive clone or interacts in some way to increase the likelihood of abnormal blood counts (fertile ground hypothesis). We aimed to explore these hypotheses by detailed genetic analysis of clinically defined patient cohorts.
Methods
Patients and samples
The Primary Thrombocythemia 1 (PT-1) study included newly diagnosed and previously treated patients, 18 years of age or older, who met the Polycythemia Vera Study Group criteria for ET. Patients were recruited into 1 of 3 multicenter studies: the Medical Research Council high-risk trial, in which high-risk patients were randomly assigned to either hydroxyurea plus aspirin or to anagrelide plus aspirin; the National Cancer Research Institute intermediate-risk study, a randomization between aspirin alone or hydroxyurea plus aspirin; or the National Cancer Research Institute low-risk study, a prospective observational study of low-risk patients given aspirin alone.12 Details obtained at trial entry included diagnostic features, such as blood counts, cytogenetics, and clinical complications at or preceding diagnosis. Samples of peripheral blood were requested at trial entry from all patients, and 776 samples were received from which DNA was extracted. These samples have been analyzed previously for V617F JAK2 and MPL mutations.5,13
Other patients who met the Polycythemia Vera Study Group or World Health Organization criteria for ET or MF were recruited from the United Kingdom, Germany, Italy, Greece, and the United States. Patients were tested for JAK2 V617F, MPL exon 10, and, if appropriate, JAK2 exon 12 mutations using allele-specific (AS) polymerase chain reaction (PCR), pyrosequencing, or high resolution melt analysis according to local procedures using DNA extracted from purified granulocytes or peripheral blood leukocytes. As controls, we used data generated from 3 population-based studies: the Wellcome Trust Case Control Consortium (WTCCC) United Kingdom blood donor cohort (n = 1500),14 the German Kooperative Gesundheitsforschung in der Augsburg cohort (KORA; n = 1814),15 and the Italian Invecchiare in Chianti cohort (InCHIANTI; n = 1200).16 We also analyzed healthy control persons from Greece (n = 108). The study was approved by the internal review boards and/or ethics committees of all participating institutions, and informed consent was provided according to the Declaration of Helsinki.
Genotyping
Quantitative genotyping assays for the 46/1 tag single nucleotide polymorphism (SNP) rs12340895, JAK2 V617F, and MPL W515K/L by pyrosequencing have been described previously.5,17 Primers for other SNPs are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). JAK2 V617F and MPL W515K/L mutations were scored as homozygous if the proportion of the mutant allele was greater than 50%, the maximum expected if a heterozygous mutant clone had expanded to include all cells in the sample. We used previously defined criteria to score homozygosity and heterozygosity for flanking SNPs in the context of skewed allele ratios brought about by acquired uniparental disomy at 1p or 9p.7 AS PCR between V617F and rs12343867 was performed as described to determine whether the V617F JAK2 mutation was in cis or trans to 46/1.
AS PCR for JAK2 exon 12 mutations
AS PCR was performed using a forward primer located in JAK2 intron 11 (JAK2_intron11_F) in combination with 1 of 2 reverse primers specific for a 46/1 tagging 5-bp insertion/deletion polymorphism rs56241661 (primers plus_TCTTA_R and del_TCTTA_R). The 398-bp products harboring the whole of JAK2 exon 12 were directly sequenced to determine whether the exon 12 mutation was present on a 46/1 or non-46/1 allele. Amplification conditions were optimized on DNA from a case carrying the H538_K539delinsL mutation and normal healthy controls: the PCR contained 25 ng of DNA, 0.5μM JAK2_intron11_F, and either plus_TCTTA_R or del_TCTTA_R, 0.5mM dNTPs, 25mM magnesium chloride, 10× AmpliTaq buffer (Applied Biosystems), 1 U of AmpliTaq Gold (Applied Biosystems), and distilled water to a final volume of 25 μL. PCR was performed on a Tetrad thermocycler (Bio-Rad), and the cycling conditions were 95°C for 15 seconds, and 34 cycles of 95°C for 30 seconds, 56°C for 45 seconds, 72°C for 45 seconds, followed by 72°C for 10 minutes. In cases where the exon 12 mutation was at low level and not clearly visible after AS PCR and sequencing, amplification of the wild-type alleles was blocked using a locked nucleic acid-oligonucleotide probe during PCR, thus facilitating the preferential amplification of mutant alleles. This PCR composed 25 ng DNA, 0.5μM JAK2_intron11_F, and either plus_TCTTA_R or del_TCTTA_R, 1μM JAK2_ex12_locked nucleic acid_oligo (Sigma-Aldrich), 0.5mM dNTPs, 75mM magnesium chloride, 10× Stoffel buffer (Applied Biosystems), 1 U of AmpliTaq DNA polymerase Stoffel fragment (Applied Biosystems), and distilled water to a final volume of 25 μL. The cycling conditions remained the same.
Genetic analysis
The LDMAP program18 was used to construct linkage disequilibrium (LD) maps measured in LD units for the entire genome using phase 2 HapMap data from the Centre d'Etude du Polymorphisme Humaine population19 and genomic locations relative to NCBI build 36.1 of the human genome sequence. For regions of interest, the PHASE program20 was used to estimate haplotypes from genotypic data derived from the WTCCC blood donor cohort.
Statistical analysis
The proportion of 46/1 alleles within each patient subgroup was compared with corresponding controls using Fisher exact test (2-tailed). To take into account variation in SNP frequency across control populations, meta-analysis was performed using local control populations for each patient group using StatsDirect, Version 2.7.6. Datasets were compared using the features built into this software to confirm that there was no evidence of noncombinability (Breslow-Day and Cochran Q tests). Deviations from expected allelic ratios of 50% were assessed by the exact binomial distribution, and mutation levels were compared between groups by the Mann-Whitney test. Pairwise univariate analyses comparing diagnostic variables and genotype were performed using the t test for continuous variables, Fisher exact test for 2 × 2 tables, and Cochran Armitage test for trend with exact P values for ordinal variables. Clinical outcome was assessed by χ2 test for trend for events preceding diagnosis and log-rank test for events after trial entry. The relationship between 46/1 haplotype, blood counts, and response to treatment was assessed using the linear mixed-effects models.
Results
Characterization of the JAK2 46/1 haplotype in the PT-1 cohort
We genotyped patients from the PT-1 cohort for rs12340895, a tag SNP that acts as a surrogate for 46/1 (G allele = 46/1; C allele = not 46/1).7 Results were obtained from 751 cases and compared initially with mutational status (Table 1). As expected, 46/1 was more frequent in patients compared with controls (P = 1.7 × 10−9), and this was accounted for principally by an excess in cases that were V617F-positive (P = 3.9 × 10−11). The 46/1 haplotype was also overrepresented in cases that tested negative for V617F (P = .025), and a relatively high 46/1 frequency was also seen in cases that were positive for MPL exon 10 variants (W515L, n = 24; W515K, n = 5; S505N, n = 3), although this did not reach statistical significance (P = .06; P = .096 if the 3 cases that were also V617F-positive are excluded).
To confirm our previous finding that V617F probably arises specifically on the 46/1 allele, we performed AS PCR for the JAK2 mutation and sequenced the products to determine the genotype of a nearby 46/1 tag SNP, rs12343867. Of the 212 V617F-positive cases that were heterozygous for 46/1, results were obtained for 207. Of these, 160 (77%) V617F alleles arose on the 46/1 haplotype (P < .001, exact binomial test), whereas only 45 (22%) residual wild-type alleles were 46/1. In addition, there were 2 V617F-positive cases where the V617F mutation was present on both 46/1 and non-46/1 haplotypes, suggesting that V617F arose at least twice in these cases.
No association between 46/1 and clinical features or outcome
If JAK2 on 46/1 is functionally different from JAK2 on other haplotypes, we speculated that 46/1 might be associated with specific disease features. We therefore compared clinical, laboratory, and demographic data with 46/1 genotype (both homozygous and heterozygous) for the PT-1 group as a whole, for V617F-positive and V617F-negative cases considered separately, and also for V617F-positive cases where the mutation was known to be on the 46/1 allele. No significant associations were seen with features at diagnosis, including age, blood counts, hemoglobin, erythropoietin, and spleen size (supplemental Table 2). Furthermore, there was no association with survival, arterial thrombosis, venous thrombosis, major hemorrhage or transformation to myelofibrosis, myelodysplastic syndrome, or acute leukemia (supplemental Table 3). There was an apparent association with transformation from ET to PV; however, the number of cases is extremely small (n = 6); furthermore, we saw no association between haplotype status and changes in hemoglobin, platelet, or white cell counts over time.
Association between 46/1 and MPL W515 mutations
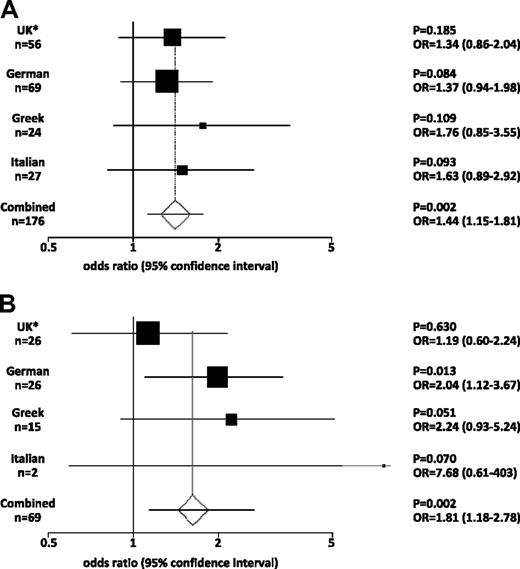
To explore the possible association of 46/1 with MPL mutations, we further investigated an independent cohort of 176 cases with W515L (n = 110), W515K (n = 58), other W515 variants (n = 4), or S505N (n = 4). All 176 cases were specifically selected as being negative for V617F using an assay with a sensitivity of 1% to 2%. As shown in Figure 1 and supplemental Table 4, the frequency of 46/1 as assessed by rs12340895 genotype was higher in MPL-mutated cases compared with controls from the same country for each of the 3 populations. Although these differences did not reach significance for each population considered in isolation, meta-analysis to control for population variation in SNP frequencies demonstrated that 46/1 was indeed more frequent in MPL-mutated cases (P = .002, odds ratio [OR] = 1.44). Even considering the worst case scenario, that is, all MPL-mutated cases compared with the control population with the highest frequency of 46/1 (Italians), the difference is still significant (P = .012). No difference in the frequency of 46/1 was seen on comparison of cases with W515L or W515K. To further confirm that 46/1 predisposes to MPL-mutated MPNs, we analyzed a second 46/1 tag SNP (rs10118390; located 20 kb downstream of JAK2). As shown in supplemental Table 5, this SNP is also significantly overrepresented in MPL-mutated, V617F-negative cases (P < .001; OR = 1.51; n = 176).
Meta-analysis of genotype data. (A) MPL-mutated cases. (B) JAK2 exon 12 mutated cases. The plots show the odds ratio (OR) and 95% confidence interval (CI) within each patient subgroup compared their corresponding population controls. The combined data show the OR and CI derived by meta-analysis of all the data after confirmation of the comparability and homogeneity of the respective datasets. *The United Kingdom MPL and JAK2 exon 12 cohorts include 1 and 2 cases, respectively, of European origin from the United States.
Meta-analysis of genotype data. (A) MPL-mutated cases. (B) JAK2 exon 12 mutated cases. The plots show the odds ratio (OR) and 95% confidence interval (CI) within each patient subgroup compared their corresponding population controls. The combined data show the OR and CI derived by meta-analysis of all the data after confirmation of the comparability and homogeneity of the respective datasets. *The United Kingdom MPL and JAK2 exon 12 cohorts include 1 and 2 cases, respectively, of European origin from the United States.
No association between MPL haplotype and MPL mutations
To investigate the possibility that MPL mutations might be associated with a specific MPL haplotype, we first determined the genetic structure of this locus using the 8 SNPs genotyped by the WTCCC in the blood donor cohort. Analysis using LDMapper and PHASE showed that the entire MPL gene resides in a single LD block, with 4 haplotypes accounting for 88% of alleles (Table 2). These 4 haplotypes could be captured by 3 SNPs (rs1199038, rs11210838, and rs839757), which we analyzed by pyrosequencing in 138 MPL-mutated cases. Because MPL mutations may be affected by acquired uniparental disomy at chromosome 1p,21 we quantified mutation levels in all cases by pyrosequencing and identified 29 cases with evidence of a homozygous clone (mutation level > 50%). We used previously defined criteria to take into account the possibility of SNP allele skewing in these persons but found no difference in the allele frequencies between cases and controls (Table 3).
As we have shown previously for V617F JAK2, the skewing of heterozygous flanking SNP ratios in cases with a homozygous mutant clone make it possible to read directly the haplotype on which the MPL mutation arose, as well as the residual wild-type haplotype.7 Of the 29 cases with a homozygous clone, we were able to assign the mutant allele to 1 of the 4 common haplotypes in 23 cases and the wild-type allele in 21 cases. As shown in Table 2, the mutant allele was not seen preferentially on any haplotype, and there was no significant difference in haplotype distribution on comparison of the mutant and wild-type alleles. Furthermore, the number of mutant and wild-type haplotypes did not differ significantly from the distribution expected from analysis of the WTCCC control cohort. These data therefore are consistent with the hypothesis that MPL mutations arise randomly on different MPL haplotypes.
Higher MPL mutation loads in MF
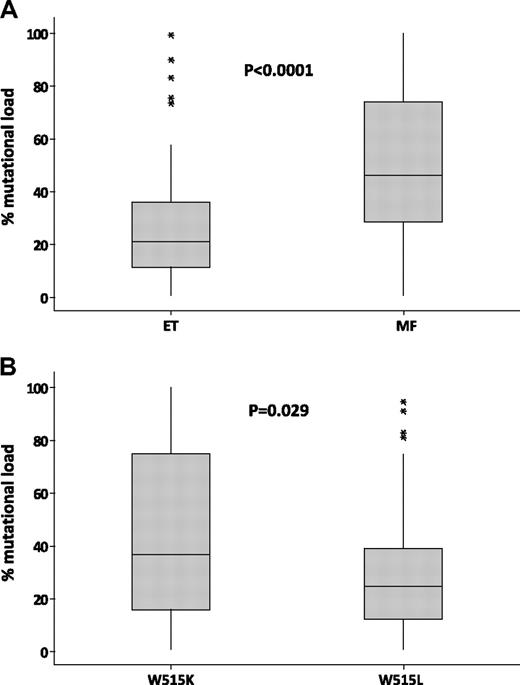
Having determined the MPL mutation load in W515K/L cases, we compared the results to clinical phenotype. Of the 138 cases (ET, n = 99; MF, n = 36; other MPN, n = 3), the median W515K/L mutation levels in ET (21%) were significantly lower than those seen in MF (46%; P < .001; Figure 2A). The 29 homozygous cases had a diagnosis of primary or secondary MF (n = 15), ET (n = 12), or accelerated/transformed MPN (n = 2); 17 cases had W515L and 12 had W515K. Overall, mutation levels in W515L cases (n = 106; median = 25%) were lower than those seen in cases with W515K (Figure 2B; n = 32; median = 37%; P = .029), confirming our previous observations in a much smaller number of cases.5,22,23
Higher MPL mutation loads in MF and cases with W515K. Box-and-whisker plots comparing the percentage of MPL W515K/L alleles in essential thrombocythemia (ET) versus myelofibrosis (MF; A) and in cases with W515K versus those with W515L (B). P values were obtained by Mann-Whitney analysis. *Outliers.
Higher MPL mutation loads in MF and cases with W515K. Box-and-whisker plots comparing the percentage of MPL W515K/L alleles in essential thrombocythemia (ET) versus myelofibrosis (MF; A) and in cases with W515K versus those with W515L (B). P values were obtained by Mann-Whitney analysis. *Outliers.
The 46/1 predisposes to JAK2 exon 12 mutations
To determine whether the 46/1 haplotype predisposes to acquisition of other MPN-associated mutations, we investigated its association with mutations in exon 12. A total of 69 cases with PV that tested positive for an exon 12 mutation but negative for V617F were genotyped for rs12340895. Cases were from Italy (n = 2), Germany (n = 26), Greece (n = 15), and the United Kingdom/United States (n = 26); and, as performed for MPL, each group was compared with local control populations (Figure 1B; supplemental Table 6). Of the 138 alleles, 54 (39%) were 46/1, and meta-analysis indicated that this was significantly higher than controls (P = .002, OR = 1.81).
Of the 69 cases, 6 were homozygous for 46/1 and 42 were heterozygous. For the heterozygous cases, we investigated whether JAK2 exon 12 mutations arose in cis or trans to the 46/1 haplotype using AS PCR. To do this, we exploited a 5-bp insertion/deletion polymorphism (rs56241661) downstream of exon 12 that we found to be tightly linked to 46/1 (supplemental Figure 1). Of the 31 informative cases, 22 (71%) exon 12 mutations arose on the 46/1 haplotype, whereas only 9 (29%) residual wild-type alleles were 46/1 (P = .029).
No differences between JAK2 on 46/1 compared with other haplotypes
We have previously demonstrated that the mRNA expression level of JAK2 on 46/1 is no different from the expression of JAK2 on other haplotypes, at least in peripheral blood leukocytes. To determine whether JAK2 on 46/1 might be functionally different, we sequenced all exons, including those encoding the 5′ and 3′ untranslated regions, in 8 MPN cases with V617F on the 46/1 allele, 2 MPN with exon 12 mutations, and 8 healthy controls that were negative for 46/1. No sequence variants were detected, except for the 2 previously described silent polymorphisms rs10429491 and/or rs2230724 in exons 6 and 19, respectively. To determine whether these exonic SNPs or other unknown intronic variants might affect splicing, we performed overlapping reverse-transcriptase PCR to cover all exon-exon junctions. Sequencing of the products did not reveal any differences between JAK2 mRNA in MPNs with V617F on 46/1 (n = 8) and controls without 46/1 (n = 8).
Role of other inherited polymorphic variants
Previous targeted and genome-wide analysis has identified other loci that may be associated with the phenotypic diversity of MPNs. Pardanani et al24 found that an SNP in the EPOR gene (rs318699) was more common in PV than controls, whereas Kilpivaara et al9 identified 3 variants (rs1524395 at 7p11, rs2279784 at 3q21, and rs12500918 at 4q31) that were more common in PV compared with ET. We sought to determine whether these associations were reproducible by analysis of these 4 SNPs in ET (n = 763), PV (n = 163), and controls. As shown in Table 4, the frequency of the EPOR SNP rs318699 was lower in PV (P = .025; OR = 0.75; 95% confidence interval [CI] = 0.59-0.96) but not ET compared with controls. No significant difference between the frequency of rs12500918 (4q31) or rs1524395 (7p11) was seen on comparison of PV and ET or either disorder and controls, although the frequency in ET versus control approached significance for rs12500918. For rs2279784 (3q21), more than 10% of the WTCCC controls were scored as failures, suggesting a genotyping problem for this SNP. We therefore determined the frequency in 224 healthy controls, which was no different from that seen in PV and ET.
Discussion
The finding that the V617F JAK2 mutation is acquired preferentially on a specific constitutional JAK2 haplotype was unexpected, and the mechanism underlying this observation remains unexplained. Two hypotheses have been postulated: (1) “hypermutability” of JAK2 on 46/1 compared with other haplotypes and (2) a functional difference of JAK2 on 46/1 that positively interacts with V617F and thus provides “fertile ground” for development of an MPN. Data from our current study give some support to both these hypotheses, but neither is able to explain all the available facts.
Hypermutability of JAK2 on 46/1 was proposed initially by Olcaydu et al8 because of their finding that V617F was seen in cis to both alleles of a nearby heterozygous SNP in 3 of 109 (3%) MPNs. In this study, we made the same observation in 2 of 207 (1%) ET cases; furthermore, occasional cases with both JAK2 V617F and JAK2 exon 12 mutations have been reported.25 Because the acquisition of V617F is a rare event and the probability of an exon 12 mutation is considerably less, the chance of acquiring 2 completely independent JAK2 mutations is miniscule. Although the fertile ground hypothesis cannot explain multiple occurrences of JAK2 mutations, there are alternative explanations that have some supporting evidence. First, other acquired or inherited genetic variants may predispose to the acquisition of JAK2 mutations, thus greatly increasing the probability of 2 separate mutations in the same person. An enhanced propensity to acquire JAK2 mutations is already accepted in the context of familial MPNs where multiple affected persons independently acquire V617F as a consequence of inheriting an as yet uncharacterized, high penetrance genetic variant.26,27 Although documented MPN families are relatively uncommon, a recent study estimated that as many as 8% of cases have a family history,28 and thus a substantial minority of affected persons may have an elevated, inherited propensity to acquire multiple JAK2 mutations. Second, V617F JAK2 has been reported to increase the rate of homologous recombination as well as the mutation rate.29 It is conceivable that the observed linkage of JAK2 to 2 SNP alleles might be explained by an initial acquisition of V617F followed by a recombination event between the mutation and the flanking SNP. Although this seems implausible over a distance of only 410 bp, it is striking that a recent study found that V617F appeared to have been acquired at least twice in 10 of 11 (91%) ET cases.30 This unexpected finding has not been independently confirmed and appears to be at odds with the fact that mitotic recombination (and thus homozygosity for V617F) is rare in ET; however, the use of an SNP that is 8 kb distal to V617F might have afforded a greater opportunity for recombination between the 2 markers.
We initially favored the fertile ground hypothesis because of the observation that 46/1 is associated with lower numbers of myeloid colony-forming units in normal persons7 and also because a 46/1 tag SNP showed robust association with Crohn disease,31 a nonmalignant disorder that is thought to have an inflammatory cause.32 Genome-wide association studies in Crohn disease also detected significant associations with genes encoding the IL-23 receptor and STAT3, thereby strongly implicating functional differences in the IL23/JAK2/STAT3 pathway in the pathogenesis of this disorder. In this study, we demonstrated that both JAK2 exon 12 and MPL mutations are associated with 46/1. The association with JAK2 exon 12 mutations confirms the findings reported by Olcaydu et al,33 and is compatible with both the fertile ground and hypermutability hypotheses. The finding of an excess of 46/1 alleles in MPL-mutated cases is difficult to explain by hypermutability of JAK2. Set against the fertile ground hypothesis, however, we did not detect any association with 46/1 and hematologic or clinical parameters, either with or without V617F. Furthermore, we found no expression difference in peripheral blood leukocytes or sequence difference of JAK2 on 46/1 compared with other haplotypes.
It was not known whether the association of JAK2 mutations with 46/1 reflects something unusual about the JAK2 locus or whether acquired driver mutations in cancer usually arise on specific inherited haplotypes. We therefore tested whether there was any association between MPL exon 10 mutations and MPL haplotype but found that the mutations appeared to arise randomly. Although this does not preclude the possibility that somatic mutations in other genes might occur preferentially on particular haplotypes, it does indicate that this is not a general phenomenon.
The mechanism by which V617F JAK2 is associated with diverse clinical phenotypes remains unexplained. Increased dosage of V617F is associated with PV as opposed to ET in both patients and mouse models34-36 ; however, this clearly does not completely explain the 2 phenotypes because many patients with PV have levels of V617F consistent with a predominant heterozygous clone (although colony analysis usually reveals the presence of both heterozygous and homozygous clones in these cases).36 In a small genome-wide association study, the frequency of 3 SNPs at 3q21, 4q31, and 7p11 was reported to differ between PV and ET, suggesting that they might contribute to phenotypic diversity.9 We were unable to replicate these findings in a larger series of cases, suggesting that these associations were either spurious or much weaker than originally proposed. In an analysis of candidate genes, Pardanani et al reported that the allele frequency of rs318699, an SNP close to the EPOR gene at 19p13, differed between PV (but not ET) and controls, suggesting that it may specifically predispose to PV.24 We also found a small but significant difference between PV, but not ET, and controls (P = .025); however, in our study, the minor allele frequency was reduced in PV, whereas Pardanani et al24 found that it was increased. We therefore consider that the relevance of this SNP is questionable.
In conclusion, our data provide some evidence that argues against the fertile ground hypothesis and some that argues against hypermutability. If a functional difference does exist, it must be very subtle, for example, variation in expression of JAK2 in a hemopoietic precursor cell. A subtle functional difference would also be consistent with the observation that 46/1 is not present in approximately 25% of JAK2-mutated MPNs,7,8 indicating that this haplotype is not required for disease development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Tim Frayling and Andy Wood (Peninsular College of Medicine & Dentistry, Exeter, United Kingdom) for helpful advice. Data from the InCHIANTI cohort were kindly provided by Dr Yuri Milaneschi, InCHIANTI Study Group, Florence, Italy. A full list of the investigators who contributed to the generation of the WTCCC data is available from www.wtccc.org.uk, funding for which was provided by the Wellcome Trust (award 07611).
This work was supported by Leukaemia Research (United Kingdom) Specialist Program (grant 0280). R.T.S. was supported by the Cancer Research and Treatment Fund Inc, New York, NY. The KORA research platform (KORA, Cooperative Research in the Region of Augsburg) was initiated and financed by the Helmholtz Zentrum München–German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research and by the State of Bavaria. The InCHIANTI study baseline (1998-2000) was supported as a targeted project (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the US National Institute on Aging (contracts 263 MD 9164 and 263 MD 821336); the InCHIANTI follow-up 1 (2001-2003) was funded by the US National Institute on Aging (contracts N01-AG-1-1 and N01-AG-1-2111); the InCHIANTI follow-up 2 and 3 studies (2004-2010) were financed by the US National Institute on Aging (contract N01-AG-5-0002), supported in part by the Intramural research program of the National Institute on Aging, National Institutes of Health, Baltimore, MD.
Authorship
Contribution: A.V.J. and N.C.P.C. designed the study; A.V.J., P.J.C., P.A.B., L.M.S., W.T., P.S., A.J.C., and N.C.P.C. performed laboratory and data analysis; and P.A.B., S.S., A.M.V., K.Z., M.J.P., M.F.M., L.M.S., R.T.S., D.O., C.N.H., H.G., A.K., and A.R.G. provided clinical samples and data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for L.M.S. is Greehey Children's Cancer Research Institute, University of Texas Health Sciences Center at San Antonio, San Antonio, TX 78229.
Correspondence: Nicholas C. P. Cross, Wessex Regional Genetics Laboratory, Salisbury District Hospital, Salisbury SP2 8BJ, United Kingdom; e-mail: ncpc@soton.ac.uk.